Abstract
Environments surrounding G-rich sequences remarkably affect the conformations of these structures. A proper evaluation system mimicking the crowded environment in a cell with macromolecules should be developed to perform structural and functional studies on G-quadruplexes. In this study, the topology and stability of a G-quadruplex formed by human telomeric repeat sequences were investigated in a macromolecule-crowded environment created by polyethylene glycol 200 (PEG200), tumor cell extract, and Xenopus laevis egg extract. The interactions between small molecules and telomeric G-quadruplexes were also evaluated in the different systems. The results suggested that the actual behavior of G-quadruplex structures in cells extract is quite different from that in the PEG crowding system, and proteins or other factors in extracts might play a very important role in G-quadruplex structures.
Keywords: G-quadruplex, Molecular crowding, Cellular environment, Stabilization, Ligands
Highlights
-
•
Cell-free system was constructed using HL60 cell extract.
-
•
Topologies and stability of G-quadruplexes were identified in different systems.
-
•
G-quadruplex’s ligands’ effects were evaluated in different systems.
1. Introduction
G-quadruplex is a special DNA secondary structure formed by tandem repetitive G-rich DNA sequences. Potential G-quadruplex-forming sequences are prevalent in functional genomic regions; these sequences may play very important regulatory roles in telomere length maintenance, gene replication, transcription, and translation processes [1], [2], [3] G-quadruplexes are promising anti-cancer drug targets because of their special structure and biological significance [4], [5]
Most in vitro studies on G-quadruplexes, such as decoding the biological functions of these structures or rational designing of targeted G-quadruplex ligands, have been performed in ionic buffer that contributes to the G-quadruplex formation [6], [7], [8] However, the actual environment within cells is rather complex, that is, nucleic acids are surrounded by numerous kinds of ions and macromolecules, especially proteins implicated in biological and biophysical functions. The overall influences of these particles on the structural conformation, thermal stability, and competitive ability on the duplex formation of G-quadruplexes may differ from those obtained through in vitro studies.
The G-quadruplex structure has been investigated under molecule-crowded conditions; this condition is considered as an efficient approach to explore actual cell environments. Crowding agents, such as polyethylene glycol 200 (PEG200), glycerol acetonitrile, polysaccharides, ethanol, dimethyl sulfoxide, and ficoll, are used to simulate the behavior of G-quadruplexes when these structures mimic cellular conditions by excluding internal cellular volume and by reducing water activity [10], [11], [12] Among these agents, PEG200 is the most widely used to simulate a molecule-crowded environment to investigate the behaviors of G-quadruplexes [9], [12], [13], [14], [15], [16] In the molecule-crowded environment created by PEG200, G-quadruplexes obtain greater thermal stability and higher competitive capacity than duplex forms; this finding has extended previous concepts regarding the behavior of G-quadruplexes in ionic buffer [15], [16] In addition to the increased stability during thermodynamic and duplex competition, the conformational transition of certain G-quadruplexes is induced by 40% PEG. The conformation polymorphisms displayed by human telomeric repeats in dilute solutions are conspicuously diminished; as a result, the parallel-stranded conformation dominates [12], [14] Thus far, molecule-crowded environments have been considered as a crucial determinant of nucleic acid structures in vivo. Conversely, the PEG200-induced conformational transition and increased stability are caused by dehydration, not molecular crowding. The same phenomenon can be observed in other low-molecular-weight solutions, such as 50% ethanol [10]. Moreover, G-quadruplex are very stable in a 40% PEG mimicking system, and the melting point reaches over 95 °C [14]. However, this phenomenon can hardly be considered biologically reasonable. If this stable condition is possible, the stability unlikely supports G-quadruplexes to perform their biological roles that regulating gene expression.
Hansel et al. [17] discovered that the G-quadruplex formed by human telomeric DNA combines the characteristics of anti-parallel folds or hybrid parallel-antiparallel conformations in physiologically relevant Xenopus laevis egg extract and not the dominant parallel conformation in PEG200 solution. Thus, PEG or other low-molecular-weight molecules may not be appropriately used to simulate molecule-crowded environments. These observations may be used as basis of further strategies to mimic the crowded environment inside a cell to determine the actual topology and stability of ubiquitous G-quadruplexes formed by promoters, telomere, or even RNAs.
We constructed a cell-free system by using a human cancer cell extract, HL-60 cell extract, which is the first recorder in the lysate obtained from mammalian cells, to overcome previous problems, to enhance the methods used to investigate G-quadruplex behaviors in crowded mammalian intracellular environments, and to provide a rational strategy to evaluate G-quadruplex ligands. We also evaluated the presence and stability of G-quadruplex and its biological function in this system. X. laevis egg extracts and PEG200 molecule-crowded system were used as controls. The results showed that G-quadruplex could form in the cell-free system and displayed the nonparallel conformation similar to that in the X. laevis egg extract, not to the parallel fold observed in the control system with the dialysis buffer containing 40% PEG200. Moreover, the G-quadruplex formed in the cell extract and X. laevis egg extract exhibited a lower thermal stability than the G-quadruplex in 40% PEG200 solutions used in a previous study [9]. The ligands in the cell-free system elicited concentration-dependent effects on the stability of the G-quadruplex structure. This phenomenon also differed from the results obtained under a PEG200 molecule-crowded condition; under this condition, the effect of ligands is reduced and even lost [18]. Further electrophoresis experiments have revealed that the different behaviors of G-quadruplex in the cell-free system are partly attributed to the interactions of this structure with proteins [19].
2. Materials and methods
2.1. Oligonucleotides
All oligonucleotides used in this study were purchased from Invitrogen (China) and the corresponding sequences were listed in Table 1. For fluorescence spectroscopy and MST, the oligonucleotides were labeled with 5′-FAM or dual-labeled with 5′-FAM and 3′-TAMRA and purified through HPLC. Oligonucleotide concentrations were determined on the basis of their absorbance at 260 nm, and extinction coefficients were calculated using the nearest neighbor method. The oligonucleotides were initially dissolved with redistilled water to obtain 0.1 mM and further diluted to certain concentrations with specific buffers depending on the experiments.
Table 1.
Oligonucleotides sequences used in the present studies.
| Oligonucleotides | Sequence (from 5′ to 3′) |
|---|---|
| HTG21 | GGGTTAGGGTTAGGGTTAGGG |
| HTG21-mu | GTGTTAGTGTTAGTGTTAGTG |
| HTG21-rev | ATCGATCGCTTCTCGTCCCTAACCCT |
| HTG21-mu-rev | ATCGATCGCTTCTCGTCACTAACACT |
2.2. Ligands
The structures of the ligands used in this paper were shown in Fig. 1. TMPyP4 [20] was obtained from a commercial source. The quindoline derivatives SYUIQ-05 [21], [22] and SYUIQ-FM05 [23], and the isaindigotone derivative ISD-05 [24] were synthesized and characterized in accordance with previously described methods. The compounds were identified through NMR, high-resolution mass spectrometry, and elemental analysis; the purity of the compound was >95%, as confirmed by these processes.
Fig. 1.
Structures of the G-quadruplex ligands used in this paper, the cationic porphyrin TMPyP4, the quindoline derivatives SYUIQ-05 and SYUIQ-FM05, and the isaindigotone derivative ISD-05.
2.3. Preparation of the cell extract
Human leukemia HL-60 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD). The cell culture was maintained in an RPMI-1640 medium supplemented with 10% fetal bovine serum at 37 °C in a humidified atmosphere with 5% CO2. The cells were collected at 80% confluence, washed with ice-cold PBS buffer, and stored at −80 °C. The whole cell extract was prepared at 4 °C in accordance with previously described protocols [25], [26]. Frozen cells were quickly thawed in a water bath at 37 °C and washed with 5 packed-cell volume (pcv) of ice-cold Buffer A, which contained 20 mM HEPES-KOH (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, and 1 tablet of protease inhibitor (Roche complete, Mini, EDTA-free protease inhibitor cocktail tablets) for 10 mL of Buffer A. The washed cell was centrifuged at 3300g for 1 min to form a tight pellet; the volume of the pellet was estimated and resuspended with 7 pcv of ice-cold Buffer A in an ice bath for 15 min. Afterward, the cells were lysed in a Dounce homogenizer with a loose pestle 15 times, and the cell lysate was centrifuged at 1000g for 10 min to remove the nuclei. The supernatant was further centrifuged with a Beckman 65.2 VTi Rotor at 100,000g at 4 °C for 1 h. The clarified supernatant was dialyzed with 2 L of dialysis buffer, which contained 20 mM HEPES-KOH (pH 7.5), 100 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, and 25% glycerol, in an ice bath overnight and then centrifuged at 18,000g at 4 °C for 10 min. The cell extract was used immediately or quickly frozen in liquid nitrogen and stored in aliquots at −80 °C. The cells with a packed volume of 1 mL may yield 5 mL of cell extract with a protein concentration of 1.38±0.3 mg/mL determined through Bradford assay.
2.4. Preparation of X. laevis egg extract
Crude cytoplasmic extracts were prepared in accordance with a previously described method [17]. Oocytes were transferred to a Petri dish containing 1 mM progesterone in Ori buffer [5 mM HEPES (pH=7.6), 110 mM NaCl, 5 mM KCl, 2 mM CaCl2, and 1 mM MgCl2] abd incubated for more than 12 h. Xenopus eggs were selected and transferred via a glass pipette to a new Petri dish and extensively washed with Ori buffer. The Petri dish was subsequently incubated in an ice bath for 20 min. The cooled eggs were transferred to a pre-cooled Petri dish containing Ori buffer. Finally, the eggs were transferred to a tube containing ice-cold intra-oocyte buffer. The cells were packed by spinning for 1 min at 400g, and the buffer above the eggs was cautiously removed. The cells were subsequently centrifuged at 12,000g for 5 min. A glass pipette was used to crush the eggs to prepare a homogenized egg extract solution in an ice bath. The homogenized eggs were centrifuged at 12,000g for 30 min to obtain the crude inter-phase extract.
2.5. CD measurements
CD measurements were recorded from 220 nm to 320 nm by using a circular diachroism spectrophotometer (Chirascan) at 25 °C controlled by a Quantum (Northwest) temperature controller. Scans were obtained at a scanning speed of 100 nm/min, a response time of 1 s, and a bandwidth of 1 nm to acquire the data. A cylindrical quartz cuvette with a path length of 0.2 mm was used, and oligonucleotide concentration was 5 μM. The spectra were signal-averaged for at least three scans, baseline-corrected by subtracting the corresponding buffer or cell extract spectrum, and smoothed by calculating the mean. Data were acquired after incubation was performed at 50 μM HTG21 DNA with different systems for 2 h at 37 °C [17].
2.6. FRET melting assay
FRET assay was performed using the same kinds of labeled DNA as described in the fluorescence experiments. The labeled DNAs were initially heated to 95 °C for 10 min and then cooled to 25 °C at a cooling speed of 3 °C/s. The DNAs were incubated with or without ligands in the different systems for 2 h at 37 °C before measurement was performed. The fluorescence melting curves were determined by using a Roche LightCycler 2.0 real-time PCR machine with a total volume of 20 µL with 0.2 μM of the labeled oligonucleotides in the different systems. Fluorescence readings were obtained at intervals of 1 °C within the range of 37–99 °C; constant temperature was maintained for 30 s before each reading was performed to ensure a stable value. Tm was calculated using GraphPad Prism®, and the fitting model was chosen as Sigmoidal fit. Experiments were performed in triplicate to calculate the standard deviation.
2.7. DNA polymerase stop assay
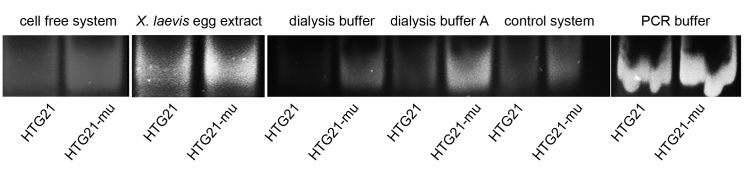
The DNA polymerase stop assay was performed in accordance with a previously described protocol with slight modifications [27]. The HTG21 sequence, its corresponding mutant sequence HTG21-mu, and complementary strands (HTG21-rev and HTG21-mu-rev) are presented in Table 1. The sequence with a final concentration of 1 μM was initially incubated at 37 °C for 1 h in the three different systems indicated in Fig. 4, Fig. 6. The complementary strands were then added. The reaction mixture was incubated at 25 °C for 1 h; afterward, 0.2 mM dNTP and 2 U of rTaq DNA polymerase per reaction were added. The mixture was further incubated at 37 °C for 30 min. The reactions were terminated by adding 1×loading buffer, and the 37 bp products were resolved on 16% native PAGE gel with a GelRed™ dye. In the ligand-stabilizing experiments, 2.5 μM ligands were added, and the mixture was incubated for another 1 h at 25 °C before annealing with the complementary strand was performed.
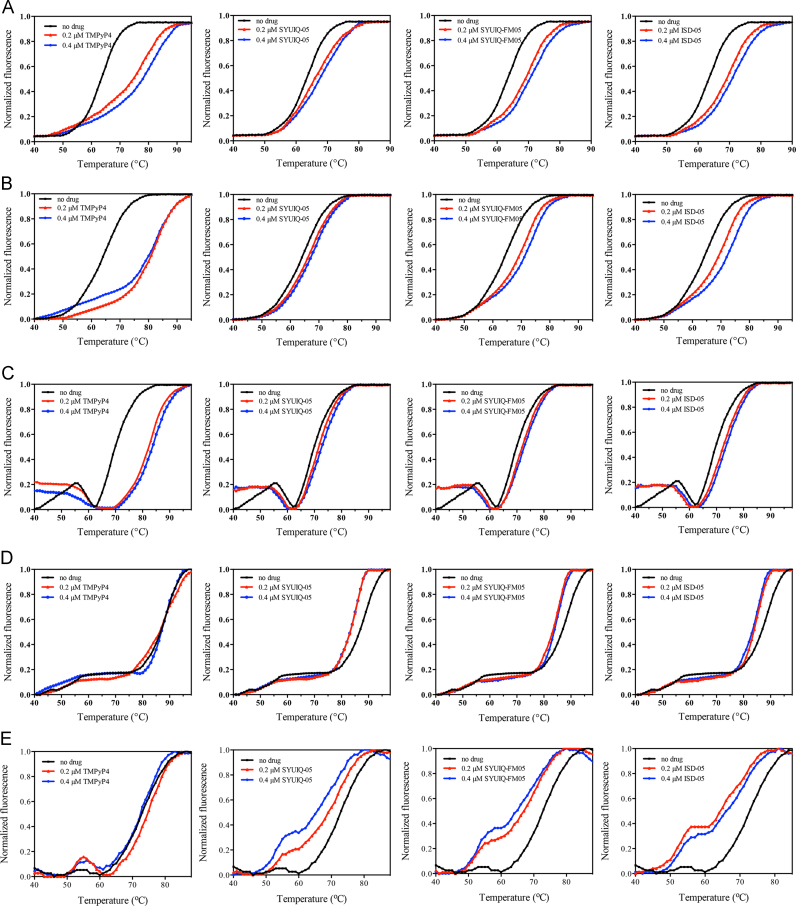
Fig. 4.
The FRET melting curves of HTG-21 G-quadruplex in ionic buffer (A), dialysis buffer (B), cell-free system (C), control system (D), and Xenopus laevis egg extracts (E), with or without 0.2 μM or 0.4 μM TMPyP4, SYUIQ-05, SYUIQ-FM05, and ISD-05.
Fig. 6.
Effects of different compounds on the polymerase stop assay with HTG21 and its corresponding mutant sequence HTG21-mu in the cell free system, Xenopus laevis egg extract, and dialysis buffer mentioned before. All of the compounds were at 2.5 μM. Data represented means of six independent experiments with standard error.
2.8. MST binding measurements
The HTG-21 sequence was labeled with FAM at their 5′-end (Invitrogen, USA). The thermophoretic movements of the fluorescent-labeled nucleic acids and compound complexes were detected by monitoring the fluorescence distributions inside the capillary by using the NT.115 MST machine (NanoTemper, Germany). The concentration of FAM-HTG21 was held constant at approximately 0.5 μM, and the compound was diluted at 1:2 from 20 μM 14 times. The samples were loaded into standard-treated MST-grade glass capillaries. The intensities of the LED and laser were 20% and 40%, respectively. Data were analyzed using NT.115.
2.9. Native gel electrophoresis
Native gel electrophoresis was run on 16% polyacrylamide gel in 0.5× TBE buffer at 4 °C for 1.5 h at a voltage of 120 V. The oligonucleotides were labeled at the 5′-end with the FAM fluorophore. The oligonucleotides were mixed with the different systems for 2 h before 2 μL of 6×loading buffer was added. The electrophoresis gel was observed to detect fluorescence emission.
3. Results
3.1. The topologies of G-quadruplex in different systems
We used the human telomeric DNA HTG21 (Table 1) as a model molecule and HL-60 cell extract to construct the cell-free system to examine the G-quadruplex behavior. The cell pellet was suspended, homogenized in a lysis buffer, ultracentrifuged, and dialyzed to construct the cell-free system. X. laevis egg extracts were obtained according to Ref. [17]. The formation of G-quadruplex in the cell-free system was confirmed by using circular dichroism (CD) spectra.
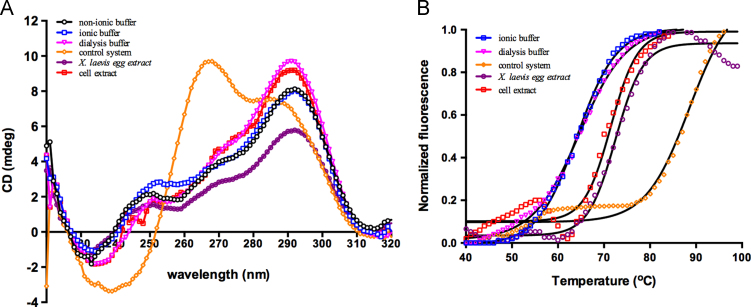
In the CD experiment, the formation and topology of human telomeric DNA HTG21 dG3(TTAG3)3 were investigated after incubation in the different systems at 37 °C for 2 h [17]. Fig. 2A showed the CD spectra of the HTG21 in the cell-free system, X. laevis egg extract, control system (dialysis buffer containing 40% PEG200), dialysis buffer, ionic buffer [10 mM Tris–HCl buffer (pH 7.4) containing 100 mM K+] and non-ionic buffer (10 mM Tris–HCl buffer, pH 7.4). HTG21 showed similar structural characteristics in the ionic buffer, non-ionic buffer, X. laevis and tumor cell extract, and dialysis buffer; indeed, HTG21 presented the expected characteristic CD signature of an anti-parallel orientation with a positive peak at 295 nm [28]. However, a notable conformational transition of the DNA sequence to the parallel conformation was induced by 40% PEG200 as the positive peak at 295 nm decreased; a strong positive peak appeared at approximately 265 nm, which is the marker of parallel topology [28]. This result was in consistent with that described in previous studies [12], [14], [17]. The telomeric sequence in the cell-free system could form a G-quadruplex, and could exhibit a non-parallel structure similar to that observed in the ionic buffer. This phenomenon was quite different from the parallel orientation in the control system.
Fig. 2.
(A), CD spectra of 5 μM HTG21 oligomer in non-ionic buffer (10 mM Tris–HCl buffer, pH 7.4), ionic buffer [10 mM Tris–HCl buffer (pH 7.4) containing 110 mM KCl], dialysis buffer, control system (dialysis buffer with 40% PEG), Xenopus laevis egg extract, and cell-free system (dialysis buffer with cell extract). (B), The FRET melting curves of HTG21G-quadruplex in ionic buffer, dialysis buffer, cell-free system, X. laevis egg extract, and control system.
3.2. The thermal stability of G-quadruplex in different systems
Fluorescence resonance energy transfer (FRET) melting assay was conducted to detect the thermal stability of the G-quadruplex. The increase in the fluorescence intensity of FAM was monitored over time by heating; in this process, the G-quadruplex unfolded. The melting curves of the G-quadruplex in the different environments were plotted and used to calculate the melting point. Fluorescein emission was normalized between 0 and 1, and melting temperature (Tm) is defined as the temperature at which the normalized emission is 0.5 [27]. Table 2 displayed Tm of HTG21 G-quadruplex in the different systems; Fig. 2B illustrated the corresponding melting curves. The G-quadruplex in the cell-free system yielded a slightly higher Tm than that in the ionic buffer; by contrast, the G-quadruplex in the cell-free system exhibited an approximately 20 °C lower Tm than that in the control system. The thermal stability of HTG21 in X. laevis egg extract with Tm of 72 °C was also significantly lower than that in the control system with Tm of 87 °C. These results suggested that the stability of the G-quadruplex in the cell free system was similar to that in the ionic buffer, but was significantly lower than that in the control system. The melting curve of the cell free system (Fig. 2B) revealed a small minor peak that could correspond to two melting transitions; therefore, a small proportion of G-quadruplexes bound to the endogenous proteins in the cell-free system. After the bound G-quadruplexes became free, Tm was calculated from the second melting transition.
Table 2.
Calculated melting temperatures (Tm) of HTG21 in ionic buffer, dialysis buffer, cell free system and control system.
| Ionic buffera | Dialysis buffer | Cell free systemb | Control systemc | X. laevis egg extract | |
|---|---|---|---|---|---|
| Tm (°C) | 64±0.8 | 66±0.6 | 68±0.9 | 87±0.6 | 72±0.5 |
Representing 10 mM Tris–HCl buffer containing 100 mM K+.
Representing dialysis buffer containing cell extract.
Representing dialysis buffer containing 40% PEG200.
DNA polymerase stop assay was also performed to compare the stabilizing effects of quadruplex DNA in the cell-free and control systems on the basis of the ability of G-quadruplexes to interfere with the polymerase elongation of specific DNAs [29]. In this study, the corresponding mutant sequence HTG21-mut without the ability to form a G-quadruplex was set as the control. The PCR buffer containing 50 mM K+ was set as another control. Dialysis buffer and dialysis buffer A were also used as controls. The two buffers were composed of the same components except 1.5 mM MgCl2 added to dialysis buffer A to facilitate PCR. Fig. 3 showed the total amount of double-stranded DNA produced through Taq polymerase amplification reaction; Table 3 listed the calculated inhibition rates in the different systems. The cell-free, X. laevis egg extract, and dialysis buffer A systems slightly affected the normal Taq polymerase amplification activity of HTG21 because HTG-21-mut could not form the quadruplex transcriptional barrier. In HTG21, the difference was observed between the cell-free system and the other systems. Among the systems, the 1× PCR buffer system used as a control showed the least inhibition of Taq polymerase probably because the low K+ concentration. The cell-free system elicited a conspicuous inhibition of amplification but caused a lower inhibition percentage than the control system. The dialysis buffer and the control system inhibited the production of double-stranded PCR products. The dialysis buffer and the control system did not efficiently facilitate the PCR.
Fig. 3.
Effects of different systems on the polymerase stop assay with HTG21 and its corresponding mutant sequence HTG21-mu in a normal PCR reaction buffer, the cell free system, Xenopus laevis egg extract, dialysis buffer, dialysis buffer A, and control system mentioned before. Data represented means of six independent experiments with standard error.
Table 3.
Calculated inhibition rates in PCR buffer, cell free system and control system. The inhibition rate was calculated by the formula, inhibition rate (%)=(ODmut–ODwt)/ODmut.
| Cell free systema | X. laevis egg extract | Control systemb | |
|---|---|---|---|
| Inhibition rate | 21.7±4.0% | 40.2±3.8% | 86.3±5% |
| Dialysis buffer | Dialysis buffer Ac | PCR buffer | |
| Inhibition rate | 100.0±1.0% | 72.2±4.3% | 12.3±5% |
Representing dialysis buffer containing cell extract.
Representing dialysis buffer containing 40% PEG200.
Representing dialysis buffer containing extra 1.5 mM MgCl2.
Thus, the G-quadruplex in the cell-free system and X. laevis egg extract likely exhibited low thermal stability and capacity to arrest Taq polymerase during amplification. These characteristics suggested that the G-quadruplex formed in the biological system may not act as a stable structure because of its inevitable contact with certain proteins. Different kinds of proteins, such as nucleolin and NM23-H2, can stabilize or unwind this unique structure; as a result, a dynamic balance occurs with the duplex DNA [20], [30] Certain proteins are also necessary to work collaboratively to bind and to help form or unwind the G-quadruplex; thus, the role of this structure as a regulator is facilitated rather than the maintenance of the stability of this structure.
3.3. Evaluation of G-quadruplex ligands in the cell-free system
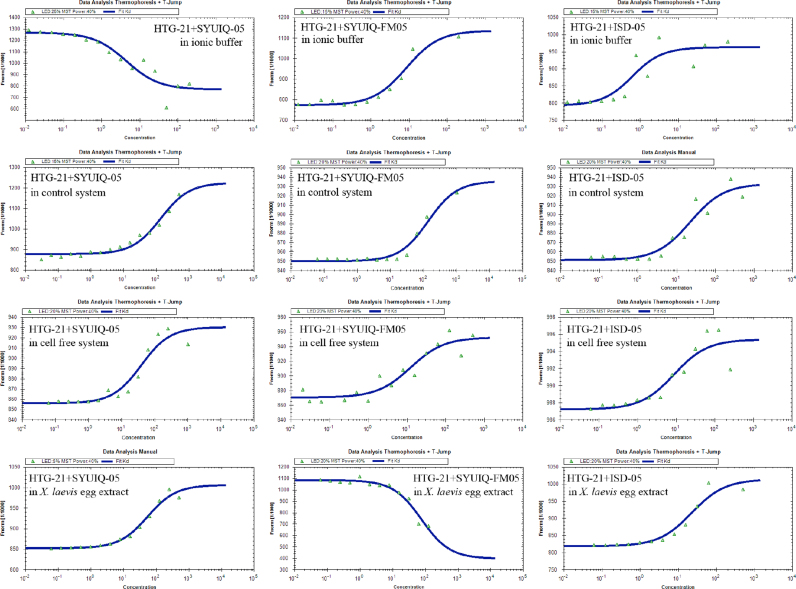
Considering that the G-quadruplex in the cell-free system exhibited stabilities that differed from the G-quadruplex in the control system, we further evaluated whether G-quadruplex ligands elicited a stabilizing effect on the cell-free system and X. laevis egg extract; we also determined whether this effect was different from that in the generally used ionic buffer and 40% PEG control systems [18]. The FRET melting assay was initially performed by co-incubating the DNAs in the different systems with four G-quadruplex binders (Fig. 1), namely, TMPyP4, [20] the quindoline derivatives SYUIQ-05 [21], [22] and SYUIQ-FM05 [23], and isaindigotone derivative ISD-05 [24]. These four compounds can effectively stabilize telomere G-quadruplex. Table 4 showed the melting temperatures of the HTG21 sequence; Fig. 4 illustrated the melting curves. In the control system, Tm of the samples incubated with the three other ligands and of the samples incubated with TMPyP4 eliciting a slight stabilizing effect was determined after the ligands were added. The results showed that Tm slightly decreased and depended on concentration. This phenomenon could be attributed to the reduced water activity, increased viscosity created by PEG200, and different structures of G-quadruplex or ligands [18]. G-quadruplexes achieved remarkable thermal stability in this system. Tm could not be easily increased and thus should not be used as a good criterion in ligand evaluation. However, G-quadruplexes folded in the ionic buffer and the cell-free system displayed an increase in thermal stability upon addition of ligands; Tm of the G-quadruplexes formed in these systems was concentration dependent. The change in Tm of HTG-21 in X. laevis egg extract exhibited a trend similar to that in the control system; the observed minor peaks could cause two melting transitions. This result indicated that a small proportion of G-quadruplexes bound to the endogenous proteins in the system or caused protein aggregated because of heat treatment.
Table 4.
Melting temperatures (Tm) of HTG21 in different systems with co-incubation with 0.2 μM or 0.4 μM ligands.
| Ionic buffera | Dialysis buffer | Cell free systemb | Control systemc | X. laevis egg extract | ||
|---|---|---|---|---|---|---|
| No drug | (μM) | 64±0.8 | 66±0.6 | 68±0.9 | 87±0.6 | 72±0.5 |
| TMPyP4 | 0.2 | 78±0.7 | 82±1.2 | 81±0.5 | 86±0.7 | 74±0.3 |
| 0.4 | 81±0.9 | 81±0.6 | 82±0.9 | 87±0.6 | 72±0.4 | |
| SYUIQ-05 | 0.2 | 69±0.7 | 68±1.0 | 71±1.2 | 83±0.1 | 70±0.5 |
| 0.4 | 70±0.5 | 70±0.6 | 73±0.6 | 83±0.2 | 68±0.8 | |
| SYUIQ-FM05 | 0.2 | 71±0.5 | 71±0.7 | 72±0.6 | 83±0.3 | 69±0.7 |
| 0.4 | 73±0.6 | 72±0.5 | 73±0.7 | 84±0.4 | 68±0.8 | |
| ISD-05 | 0.2 | 71±0.1 | 71±0.2 | 73±0.4 | 84±0.3 | 67±1.0 |
| 0.4 | 74±0.2 | 73±0.3 | 75±0.3 | 83±0.6 | 68±0.8 |
Representing 10 mM Tris–HCl buffer containing 100 mM K+.
Representing dialysis buffer containing cell extract.
Representing dialysis buffer containing 40% PEG200.
Thus, we applied a new technology to evaluate the interactions between HTG-21 G-quadruplex and the ligands. Microscale thermophoresis (MST) is employed to analyze the interaction of biomolecules in solutions, such as serum and cell lysate. We can also obtain the thermal binding characteristics between biomolecules through MST [31]. However, TMPyP4 cannot be applied in this assay because its fluorescence likely affects the detection of the FAM-labeled HTG-21 in MST. Table 5 and Fig. 5 illustrated that the detected ligands bound to the FAM-labeled HTG-21 in the ionic buffer to the greatest extent. The dissociation constants (Kd) significantly increased in the control system, the cell free system, and the X. laevis egg extract. In addition, the binding of ligands to the HTG-21 G-quadruplex in the cell-free system and X. laevis egg extract was stronger than that in the control system; this finding was consistent with that revealed through FRET assay.
Table 5.
Calculated dissociation constants (Kd) of SYUIQ-05, SYUIQ-FM05 and ISD-05 in different solutions.
| Compounds | Ionic buffer (μM) | Control system (μM) | Cell free system (μM) | X. laevis egg extract (μM) |
|---|---|---|---|---|
| SYUIQ-05 | 4.3±0.5 | 138±6.8 | 38.7±3.8 | 54.4±2.6 |
| SYUIQ-FM05 | 8.4±0.7 | 137±7.9 | 12.3±1.6 | 79.6±6.4 |
| ISD-05 | 0.5±0.1 | 22.3±2.6 | 8.6±1.1 | 19.7±1.1 |
Fig. 5.
Fitted curves obtained by MST measurements of 5’-FAM-labeled HTG-21 with ligands diluted 1:2 from 20 μM for 14 times.
The stabilizing effects of the ligands on the G-quadruplexes in the different systems were further validated through the DNA polymerase stop assay. The ligands at 2 μM in the cell-free system, X. laevis egg extract, and dialysis buffer also induced stabilizing effects on the G-quadruplex structure (Fig. 6). The inhibition rate of each ligand was calculated (Table 6). Most of the evaluations revealed similar structure–activity relationships; in particular, TMPyP4 was the strongest among the G-quadruplex ligands; ISD-05 exhibited a stronger interaction than SYUIQ-05 and SYUIQ-FM05. However, the evident increased inhibition in X. laevis egg extract was inconsistent with the measured Tm. This phenomenon was probably attributed to the complex protein environment or the different ligand concentrations. In simple system, such as Tris–HCl buffer, the increased thermal stability caused the increased inhibition in the rTaq DNA polymerase assay. In a complex system, the efficiency of rTaq is dependent not only on the secondary structure of the template DNA but also on the solution environment. Thus, the inhibition of ligands on rTaq elongation was partially caused by the stabilizing abilities on G-quadruplex structures.
Table 6.
Calculated inhibition rate of 2.5 μM TMPyP4, SYUIQ-05, SYUIQ-FM05 and ISD-05 in the cell free system, X. laevis egg extract, and the dialysis buffer. The inhibition rate was calculated by the formula, inhibition rate (%)=(ODmut–ODwt)/ODmut.
| No drug | TMPyP4 | SYUIQ-05 | SYUIQ-FM05 | ISD-05 | |
|---|---|---|---|---|---|
| Inhibition rates in the cell free system | 21.7±4.0% | 87.5±8.0% | 58.6±3.2% | 79.5±7.0% | 75.5±9.3% |
| Inhibition rates in the X. laevis egg extract | 40.2±3.8% | 82.3±4.0% | 60.2±3.5% | 59.5±3.8% | 67.3±3.3% |
| Inhibition rates in the dialysis buffer A | 72.2±4.3% | 92.6±5.3% | 90.8±5.6% | 62.4±5.0% | 65.3±4.4% |
3.4. G-quadruplex–protein interaction may contribute to the stability and biological functions within cells
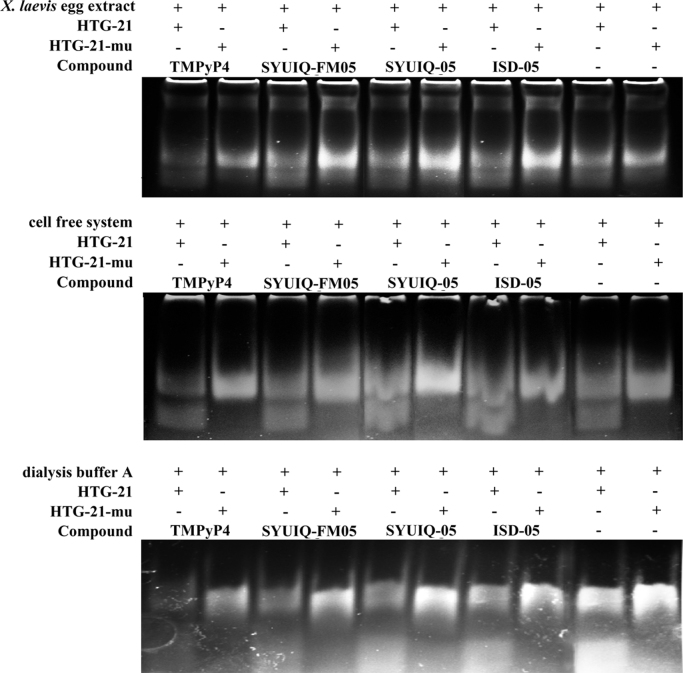
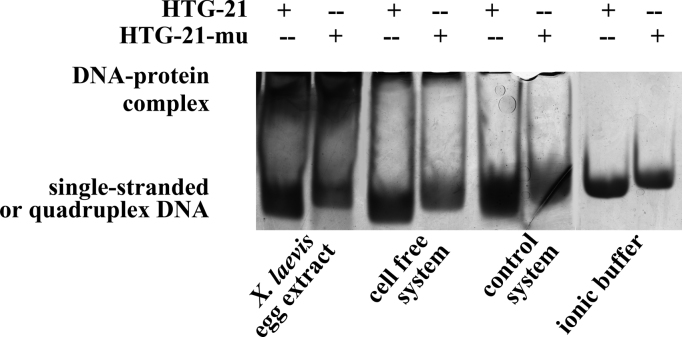
We have yet to determine the reasons accounted for the differences in stabilizing effects between the cell-free system and the other systems. The most possible reason could be the components of the cell extract, including proteins and other biological molecules produced by the cells. To test this hypothesis, we conducted native gel electrophoresis. The sequence of HTG-21-mu that could not form a G-quadruplex was used as a control to help distinguish the single-strand DNA and G-quadruplexes. The gel evidently showed that the control system provided the same pattern as the ionic buffer (Fig. 7); in the ionic buffer, HTG-21 more rapidly migrated than HTG-21-mu. However, an ideal protocol to separate different conformations was difficult to establish because of different systems running in the same gel. The gel revealed an evident slow-migrating DNA band in the cell-free system and X. laevis egg extract lanes. No protein-binding bands were observed in the other systems. This phenomenon could be explained as a combination of specific proteins binding to the G-quadruplex in the cell-free system. However, this study failed to confirm whether this protein binding affects the G-quadruplex stability and function in vivo or in vitro. Hence, we assumed that this protein–G-quadruplex interaction may affect the stability of G-quadruplexes formed in the cell-free system.
Fig. 7.
Gel electrophoresis of HTG21 in ionic buffer [10 mM Tris–HCl buffer (pH 7.4) containing 110 mM KCl], cell free system, Xenopus laevis egg extract, and control system (the dialysis buffer containing 40% PEG).
4. Discussion
In the present study, a cell-free system was constructed and prepared from the HL-60 cell extract. This cell-free system was used to investigate the stability and biological functions of G-quadruplexes to enhance the approach used to mimic the crowded environment inside cells with macromolecules containing G-quadruplex-associated proteins. X. laevis egg extract, which is an established system, was also used as the control. The results showed that the G-quadruplex could be stably formed and could achieve a non-parallel topology in the cell-free system, not the parallel topology observed in PEG200 solutions. The G-quadruplex in the cell-free system was less stable than that in the 40% PEG system; therefore, G-quadruplex may not be relatively stable in vivo [9]. Ligands were also evaluated experimentally. The results demonstrated the normal effects of stabilizing the G-quadruplex structure, and the effects were dependent on concentration. The G-quadruplex should not maintain a very stable structure in vivo because of the roles of G-quadruplexes within biological systems; some of these functions include sustaining the telomere structure and regulating gene transcription. Instead, the structure should remain in a dynamic state at which the structure can be unwound by functional proteins [1], [4], [25] The formation of G-quadruplexes could only be regulated in terms of place and time; this process may be a reasonable mechanism of the regulatory processes within cells. Moreover, the physiologically relevant protein–G-quadruplex interaction cannot be simply simulated by PEG200.
However, our cell-free system exhibits intrinsic limitations. First, oligonucleotides are exposed to cytosolic and nuclear proteins, not to nuclear protein alone. Second, confinement is lost without the nucleus. Third, dialysis removed low-molecular-weight compounds to maintain physiological ion concentrations. This process plays more significant roles than molecular crowding in determining the conformation of the G-quadruplex. We suggest that the homogeneous molecule-crowded condition simulated by PEG200 in vitro is not the optimum condition to evaluate the G-quadruplex behavior; as such, further studies should be performed. Moreover, this physicochemical characteristic may not be the most important factor affecting the activities of G-quadruplexes. Our results, along with those of Hansel et al. [17], indicated that the prevailing use of PEG200 to mimic the cellular molecular environment should be reconsidered. Although PEG is not an appropriate material to create a physiologically crowded intracellular environment to examine the behaviors of G-quadruplexes, PEG can be used as a good crowding agent to explore the equilibrium and the rate of biomedical reactions involving macromolecules [35].
The behaviors of G-quadruplexes and ligands in a crowded system have been studied extensively; our study constructed a cell-free system from tumor cells to mimic the actual cellular environment. The cell-free system is more similar to the actual cellular environment than the PEG system and X. laevis egg extract. The cell-free system is also easier to construct than X. laevis egg extract because the latter requires animal experiments. However, limited technology could be used to examine the effects of ligands on G-quadruplexes in complex systems, such as X. laevis egg extract or cell-free system. In this study, MST was applied to obtain the quantitative data, such as binding constant, related to quadruplex–ligand binding. Thus, this study demonstrated MST as a novel technology to evaluate the quadruplex–ligand interaction in a complicated but easy-to-establish cell-free system mimicking actual cellular environments.
Acknowledgments
We express our gratitude for the financial support provided by the National Natural Science Foundation of China (Grant nos. 21372263, 91213302 and 81330077), the Fundamental Research Funds for the Central Universities (Grant no. 11ykzd04), the Zhujiang Nova Program (Grant no. 2011J2200075), and the Foundation for Distinguished Young Talents in Higher Education of Guangdong (Grant no. Yq2013002).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.09.022.
Appendix A. Supplementary material
Supplementary material
References
- 1.Dexheimer T.S., Carey S.S., Zuohe S., Gokhale V.M., Hu X., Murata L.B., Maes E.M., Weichsel A., Sun D., Meuillet E.J., Montfort W.R., Hurley L.H. NM23-H2 may play an indirect role in transcriptional activation of c-myc gene expression but does not cleave the nuclease hypersensitive element III(1) Mol. Cancer Ther. 2009;8:1363–1377. doi: 10.1158/1535-7163.MCT-08-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumari S., Bugaut A., Huppert J.L., Balasubramanian S. An RNA G-quadruplex in the 5’ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackburn E.H. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 4.Balasubramanian S., Hurley L.H., Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasubramanian S., Neidle S. G-quadruplex nucleic acids as therapeutic targets. Curr. Opin. Chem. Biol. 2009;13:345–353. doi: 10.1016/j.cbpa.2009.04.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reed J.E., Arnal A.A., Neidle S., Vilar R. Stabilization of G-quadruplex DNA and inhibition of telomerase activity by square-planar nickel(II) complexes. J. Am. Chem. Soc. 2006;128:5992–5993. doi: 10.1021/ja058509n. [DOI] [PubMed] [Google Scholar]
- 7.Luu K.N., Phan A.T., Kuryavyi V., Lacroix L., Patel D.J. Structure of the human telomere in K+ solution: an intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambrus A., Chen D., Dai J., Bialis T., Jones R.A., Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kan Z.Y., Yao Y., Wang P., Li X.H., Hao Y.H., Tan Z. Molecular crowding induces telomere G-quadruplex formation under salt-deficient conditions and enhances its competition with duplex formation. Angew. Chem. 2006;45:1629–1632. doi: 10.1002/anie.200502960. [DOI] [PubMed] [Google Scholar]
- 10.Miller M.C., Buscaglia R., Chaires J.B., Lane A.N., Trent J.O. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3’ sequence of d(AG3(TTAG3)3. J. Am. Chem. Soc. 2010;132:17105–17107. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- 11.Kumar N., Maiti S. The effect of osmolytes and small molecule on Quadruplex-WC duplex equilibrium: a fluorescence resonance energy transfer study. Nucleic Acids Res. 2005;33:6723–6732. doi: 10.1093/nar/gki961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heddi B., Phan A.T. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 13.Zheng K.W., Chen Z., Hao Y.H., Tan Z. Molecular crowding creates an essential environment for the formation of stable G-quadruplexes in long double-stranded DNA. Nucleic Acids Res. 2010;38:327–338. doi: 10.1093/nar/gkp898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue Y., Kan Z.Y., Wang Q., Yao Y., Liu J., Hao Y.H., Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 15.Miyoshi D., Karimata H., Sugimoto N. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006;128:7957–7963. doi: 10.1021/ja061267m. [DOI] [PubMed] [Google Scholar]
- 16.Miyoshi D., Matsumura S., Nakano S., Sugimoto N. Duplex dissociation of telomere DNAs induced by molecular crowding. J. Am. Chem. Soc. 2004;126:165–169. doi: 10.1021/ja036721q. [DOI] [PubMed] [Google Scholar]
- 17.Hansel R., Lohr F., Foldynova-Trantirkova S., Bamberg E., Trantirek L., Dotsch V. The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 2011;39:5768–5775. doi: 10.1093/nar/gkr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Z., Zheng K.W., Hao Y.H., Tan Z. Reduced or diminished stabilization of the telomere G-quadruplex and inhibition of telomerase by small chemical ligands under molecular crowding condition. J. Am. Chem. Soc. 2009;131:10430–10438. doi: 10.1021/ja9010749. [DOI] [PubMed] [Google Scholar]
- 19.Fry M. Tetraplex DNA and its interacting proteins. Front. Biosci.: J. Virtual Libr. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 20.Izbicka E., Wheelhouse R.T., Raymond E., Davidson K.K., Lawrence R.A., Sun D., Windle B.E., Hurley L.H., Von Hoff D.D. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 1999;59:639–644. [PubMed] [Google Scholar]
- 21.Ou T.M., Lu Y.J., Zhang C., Huang Z.S., Wang X.D., Tan J.H., Chen Y., Ma D.L., Wong K.Y., Tang J.C., Chan A.S., Gu L.Q. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives. J. Med. Chem. 2007;50:1465–1474. doi: 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J.L., Lu Y.J., Ou T.M., Zhou J.M., Huang Z.S., Zhu X.F., Du C.J., Bu X.Z., Ma L., Gu L.Q., Li Y.M., Chan A.S. Synthesis and evaluation of quindoline derivatives as G-quadruplex inducing and stabilizing ligands and potential inhibitors of telomerase. J. Med. Chem. 2005;48:7315–7321. doi: 10.1021/jm050041b. [DOI] [PubMed] [Google Scholar]
- 23.Lu Y.J., Ou T.M., Tan J.H., Hou J.Q., Shao W.Y., Peng D., Sun N., Wang X.D., Wu W.B., Bu X.Z., Huang Z.S., Ma D.L., Wong K.Y., Gu L.Q. 5-N-methylated quindoline derivatives as telomeric g-quadruplex stabilizing ligands: effects of 5-N positive charge on quadruplex binding affinity and cell proliferation. J. Med. Chem. 2008;51:6381–6392. doi: 10.1021/jm800497p. [DOI] [PubMed] [Google Scholar]
- 24.Tan J.H., Ou T.M., Hou J.Q., Lu Y.J., Huang S.L., Luo H.B., Wu J.Y., Huang Z.S., Wong K.Y., Gu L.Q. Isaindigotone derivatives: a new class of highly selective ligands for telomeric G-quadruplex DNA. J. Med. Chem. 2009;52:2825–2835. doi: 10.1021/jm801600m. [DOI] [PubMed] [Google Scholar]
- 25.Liu X., Kim C.N., Yang J., Jemmerson R., Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 26.Leoni L.M., Chao Q., Cottam H.B., Genini D., Rosenbach M., Carrera C.J., Budihardjo I., Wang X., Carson D.A. Induction of an apoptotic program in cell-free extracts by 2-chloro-2’-deoxyadenosine 5’-triphosphate and cytochrome c. Proc. Natl. Acad. Sci. USA. 1998;95:9567–9571. doi: 10.1073/pnas.95.16.9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Cian A., Guittat L., Kaiser M., Sacca B., Amrane S., Bourdoncle A., Alberti P., Teulade-Fichou M.P., Lacroix L., Mergny J.L. Fluorescence-based melting assays for studying quadruplex ligands. Methods. 2007;42:183–195. doi: 10.1016/j.ymeth.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Kypr J., Kejnovska I., Renciuk D., Vorlickova M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han H., Hurley L.H., Salazar M. A DNA polymerase stop assay for G-quadruplex-interactive compounds. Nucleic Acids Res. 1999;27:537–542. doi: 10.1093/nar/27.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thakur R.K., Kumar P., Halder K., Verma A., Kar A., Parent J.L., Basundra R., Kumar A., Chowdhury S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37:172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wienken C.J., Baaske P., Duhr S., Braun D. Thermophoretic melting curves quantify the conformation and stability of RNA and DNA. Nucleic Acids Res. 2011;39:e52. doi: 10.1093/nar/gkr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material