Abstract
Osteoclasts are multinucleated cells with bone resorption activity that is crucial for bone remodeling. RANK‐RANKL (receptor activator of nuclear factor κB ligand) signaling has been shown as a main signal pathway for osteoclast differentiation. However, the molecular mechanism and the factors regulating osteoclastogenesis remain to be fully understood. In this study, we performed a chemical genetic screen, and identified a Cdks/GSK-3β (cyclin-dependent kinases/glycogen synthase kinase 3β) inhibitor, kenpaullone, and two Cdks inhibitors, olomoucine and roscovitine, all of which significantly enhance osteoclastogenesis of RAW264.7 cells by upregulating NFATc1 (nuclear factor of activated T cells, cytoplasmic 1) levels. We also determined that the all three compounds increase the number of osteoclast differentiated from murine bone marrow cells. Furthermore, the three inhibitors, especially kenpaullone, promoted maturation of cathepsin K, suggesting that the resorption activity of the resultant osteoclasts is also activated. Our findings indicate that inhibition of GSK-3β and/or Cdks enhance osteoclastogenesis by modulating the RANK–RANKL signaling pathway.
Keywords: Cdks inhibitor, GSK-3β inhibitor, Osteoclast
Highlights
-
•
We performed a chemical genetic screen to identify drugs which modulate osteoclastogenesis.
-
•
The screening determined a Cdk/GSK-3β inhibitor, kenpaullone, and two Cdk inhibitors, olomoucine and roscovitine, as activators of osteoclastogenesis.
-
•
The kenpaullone, olomoucine, and roscovitine induce an enhanced osteoclastogenesis by upregulating NFATc1 and mature cathepsin K levels.
1. Introduction
Bone homeostasis is tightly controlled by osteoblasts and osteoclasts which are involved in bone formation and resorption, respectively [1], [2], [3]. The imbalance between bone formation and resorption leads to impaired bone remodeling and development of bone disorders. Enhanced bone resorption by osteoclasts weakens bone structure and can cause osteoporosis over time whereas defects in the function of osteoclasts lead to osteopetrosis [1]. Osteoclasts are giant multinucleated cells derived from monocyte/macrophage lineage precursor cells through the differentiation process primarily induced by two cytokines, macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor κB ligand (RANKL), which are produced by osteoblasts [4]. The M-CSF supports proliferation and survival of osteoclast precursor cells, and upregulates RANK expression. The RANKL and RANK interaction recruits adaptor protein TRAF6, which in turn assembles with TAB2-TAK1 to activate mitogen-activated kinases (MAPKs) such as extracellular signal-regulated kinase (ERK), Jun N-terminal kinase (JNK), and p38 as well as NF-κB pathways [5]. NF-κB is required for initial induction of NFATc1, a key transcription factor for osteoclast differentiation. Then, MAPKs activate AP-1 (c-Fos/c-Jun), that further amplifies NFATc1 [6]. The activity of NFATc1 is regulated by calcium signaling that is induced by activation of the immunoglobulin-like receptors associated with the immunoreceptor tyrosine-based activation motif (ITAM)-harboring adaptor proteins, including DAP12 and FcRγ [7]. The activated calcineurin dephosphorylates NFATc1, which subsequently translocates to nucleus and cooperatively induces osteoclast-related genes with other transcription factors such as MITF, PU.1, CREB, and AP-1 [8]. Thus, the RANK–RANKL signaling activates various downstream signaling pathways required for the osteoclastogenesis [9]. Recent studies have significantly advanced our knowledge about the regulatory mechanism of osteoclastogenesis pathway, but the whole osteoclastic signaling network is yet unknown. Therefore, we performed a chemical genetic screen to identify novel pathways and factors which controls osteoclastogenesis by using annotated small compounds LOPAC1280 (Sigma). Our screen identified a Cdk/GSK-3β inhibitor, kenpaullone, and two Cdk inhibitors, olomoucine and roscovitine, as activators of the osteoclastogenesis. Our data showed that the inhibition of Cdks and/or GSK3-β significantly upregulates NFATc1 and subsequently enhances the formation of functional osteoclasts.
2. Material and methods
2.1. Cells and reagents
For the osteoclastogenic culture, RAW264.7 and pNFAT/Luc-RAW cells were cultured in α-MEM medium containing 10% fetal bovine serum (FBS), 100 ng/ml soluble RANKL (sRANKL, Peprotech and Oriental Yeast), 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin [10]. For osteoclast formation by using primary pre-osteoclast cultures, murine bone marrow cells were obtained from femurs and tibiae of 7-week-old ddY mice (Japan SLC, Inc.). 4.0×105 cells were cultured in α-MEM medium containing 10% fetal bovine serum (FBS), 100 ng/ml sRANKL, 10 ng/ml M-CSF (Wako, JPN), 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin in 24-well plates. Kenpaullone (Sigma), olomoucine (Sigma), roscovitine (Sigma) are used at a final concentration of 50 nM. The culture medium in each well was replaced by fresh medium containing M-CSF and sRANKL every 2 days. TRAP staining was performed after 5 days of the induction.
2.2. Cell-based screening of the small-compound library
The pNFAT/Luc-RAW cells were used for screen small compounds, LOPAC1280 (Sigma) that contains 1280 compounds of marketed drugs and pharmaceutically relevant structural derivatives. These compounds are annotated with biological activities and classified as follows: cell signaling (9%), phosphorylation (8%), cell stress (4%), lipids (4%), ion channels (6%), G proteins (3%), apoptosis/cell cycle (2%), gene regulation (3%), hormone related (3%), and neuroscience related (58%). The pNFAT/Luc-RAW cells (5000 cells/well) were plated into 96-well plates in 100 μl of α-MEM medium with 10% FBS, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. The culture medium was exchanged the next day for fresh medium containing sRANKL (100 ng/ml), followed by the addition of each compound in the library to the cells at 10 μM. The luciferase activity of each well was measured after 24 h using the ONEGlo™ luciferase assay system (Promega) and a microplate reader (GloMax-Multi Detection System, Promega).
2.3. TRAP staining
TRAP staining was performed as described previously [11]. Briefly, the cells were fixed with 10% glutaraldehyde for 15 min at 37 °C, and subsequently incubated for 10 min at 37 °C in TRAP buffer, which consisted of 0.1 M sodium acetate, 0.1 M acetic acid, 10 mg/ml naphthol AS-MX phosphate (Sigma), 0.1% Triton X-100 (Sigma), 0.3 M potassium tartrate (Sigma) and 0.3 mg/ml Fast Red Violet LB Salt (Sigma). TRAP-positive dark-red cells with more than three nuclei were counted under light microscope as multinucleated osteoclasts.
2.4. Quantitative real-time PCR
Total RNA was extracted with an RNeasy Mini Kit (QIAGEN, Hilden, Germany). After DNase I treatment (Ambion, Austin, TX), cDNAs were synthesized from 2 μg of total RNA using Super ScriptIII reverse transcriptase (Invitogen, Carlsbad, CA). Quantitative TaqMan® real-time PCR analysis for expression of NFATc1 was performed using the AB 7300 real-time PCR system (Applied Biosystems, Foster City, USA). The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as an internal control. The TaqMan® primer and probe sets used were Mm00479445_ml (NFATc1) and 4352339E (GAPDH).
2.5. Western blotting
The cells were harvested and lysed in Laemmli sample buffer [pH6.8; 50 mM Tris–HCl, 2% sodium dodecyl sulfate (SDS), 10% glycerol, 5% 2-mercaptoethanol] and heated at 95 °C for 5 min. Equal amounts of whole cell lysates were loaded and resolved via SDS-PAGE, and subjected to Western blotting analysis by using anti-NFATc1 monoclonal (7A6, Santa Cruz Biotechnology, Santa Cruz, CA), anti-cathepsin K polyclonal (ab19027, Abcam), and anti-β-actin monoclonal antibodies (A5441, Sigma). The signals were detected with an enhanced chemiluminescence (ECL) plus kit (GE Healthcare).
2.6. MTT assay
RAW264.7 cells were seeded onto 96-well plates at 50,000 cells/ml in 90 μl α-MEM complete medium. The cultures were incubated in the presence of 0.1% DMSO, 5 μM kenpaullone, 5 μM olomoucine, or 5 μM roscovitine for 0 (6 h), 1, 2, and 3 days followed by the addition of 10 μl of 5 mg/ml MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide) (Sigma). After 4 h of incubation, 100 μl of SDS-HCl solution was added and incubated for 24 h to dissolve the formazan produced by the cells. Optical densities (OD) were measured at a wavelength of 571 nm.
3. Results
3.1. Screen of annotated small compounds modulating osteoclastogenesis of RAW264.7
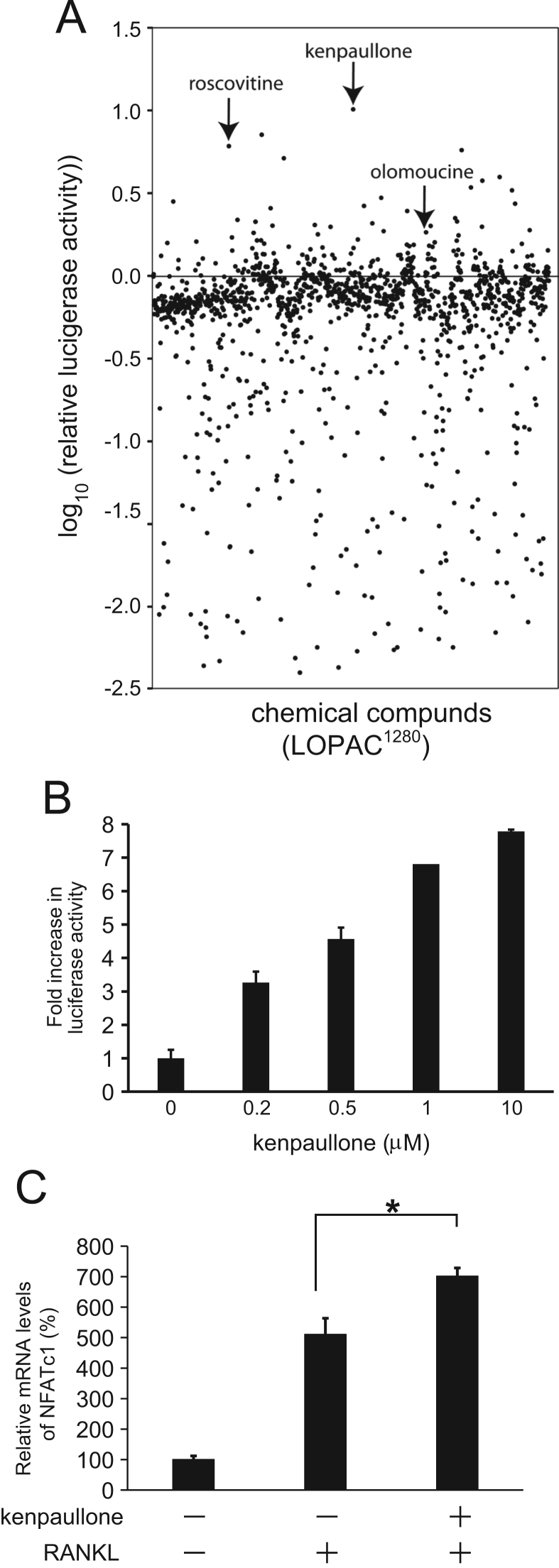
Previously, we constructed RAW264.7 reporter cell line (pNFAT/Luc-RAW) that stably expresses luciferase under the control of NFAT-response element activation-dependent promoter [10]. By using this cell line, we screened the small compound Library of Pharmacologically Active Compounds (LOPAC1280, Sigma) to identify drugs which modulate osteoclastogenic pathway. The cells were treated with sRANKL in the presence of 10 μM of each library compound or DMSO as a control, and the luciferase activity was measured after 24 h of incubation (n=3). Relative luciferase activity is defined as log10 ratio of the activity in the presence of compound vs. the solvent (DMSO). As shown in Fig. 1A, a number of compounds which significantly enhance or inhibit the NFAT-luciferase activity were identified. In this study, we focused our analysis on the activators for osteoclastogenesis. The top 30 activator candidates for osteoclastogenesis are listed in Table 1. Kenpallone (9-bromo-7,12-dihydroindolo[3,2-d][1]benzazepin-6(5H)-one), a cyclin-dependent kinases (Cdks)/glycogen synthase kinase 3β (GSK-3β) inhibitor, showed the most significant activity in this screen. As shown in Fig. 1B, the kenpaullone enhanced NFAT-luciferase activity in a concentration-dependent manner in the range of which we tested (0–10 μM). Furthermore, to see the effect of kenpaullone on endogenous NFATc1 mRNA expression levels, we induced osteoclast formation in RAW264.7 cells with sRANKL in the presence (5 μM) or absence of kenpaullone for 4 days, and subsequently analyzed the NFATc1 mRNA levels by real-time RT-PCR. The mRNA levels were normalized by the levels of control sample (no sRANKL). As shown in Fig. 1C, the NFATc1 mRNA expression was significantly enhanced in the presence of kenpaullone compared to the cells without the compound.
Fig. 1.
Screening of compounds which modulate osteoclastogenesis. (A) Screening of LOPAC1280 by using cell-based NFAT-luciferase reporter assay. The library compounds in a 96-well format were incubated with pNFAT/Luc-RAW cells at concentrations of 10 μM in the presence of 100 ng/ml sRNAKL. Luciferase activity was measured after 24 h of the treatment. The data represents mean values (n=3) of log10 ratio of relative luciferase activity for each compound. Arrows indicate kenpaullone, roscovitine, and olomoucine. (B) Kenpaullone enhances NFAT promoter. Promoter activity of NFAT-response element in pNFAT/Luc-RAW cells was measured by luciferase reporter assay at 24 h after treatment with Kenpaullone. The results are expressed as the mean values±S.E. (n=3). (C) Kenpaullone upregulates NFATc1 mRNA levels in the presence of sRANKL. NFATc1 mRNA levels in the presence or absence of 5 μM kenpaullone with sRANKL were determined by real-time RT-PCR and the relative mRNA expression levels were compared. The data represent mean values±S.E. with triplicate samples (*P<0.01: ANOVA with Tukey's multiple comparison test).
Table 1.
Top 30 compound hits which upregulate the expression of NFAT-luciferase reporter.
| Compounds | Targets | Mean values of relative luciferase activity (log10) | |
|---|---|---|---|
| 1 | Kenpaullone | Cdks/GSK3β | 1.012 |
| 2 | CK2 Inhibitor 2 | Casein kinase II | 0.858 |
| 3 | Roscovitine | Cdks | 0.789 |
| 4 | Phenamil methanesulfonate | Amiloride-sensitive Na+channels | 0.765 |
| 5 | 3-Deazaadenosine | S-adenosyl homocysteine hydrolase | 0.716 |
| 6 | SU 4312 | VEGF receptor-2 | 0.602 |
| 7 | SB-366791 | TRPV1 receptor | 0.580 |
| 8 | PD 98059 | MEK | 0.539 |
| 9 | SIB 1757 | mGluR5 | 0.521 |
| 10 | 6-Methyl-2-(phenylethynyl)pyridine hydrochloride | mGluR5 | 0.476 |
| 11 | Apigenin | Casein kinase II | 0.454 |
| 12 | SIB 1893 | mGluR5 | 0.440 |
| 13 | 1-(5-Isoquinolinylsulfonyl)-2-methylpiperazine dihydrochloride | Ser/Thr kinase inhibitor | 0.427 |
| 14 | Tyrphostin AG 1296 | PDGF receptor kinase | 0.413 |
| 15 | H-8 dihydrochloride | PKA | 0.396 |
| 16 | BF-170 hydrochloride | Tau | 0.354 |
| 17 | 9-Cyclopentyladenine | Adenylyl cyclase | 0.343 |
| 18 | 1,4-PBIT dihydrobromide | iNOS | 0.333 |
| 19 | N6-Cyclohexyladenosine | Adenosine A1 receptor | 0.332 |
| 20 | Lansoprazole | Proton pump | 0.322 |
| 21 | Phenserine | Acetylcholinesterase | 0.312 |
| 22 | D-ribofuranosylbenzimidazole | RNA polII | 0.308 |
| 23 | 1-Phenyl-3-(2-thiazolyl)-2-thiourea | Dopamine β-hydroxylase | 0.307 |
| 24 | Riluzole | Sodium channels | 0.302 |
| 25 | Ciproxifan hydrochloride | H3 receptor | 0.301 |
| 26 | AMG 9810 | TRPV1 receptor | 0.281 |
| 27 | Clorgyline hydrochloride | Monoamine oxidase | 0.274 |
| 28 | Olomoucine | Cdks | 0.268 |
| 29 | Chloro-IB-MECA | Adenosine A3 receptor | 0.264 |
| 30 | N-Methyl-beta-carboline-3-carboxamide | Benzodiazepine receptor | 0.255 |
3.2. Enhancing effect of kenpaullone, olomoucine, and roscovitine on osteoclastogenesis
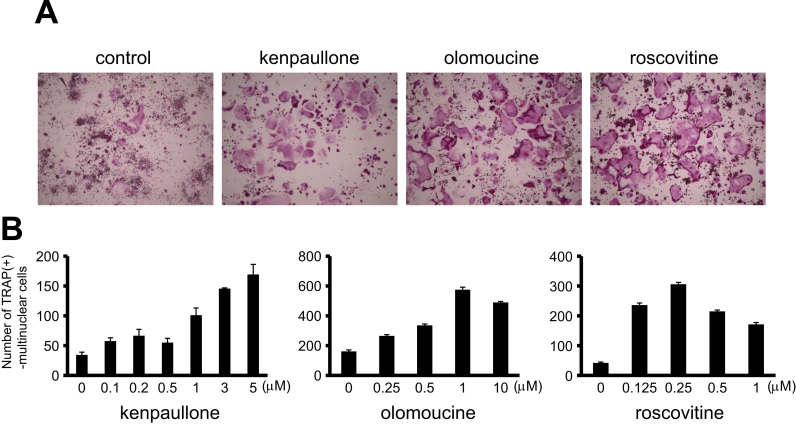
To determine the effect of kenpaullone on osteoclastogenesis, RAW264.7 cells were treated with sRANKL in the presence of kenpaullone. After 4 days of the induction, TRAP staining was performed. As shown in Fig. 2A, 5 μM kenpaullone significantly increased a number of multinuclear mature osteoclasts compared to the control. As shown in Table 1, another two Cdk inhibitors, roscovitine and olomoucine, are in the list of top 30 activator candidates. Therefore, we also examined the effect of roscovitine and olomoucine on osteoclastogenesis. As with the kenpaullone, 5 μM of olomoucin and roscovitine enhanced the formation of osteoclasts (Fig. 2A). Then, we determined the number of TRAP-positive mature osteoclasts with different concentrations of each compound. All compounds, kenpaullone, olomoucine, and roscovitine, remarkably increased the osteoclast differentiation in a concentration-dependent manner until 5 μM, 1 μM, and 0.25 μM, respectively (Fig. 2B). To confirm the effect of the compounds on osteoclastogenesis, we performed the experiment by using primary pre-osteoclast cultures derived from mice. Since higher concentration, in the range of μM, of the compounds showed toxicity to the bone marrow cells (data not shown), we reduced the concentration of all compounds to 50 nM. After 5 days of osteoclastogenic induction by M-CSF and sRANKL with or without each compound, we observed an increased number of TRAP-positive mature osteoclasts in the presence of kenpaullone, olomoucine, or roscovitine compared to the control (DMSO) (Fig. 3A). Then, we determined the number of the multinuclear osteoclasts. Consistent with the result of using RAW264.7 cells (Fig. 2), the all compounds, kenpullone, olomoucine, and roscovitine, showed ~3-fold increase in the number of osteoclast induced from primary bone marrow cells (Fig. 3B). Taken together, kenpaullone, olomoucine, and roscovitine significantly enhanced the differentiation of both RAW264.7 and primary pre-osteoclast cells (Fig. 2, Fig. 3), indicating that GSK-3β and/or Cdks negatively regulate osteoclastogenesis.
Fig. 2.
Kenpaullone, olomoucine, and roscovitine enhance osteoclastogenesis of RAW264.7. (A) The images of TRAP-stained osteoclasts differentiated from RAW264.7 cells are shown. RAW264.7 cells were induced osteoclastogenic differentiation by 100 ng/ml sRANKL in the presence of 5 μM of kenpaullone, olomoucine, or roscovitine. (B) The number of generated multinuclear osteoclasts in the presence of various concentrations of the compounds, kenpaullone, olomoucine, and roscovitine, were evaluated after 4 days of the induction. The data represent mean values±S.E. with triplicate samples.
Fig. 3.
Kenpaullone, olomoucine, and roscovitine enhanced osteoclastogenic differentiation of bone marrow-derived pre-osteoclasts. (A) The images of TRAP-stained osteoclasts differentiated from murine bone marrow cells are shown. The primary bone marrow cells were induced osteoclastogenic differentiation by 100 ng/ml sRANKL in the presence of 50 nM kenpaullone, olomoucine, or roscovitine. (B) The number of generated multinuclear osteoclasts in the presence of the compounds, kenpaullone, olomoucine, and roscovitine, were evaluated after 5 days of the induction. The data represent mean values±S.D. with triplicate samples.
3.3. Kenpaullone, olomoucine, and roscovitine increase the levels of mature cathepsin K in the resultant osteoclasts
To determine whether the resultant mature osteoclasts in the presence of the compounds are functional or not, we analyzed the levels of cathpsin K, which is an essential enzyme for bone resorption activity of the mature osteoclast [3], [4], [12]. Cathepsin K is processed from premature (approximately 40 kDa) to mature form (approximately 29 kDa) during osteoclastogenesis. RAW264.7 cells were induced osteoclastogenic differentiation by RANKL in the presence or absence of the compounds, kenpuallone, olomoucine, or roscovotine. After 4 days of the incubation, levels of cathepsin K in the soluble cell extracts were analyzed by Western blot. As shown in Fig. 4, the ratio of the levels of mature cathepsin K to premature cathepsin K in the presence of 5 μM of each compound, kenpaullone, olomoucine, or roscovitine, was significantly higher compared to the control. This result suggests that the GSK-3β and/or Cdks inhibitors enhance the resorption activity of the resultant osteoclasts through promoting osteoclastogenesis.
Fig. 4.
Kenpaullone, olomoucine, and roscovitine increase the levels of mature cathepsin K. RAW264.7 cells were incubated with 100 ng/ml sRANKL in the presence of 5 μM of kenpaullone, olomoucine, or roscovitine. After 4 days of the induction, total cell lysates were subjected to Western blot analysis with anti-cathepsin K and anti-actin antibodies. Premature and mature forms of cathepsisn K are approximately 40 kDa and 29 kDa, respectively.
3.4. Anti-proliferative effect of kenpaullone, olomoucine, and roscovitine on RAW264.7 cells
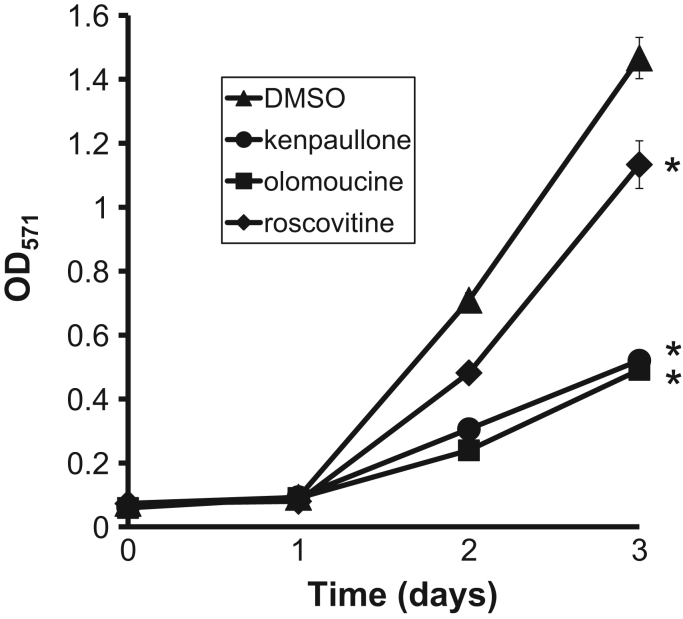
Kenpaullone is known to have an anti-proliferative effect on MCF-10A cells which are immortalized, non-transformed epithelial cells derived from human fibrocystic tissue [13]. Also, roscovitine and olomoucine cause cell-cycle arrest on mouse lymphocytic leukemia cells, L1210, and small cell lung cancer cell lines, NCI-H417 and DMS153 [14]. To investigate the effect of the three compounds on cell proliferation of RAW264.7 cells, we performed MTT assay. As shown in Fig. 5, the all three compounds caused retardation to the cell proliferation: kenpaullone and olomoucine inhibited to a greater extent than roscovitine. This result suggest that the decrease in cell cycle progression triggered by GSK-3β and/or Cdks inhibition could facilitate the direction of the pre-osteoclast cells toward osteoclastogenic differentiation.
Fig. 5.
Kenpaullone, olomoucine, and roscovitine affect cell proliferation of RAW264.7. RAW264.7 cells were seeded onto 96-well plates and cultured in the presence of 0.1% DMSO (▲), 5 μM kenpaullone (●),5 μM olomoucine (■), or 5 μM roscovitine (♦) for 0 (6 h), 1, 2, and 3 days followed by the addition of MTT. Optical densities (OD) were measured at a wavelength of 571 nm. The data represent mean values±S.D. with triplicate samples (*P<0.01: ANOVA with Tukey's multiple comparison test).
4. Discussion
We screened chemical compounds which modulate osteoclastogenesis and identified that Cdks/GSK3-β inhibitor, kenpaullone, as well as Cdks inhibitors, roscovitine and olomoucine, have enhancing effect on osteoclast differentiation of RAW264.7 through upregulating NFATc1 expression (Fig. 1, Fig. 2). Then, we confirmed that the all three inhibitors activates osteoclasogenesis of primary pre-osteoclasts derived from murine bone marrow cells (Fig. 3). Furthermore, the cathepsin K processing during the osteoclastogenesis was promoted by the compounds, suggesting that the bone absorption activity could be also increased (Fig. 4).
Kenpaullone is a well-known Cdk/GSK-3β inhibitor [13], [15]. Recent studies have shown that GSK-3β negatively regulates osteoclastogenesis by directly phosphorylating NFATc1 [16]. The phosphorylated NFATc1 is no longer able to bind DNA and is transported to cytoplasm resulting in downregulation of NFATc1-dependent osteoclast-related genes [17]. It has been shown that GSK-3β inhibitors such as SB216763, SB415286, and kenpaullone remarkably enhance osteoclastogenesis [18]. We also confirmed that GSK-3β inhibitors, SB415286 and CHIR99021, significantly enhanced the differentiation of RAW264.7 (data not shown). These results indicate that the GSK-3β inhibitors promote osteocalstogenesis. Among them, kenpaullone has one of the strongest inhibitory function on GSK-3β (IC50=0.023 μM) [15]. Kenpaullone also has a potent inhibitory effect on Cdks with the following IC50 values: Cdk1/cyclin B (IC50=0.4 μM), Cdk2/cyclin A (IC50=0.68 μM), Cdk5/p35 (IC50=0.85 μM) [13]. Intriguingly, our chemical screen identified Cdks inhibitors, olomoucine and roscovitine, as activators of osteoclastogenesis. Olomoucine inhibits Cdk2/cyclin A at IC50=7 μM and Cdk5/P35 at IC50=3 μM [19], while roscovitine has more potency than olomoucine: Cdk2/cyclin A (IC50=0.7 μM) and Cdk5/P35 (IC50=0.16 μM) [20]. Note that roscovitine is unable to inhibit GSK-3β [15]. Thus, olomoucine and roscovitine could enhance osteoclastogenesis by inhibiting Cdks but not GSK-3β while kenpaullone promotes osteoclast differentiation by inhibiting both Cdks and GSK-3β. Hence, our data indicates that the both Cdks and GSK-3β negatively regulate osteoclastogenesis.
It has been suggested that cell proliferation and osteoclast differentiation are regulated by cyclin, Cdk, and Cdk inhibitor proteins [21], [22], [23]. RANKL stimulation triggers dynamic changes in the expression of cell cycle regulatory proteins. Cyclin D2 and D3, positive regulators of G1/S transition, as well as Cdk6 are all upregulated whereas Cdk2 levels are repressed, resulting in S phase delay [21]. Also, Cdk inhibitor proteins, p21CIP1 and p27KIP1 are significantly upregulated at initial stage of the osteoclastogenesis and decreased at 2 days post-RANKL induction in mouse bone marrow-derived macrophages [22], indicating that the cell cycle stalling in osteoclast precursors is a prerequisite for differentiation into osteoclasts. Another study identified a cell cycle-arrested quiescent osteoclast precursors (QuOPs) in vivo which are maintained as committed osteoclast precursors by downregulating Cdks and cyclins and upregulating p27KIP1 [23]. We also observed that the kenpaullone, olomoucine, and roscovitine stall the cell proliferation of the pre-osteoclast cells (Fig. 5). These results indicate that the cell cycle regulatory system plays an important role in osteoclastogenesis pathway, although it remains unclear the molecular mechanism. Further mechanistic studies will be required to understand the physiological role of the cell cycle regulators in osteoclast differentiation and bone remodeling.
Acknowledgments
We thank Ms. Misa Saikawa, Dr. Naoaki Saito, Mr. Yuki Kiyokawa, and Mr. Naoyuki Yamashita (Niigata University, Japan) for assisting experiments and data analyses, and Dr. Jennifer Huen (Academia Sinica, Taiwan) for critical reading of the manuscript. This work was supported by the JSPS KAKENHI Grant Number 15K15683.
Footnotes
Transparency Document associated with this article can be found in the online version at 10.1016/j.bbrep.2015.12.011.
Appendix A. Transparency Document
Transparency Document
Transparency Document
Transparency Document
Transparency Document
Transparency Document
Transparency Document
Transparency Document
Transparency Document
References
- 1.Zaidi M. Skeletal remodeling in health and disease. Nat. Med. 2007;13:791–801. doi: 10.1038/nm1593. [DOI] [PubMed] [Google Scholar]
- 2.Sims N.A., Gooi J.H. Bone remodeling: multiple cellular interactions required for coupling of bone formation and resorption. Semin. Cell Dev. Biol. 2008;19:444–451. doi: 10.1016/j.semcdb.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 3.Raggatt L.J., Partridge N.C. Cellular and molecular mechanisms of bone remodeling. J. Biol. Chem. 2010;285:25103–25108. doi: 10.1074/jbc.R109.041087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teitelbaum S.L. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 5.Blair H.C., Robinson L.J., Zaidi M. Osteoclast signalling pathways. Biochem. Biophys. Res. Commun. 2005;328:728–738. doi: 10.1016/j.bbrc.2004.11.077. [DOI] [PubMed] [Google Scholar]
- 6.Ikeda F., Nishimura R., Matsubara T., Tanaka S., Inoue J., Reddy S.V. Critical roles of c-Jun signaling in regulation of NFAT family and RANKL-regulated osteoclast differentiation. J. Clin. Investig. 2004;114:475–484. doi: 10.1172/JCI19657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takayanagi H. The role of NFAT in osteoclast formation. Ann. N. Y. Acad. Sci. 2007;1116:227–237. doi: 10.1196/annals.1402.071. [DOI] [PubMed] [Google Scholar]
- 8.Nakashima T., Takayanagi H. Osteoimmunology: crosstalk between the immune and bone systems. J. Clin. Immunol. 2009;29:555–567. doi: 10.1007/s10875-009-9316-6. [DOI] [PubMed] [Google Scholar]
- 9.Wada T., Nakashima T., Hiroshi N., Penninger J.M. RANKL–RANK signaling in osteoclastogenesis and bone disease. Trends Mol. Med. 2006;12:17–25. doi: 10.1016/j.molmed.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Egusa H., Doi M., Saeki M., Fukuyasu S., Akashi Y., Yokota Y. The small molecule harmine regulates NFATc1 and Id2 expression in osteoclast progenitor cells. Bone. 2011;49:264–274. doi: 10.1016/j.bone.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Egusa H., Kaneda Y., Akashi Y., Hamada Y., Matsumoto T., Saeki M. Enhanced bone regeneration via multimodal actions of synthetic peptide SVVYGLR on osteoprogenitors and osteoclasts. Biomaterials. 2009;30:4676–4686. doi: 10.1016/j.biomaterials.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 12.Park Y.G., Kim Y.H., Kang S.K., Kim C.H. cAMP–PKA signaling pathway regulates bone resorption mediated by processing of cathepsin K in cultured mouse osteoclasts. Int. Immunopharmacol. 2006;6:947–956. doi: 10.1016/j.intimp.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Zaharevitz D.W., Gussio R., Leost M., Senderowitz A.M., Lahusen T., Kunick C. Discovery and initial characterisation of the paullones, a novel class of small-molecule inhibitors of cyclin-dependent kinases. Cancer Res. 1999;59:2566–2569. [PubMed] [Google Scholar]
- 14.Hamilton G., Klameth L., Rath B., Thalhammer T. Synergism of cyclin-dependent kinase inhibitors with camptothecin derivatives in small cell lung cancer cell lines. Molecules. 2014;19:2077–2088. doi: 10.3390/molecules19022077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bain J., McLauchlan H., Elliott M., Cohen P. The specificities of protein kinase inhibitors: an update. Biochem. J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beals C.R., Sheridan C.M., Turck C.W., Gardner P., Crabtree G.R. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 17.Neal J.W., Clipstone N.A. Glycogen synthase kinase-3 inhibits the DNA binding activity of NFATc. J. Biol. Chem. 2001;276:3666–3673. doi: 10.1074/jbc.M004888200. [DOI] [PubMed] [Google Scholar]
- 18.Jang H.D., Shin J.H., Park D.R., Hong J.H., Yoon K., Ko R. Inactivation of glycogen synthase kinase-3β is required for osteoclast differentiation. J. Biol. Chem. 2011;286:39043–39050. doi: 10.1074/jbc.M111.256768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veselý J., Havlicek L., Strnad M., Blow J.J., Donella-Deana A., Pinna L. Inhibition of cyclin-dependent kinases by purine analogues. Eur. J. Biochem. 1994;224:771–786. doi: 10.1111/j.1432-1033.1994.00771.x. [DOI] [PubMed] [Google Scholar]
- 20.Meijer L., Borgne A., Mulner O., Chong J.P., Blow J.J., Inagaki N. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2 and cdk5. Eur. J. Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- 21.Meiyanto E., Hoshijima M., Ogawa T., Ishida N., Takeya T. Osteoclast differentiation factor modulates cell cycle machinery and causes a delay in S phase progression in RAW264 cells. Biochem. Biophys. Res. Commun. 2001;283:278–283. doi: 10.1006/bbrc.2001.4564. [DOI] [PubMed] [Google Scholar]
- 22.Okahashi N., Murase Y., Koseki T., Sato T., Yamato K., Nishihara T. Osteoclast differentiation is associated with transient upregulation of cyclin-dependent kinase inhibitors p21WAF1/CIP1 and p27KIP1. J. Cell. Biochem. 2000;80:339–345. [PubMed] [Google Scholar]
- 23.Takahashi N., Muto A., Arai A., Mizoguchi T. Identification of cell cycle-arrested quiescent osteoclast precursors in vivo. Adv. Exp. Med. Biol. 2010:21–30. doi: 10.1007/978-1-4419-1050-9_3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency Document
Transparency Document
Transparency Document
Transparency Document
Transparency Document
Transparency Document
Transparency Document
Transparency Document