Abstract
Background
GASP-2 is a secreted multi-domain glycoprotein known as a specific inhibitor of myostatin and GDF-11. Here we investigate the role of GASP-2 on myogenesis and the effect of its glycosylation on its activity.
Methods
GASP-2 overexpression or knockdown by shRNAs were carried out on C2C12 myoblasts cells. In silico analysis of GASP-2 protein was performed to identify its glycosylation sites. We produced a mouse recombinant GASP-2 protein in a prokaryotic system to obtain a fully deglycosylated protein allowing us to study the importance of this post-translational modification on GASP-2 activity.
Results
Both mature and deglycosylated GASP-2 proteins increase C2C12 proliferation and differentiation by inhibiting the myostatin pathway. In silico and western-blot analyses revealed that GASP-2 presents one consensus sequence for N-glycosylation and six potential sites of mucin-type O-glycosylation.
Conclusions
GASP-2 promotes myogenesis and thus independently of its glycosylation.
General significance
This is the first report demonstrating that GASP-2 promotes proliferation and differentiation of myoblasts by inhibiting the canonical pathway of myostatin.
Keywords: GASP-2, Myostatin, Myogenesis, Glycosylation
Highlights
-
•
Myostatin is a key negative regulator of muscle development.
-
•
GASP-2 is a secreted glycoprotein known to interact with myostatin.
-
•
GASP-2 overexpression promotes proliferation and differentiation of myoblasts.
-
•
GASP-2 Knockdown decreases proliferation and differentiation of myoblasts.
-
•
Glycosylation is not required for GASP-2 inhibitor function of myostatin actions.
1. Introduction
GASPs (Growth and Differentiation Factor Associated Serum Protein) are secreted glycoproteins known to interact with myostatin (MSTN), a key regulator of muscle development [1], [2]. These proteins contain multiple protease inhibitors domains [3], [4]. Thus, the two members, GASP-1 and GASP-2 (also named WFIKKN2 and WFIKKN1) are made of four serine proteases inhibitor modules (WAP, Follistatin/Kazal and two Kunitz), a Netrin domain which is implicated in inhibition of metalloproteinases and an Ig domain involved in protein folding (Igc2) [3], [4]. These domains are highly conserved among mammals especially the Follistatin/Kazal, the second Kunitz and the Netrin domains [5]. This conservation strongly suggests the importance of their functions [5]. Since 2003 and its discovery [6], most of the studies have focused on the role of GASP-1 as a myostatin antagonist. It was shown that GASP-1 interacts with and inhibits myostatin via its Follistatin domain [6], [7], [8], [9]. The myostatin is a member of TGFβ superfamily that negatively regulates myogenesis mainly via the SMAD2/3 pathway [10]. The disruption of the MSTN gene in mice leads to a dramatic increase of skeletal muscle mass due to both hyperplasia and hypertrophy [1]. Overexpression of myostatin inhibitors, like Follistatin and FSTL3, leads to an increase of myoblasts proliferation and differentiation and an increase of muscle mass in mice [11], [12]. Recently, we also described in mice overexpressing Gasp-1 a hypermuscular phenotype owing only to hypertrophy, without hyperplasia of the myofibers [13]. We demonstrated that in addition to the inhibition of its canonical pathway, myostatin is up-regulated in these mice leading to the absence of hyperplasia [14].
Unlike GASP-1, GASP-2 was never found associated with myostatin in serum although their interaction is well known [8]. As GASP-1 and GASP-2 share 54% of identity and are expressed, at few exceptions, in similar tissues including skeletal muscle [4], we asked if GASP-2 could also be involved in the regulation of myostatin during myogenesis. In this paper, we analyzed at cellular and molecular levels the consequences on myoblasts proliferation and differentiation of Gasp-2 over or knockdown gene expression. Like most of secreted proteins, GASP-2 is glycosylated. In addition to play a central role on the solubility and half-life of the protein, glycosylation is well known to modulate its function. We also analyzed the involvement of this post-translational modification on GASP-2 activity, focusing our study on its role as an inhibitor of myostatin.
2. Materials and methods
2.1. Cell culture
Mouse C2C12 myoblasts [15] were obtained from American Type Culture Collection (ATCC). C2C12 myoblasts were grown in Growth Medium (GM) consisting in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen) supplemented with 10% Fetal Bovine Serum (FBS, Invitrogen), 100 units ml−1 penicillin and 100 μg ml−1 streptomycin (Invitrogen). C2C12 differentiation was induced upon 70% confluence by serum withdrawal (DMEM with 2% horse serum (HS, Invitrogen)). For proliferation assay, C2C12 were grown or not with 1 μg ml−1 of hGASP-2 or mGASP-2 deglycosylated (deglyco-mGASP-2).
2.2. Bioinformatics analyses
GASP-2 orthologs were retrieved from databases using BLAST (http://www.ncbi.nlm.nih.gov/BLAST) and Ensembl Genome Browser (www.ensembl.org). Alignment was performed using ClustalW program and analyzed with WebLogo. Asparagine and serine/threonine residues potentially glycosylated were identified using respectively NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/) and NetOGlyc 3.1 Server [16].
2.3. Transient transfections of C2C12 cells
Mouse C2C12 myoblasts at 70% confluence were transiently transfected using the XtremeGENE 9 Transfection Reagent (Roche Applied Science) with 5 μg of pcDNA3.1-Gasp-2 or pcDNA3.1-empty in DMEM serum-free media. After 16 h of transfection, the medium was replaced and myoblasts were allowed to proliferate and differentiate for further analyses.
2.4. Production and purification of murine GASP-2 in prokaryotic system
The mouse Gasp-2cDNA coding sequence without peptide signal were amplified and cloned into the pGEX-4-T1 plasmid (Sigma Aldrich). Production was carried out in E.coli BL21 (DE3) in LB (Luria Bertani) supplemented with 0.2% glucose and 100 μg ml−1 ampicillin. Bacteria were grown to reach OD600 at 0.5 and then induced with 0.025 mM IPTG at 20 °C for 14 h at 250 rpm. Bacterial growth was stopped by centrifugation (6000 g, 15 min; 4 °C). Bacteria were lysed in 20 ml of lysis buffer (20 mM Tris–HCl, pH7.5, 250 mM NaCl, 1 mM EDTA, 0.05% Triton X-100, 10 µg ml−1 lysozyme) and centrifuged (48,000g, 20 min, 4 °C). Purification was carried out with the ÄKTAprime system (Amersham-Biosciences). Supernatant was loaded into a GST-Trap HP 5 ml column (GE Healthcare) equilibrated in 20 mM sodium phosphate, pH7.3, 0.15 M NaCl. Elution was performed at a rate of 1 ml min−1 with 50 mM Tris–HCl, pH8, 10 mM reduced glutathione. Cleavage of the GST Tag located at the N-terminal region of the fusion protein was performed with 10 units mg−1 of recombinant thrombin protein (Sigma T68884) by incubation for 16 h at 4 °C. The sample was then concentrated on an Amicon Ultra 30 K filter (Millipore) by centrifugation (4000g, 4 °C).
2.5. Quantitative real-time PCR (qPCR)
Total RNA from cells and tissues was isolated using RNeasy midi kit (Qiagen). Synthesis of cDNA was performed with the High Capacity cDNA Archive Kit (Applied Biosystems) to convert 2 μg of total RNA into single-stranded cDNA. Real-time PCR was performed in triplicate using 50 ng of cDNA. Relative amounts of transcripts were determined using Taqman probes specific for Gasp-2 (Mm01308311_m1), β2m (Mm00437762_m1), Dffa (Mm00507317) and MyoG (Mm00446194_m1), on an ABI PRISM© 7900 System. Relative mRNA expression values were calculated by the ΔΔCt method with normalization of each sample to the average change in cycle threshold (Ct) value of the controls. For all analyses, three independent experiments have been performed, each assay corresponding to 3 wells /condition/ probe.
2.6. Proliferation assay
Three independent experiments have been carried out. Each experiment corresponds to the analysis of 12 wells with 2000 cells/well at t=0 h. The CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega) was used to evaluate the cell proliferation during 72 h. 20 µl de MTS solution was added to the cells for 3 h and absorbance at 490 nm was then recorded with a 96-well plate reader.
2.7. Fusion index
The fusion index corresponds to the proportion of nuclei present within myotubes that contain two or more nuclei. Cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (PFA) in PBS for 15 min. The fusion index was analyzed after hematoxylin and eosin (H&E) staining.
2.8. Lentiviral-mediated knockdown of Gasp‐2
Individual shRNA constructs specifically designed to target the Gasp-2 were purchased from Sigma Aldrich (Gasp-2_sh1: XM_128578.3-784s1c1; Gasp-2_sh2: XM_128578.3-500s1c1). Lentiviral particles, consisting of the shRNA transfer vector PLKO.1-puro (which contains the sequence of shRNA as well as the cis acting sequences necessary for RNA production and packaging) and the mission lentiviral packaging (Sigma Aldrich). Mission lentiviral particles were generated from different components. The packaging vector contains the minimal set of lentiviral genes required to generate the virion structural proteins and packaging functions. The pCMV-VSV-G envelope vector provides the envelope vector for pseudo-typing. The PLKO.1-puro Gasp-2 shRNA or empty pLKO.1 and pCMV-VSVG vectors were transfected into HEK293Tcells using a Lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s instructions. Briefly, HEK293T cells were grown in GM at 70-80% confluence and were transfected with 1 µg of plasmid PLKO.1-puro with the mission lentiviral packaging mix. After 16 h of transfection, the medium was replaced with fresh GM and cells were incubated for a further 60 h. The supernatant was then collected as a source of viral particles. C2C12 myoblasts were infected with lentivirus-containing media for 24 h and selected with 2 μg ml−1 puromycin.
2.9. SDS-page and western blot analyses
Cells and tissues were collected in lysis buffer (50 mM Tris, pH8, 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% sodium deoxycholate, and protease inhibitors) followed by centrifugation at 12,000 g, 20 min, 4 °C. Protein quantification was performed with a Bradford assay. Proteins were separated under denaturing conditions into a 10% SDS-polyacrylamide gel before being transferred during 1.5 h at 40 mA to a nitrocellulose membrane. Membranes were blocked in TBS (Tris-Buffered Saline) with 0.1% Tween 20% and 5% non-fat dry milk. Membranes were incubated overnight with a monoclonal mouse CDK2 (1:500, sc-6248, Santa Cruz), a mouse monoclonal p21 (1:400, sc-53870, Santa Cruz), a goat polyclonal SMAD2/3 (1:2000, AF3797, R&D Systems), a polyclonal rabbit phospho–SMAD3 (1:500, AB3226, R&D Systems), a polyclonal goat GASP-2 (1:1000, AF2136, R&D Systems), a polyclonal mouse MyHC (Myosin Heavy Chain, 1:4000, M4276, Sigma Aldrich) and polyclonal goat anti-mouse GAPDH antibody, (1:2000, AF5718, R&D Systems) diluted in 2% non-fat dry milk. After 4 washes in TBS-Tween 0.1% (v/v), membranes were incubated for 1 h at room temperature with 1:1000 dilution of secondary antibodies: anti-goat IgG horse-radish peroxidase (HRP) conjugate (P0449, Dako), anti-mouse IgG HRP conjugate (P0447, Dako) or anti-rabbit IgG HRP conjugate (P0399, Dako). After 4 more washes in TBS-Tween 0.1% (v/v), immunoblots were developed by enhanced chemiluminescence. The films were analyzed using ImageQuant TL software (GE Healthcare).
2.10. Glycosylation analysis
N-deglycosylation: 250 ng of purified proteins were denatured at 95 °C for 10 min in 10 mM potassium phosphate and 0.2% SDS. The samples were then incubated overnight at 37 °C in a buffer (10 mM potassium phosphate, pH8, 10 mM EDTA, 0.5% Triton X-100, 0.2% SDS, 1% β-mercaptoethanol) containing 0.1–1 U of PNGase F (Roche Applied Science).
O-deglycosylation: The O-deglycosylation was performed with the EDEGLY kit (Sigma-Aldrich).
250 ng of purified proteins were denatured at 100 °C for 5 min in 2.5 μl of denaturation solution (Sigma-Aldrich) and 10 μl of reaction buffer 5X. Samples were incubated overnight at 37 °C with 1 μl of each enzyme (PNGase F, O-Glycosidase, α-(2→3,6,8,9)-Neuraminidase, β-N-Acetylglucosaminidase, β-(1→4)-Galactosidase).
3. Results
3.1. GASP-2 regulates myogenesis
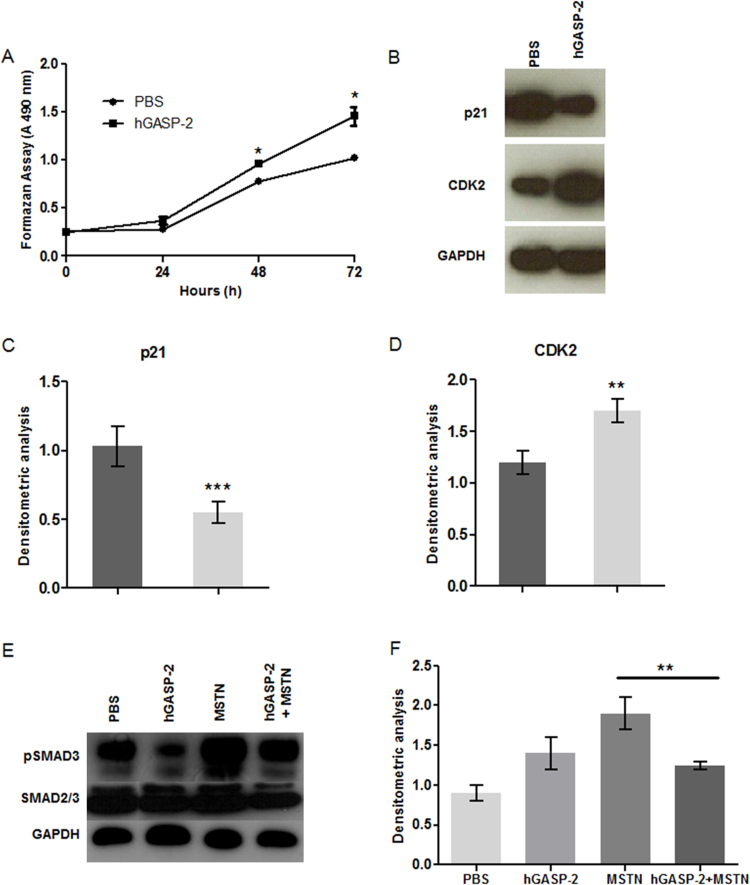
To study the role of GASP-2 in myogenesis, we treated C2C12 myoblasts cells with 1 µg ml−1 of hGASP-2 protein. Cell proliferation assay showed that hGASP-2 treatment improves significatively myoblast proliferation rate compared to control cells (29% at 48 h; 46% at 72 h) (Fig. 1A). We also analyzed the expression of cyclin-dependent kinase 2 (CDK2) that positively regulates cell-cycle progression. Western blot analyses showed an increase of CDK2 in C2C12 myoblasts treated with hGASP-2 compared to the control, consistent with an increased proliferation rate, while p21 expression, a cyclin-dependent kinase inhibitor (CKI), decreases in treated myoblasts (Fig. 1B–D). These results revealed that hGASP-2 treatment promotes cell-cycle progression. To determine whether the increase of C2C12 proliferation rate in presence of hGASP-2 was due to an inhibition of myostatin (MSTN), we evaluated the phospho-SMAD3 (pSMAD3) levels of C2C12 cells treated with MSTN and/or hGASP-2 (Fig. 1E–F). As expected C2C12 myoblasts treatment with MSTN leads to an increase of the pSMAD3 rate compared to the untreated myoblasts. However, hGASP-2 treatment decreases the pSMAD3 signal confirming that hGASP-2 inhibits the MSTN pathway. No change was shown in the total SMAD2/3 rates (Fig. 1E).
Fig. 1.
Enhancement of C2C12 cells proliferation by GASP-2 overexpression. (A) Proliferation analysis of C2C12 cells cultured for 72 h and treated in the absence (PBS) or presence of 1 µg ml−1 of hGASP-2 as measured by formazan assay. Each point corresponds to the mean ± S.D. of three independent experiments. (B-D) Total proteins extracted from C2C12 treated with PBS orhGASP-2 (1 µg ml−1) were resolved by SDS-PAGE. Membranes were immunoblotted with specific anti-CDK2 and anti-p21 antibodies. (E-F) Total proteins extracted from C2C12 treated with MSTN (250 ng ml−1) and/or hGASP-2 (1 µg ml−1) were resolved by SDS-PAGE. Membranes were immunoblotted with specific anti-pSMAD3 and anti-SMAD2/3 antibodies. (B-F) Nitrocellulose membranes were also probed with anti-GAPDH antibodies to show equal loading. The graphs were obtained using Image J software to quantify CDK2, p21, pSMAD3 and SMAD2/3 signals normalized with GAPDH signals of three different experiments.
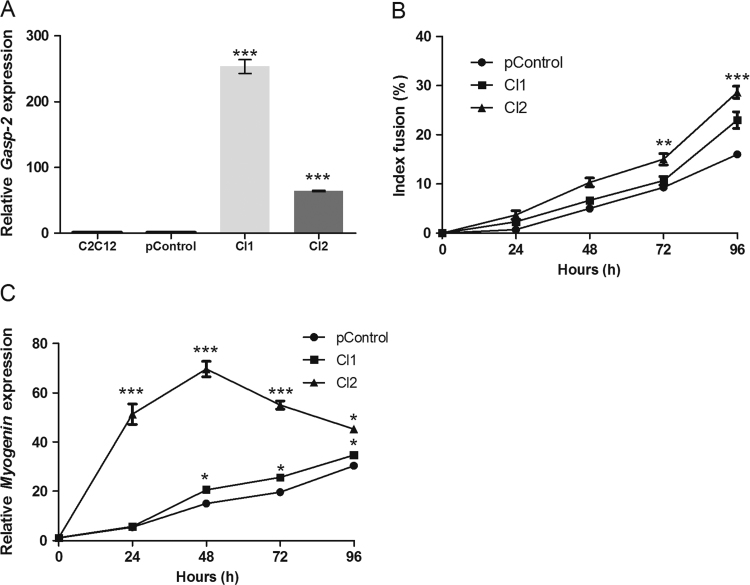
Then, we analyzed the effect of Gasp-2 during C2C12 myoblast differentiation. We transiently transfected the C2C12 cells with a vector containing the Gasp-2 cDNA sequence. Myoblast differentiation was assessed with two clones, Cl1 and Cl2 that overexpress Gasp-2 about 250 times and 80 times respectively (Fig. 2A). Cl2 presents an increase of 15% of myotube formation in 72 h and 17% at 96 h (Fig. 2B). Fusion index is also significantly increased in Cl1 at 96 h compared to C2C12 and control cells (Fig. 2B). No variation in myotubes size was observed in both clones. Consistent with the increase of C2C12 differentiation, myogenin expression, a Myogenic Regulatory Factor (MRF) required for myoblast differentiation, was up-regulated 10 times at 24 h and 48 h of differentiation in the Cl2 myotubes when compared to the control cells (Fig. 2C). Myogenin expression was also slightly up-regulated in Cl1 compared to control cells (Fig. 2C). Differences in transcript levels of Gasp-2 in both clones (Fig. 2A) and the observed Gasp-2 overexpression effect on differentiation (Fig. 2B and C) suggests that there is no dose-dependent consequence on cell differentiation.
Fig. 2.
Enhancement of C2C12 cells differentiation by GASP-2 overexpression. (A) qPCR analysis of Gasp-2 expression in C2C12 transfected with pcDNA3.1-Gasp-2 (Cl1 and Cl2) or pcDNA3.1-empty (pControl). Each histogram corresponds to the mean±S.D. of three independent experiments. The graph represents fold change normalized to Dffa and β2m expression.(B) Quantification of fusion index C2C12 myotubes transfected with pcDNA3.1-Gasp-2 (Cl1 and Cl2) or pcDNA3.1-empty (pControl) and cultured for 96 h of differentiation. Each point corresponds to the mean ± S.D. of three experiments. (C) qPCR analysis of Myogenin expression during 96 h of differentiation in C2C12 myotubes transfected with pcDNA3.1-Gasp-2 (Cl1 and Cl2) or pcDNA3.1-empty (pControl). Each point corresponds to the mean ± S.D. of three independent experiments. The graph represents fold change normalized to Dffa and β2m. Statistical significance was determined using a t-test analysis. *: p<0.05; **: p<0.01; ***: p value<0.005.
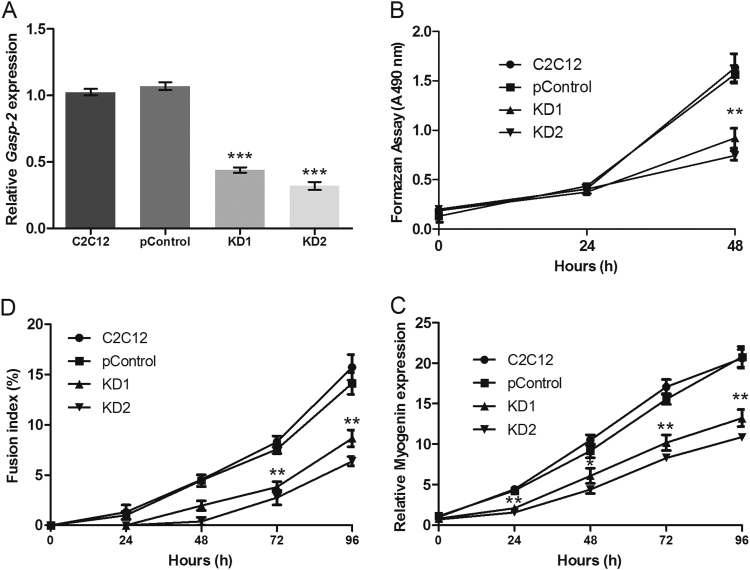
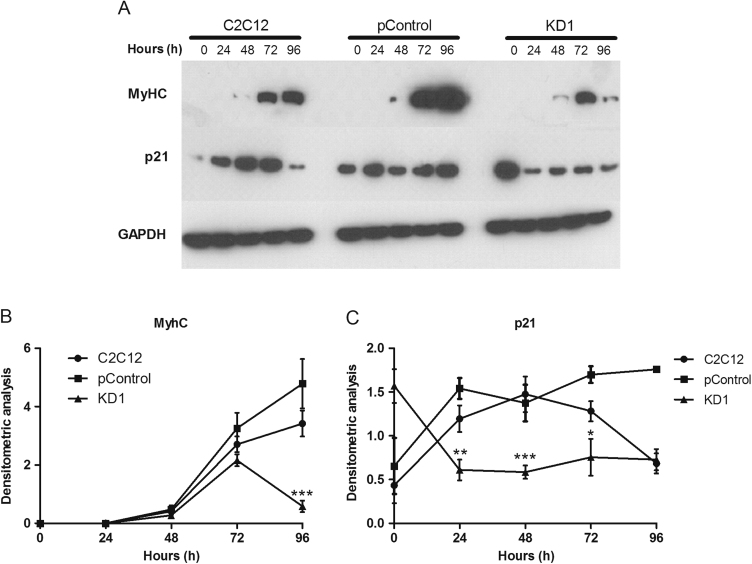
In addition, we generated cell lines that underexpress Gasp-2 using two different lentivirus based-shRNA. These cell lines were named KD1 and KD2. Subsequent qPCR analyses revealed a significant reduction of Gasp-2 expression in both cell lines, around 60% for KD1 and 65% for KD2 (Fig. 3A). Next, we assessed proliferation and differentiation of both clones. KD1 and KD2 cell lines present a reduced myoblast proliferation as the number of myoblasts is significantly decreased after 48 h of culture (Fig. 3B). Furthermore, Gasp-2 knockdown also leads to a decreased differentiation (Fig. 3C). Indeed, the fusion index of both clones is decreased about 40% at 72 h and 60% at 96 h compared to the fusion index of C2C12 and control cells (Fig. 3C). No variation in myotubes size was observed in both clones. Myogenin expression is also down-regulated by 50% at 72 h and 96 h of differentiation in both Gasp-2 knockdown clones (Fig. 3D). Finally, we also observed a global decrease of MyHC expression and a global decrease of p21 (a cyclin-dependent kinase inhibitor) in KD1 cell lines compared to the control cells (Fig. 4A–C). Same results were observed in KD2 cell lines (data not shown). All these results correlated with an impairment of differentiation when Gasp-2 is down-regulated. Taken together, these data suggest that Gasp-2 plays an important role during the normal progression of myogenesis, specifically through regulation of both myoblast proliferation and differentiation.
Fig. 3.
Inhibition of C2C12 cells proliferation and differentiation by Gasp-2 knockdown. (A) qPCR analysis of mGasp-2 transcriptional expression in C2C12 cells infected with lentivirus containing either empty shRNA (pControl) or shRNAs (KD1 and KD2) designed to specifically target and repress mGasp-2. Each histogram corresponds to the mean ± S.D. of three independent experiments. The graph represents fold change normalized to Dffa and β2m. (B) Proliferation analysis of C2C12 cells cultured for 48 h not infected or infected with lentivirus containing either empty shRNA (pControl) or shRNAs (KD1 and KD2) as measured by formazan assay. Each histogram corresponds to the mean ± S.D. of three independent experiments. (C) Quantification of fusion index C2C12 myotubes infected with lentivirus containing either empty shRNA (pControl) or shRNAs (KD1 and KD2) and cultured for 96 h of differentiation. Each point corresponds to the mean ± S.D. of three experiments. (D) qPCR analysis of Myogenin expression during 96 h of differentiation in C2C12 myotubes infected with lentivirus containing either empty shRNA (pControl) or shRNAs (KD1 and KD2). Each point corresponds to the mean ± S.D. of three independent experiments. The graph represents fold change normalized to Dffa and β2m. Statistical significance was determined using a t-test analysis. *: p<0.05; **: p<0.01; ***: p<0.005.
Fig. 4.
Expression of MyHC and p21 in Gasp-2 knockdown cells. (A) Immunoblot analysis of MyHC and p21 from 0 h to 96 h of differentiation C2C12 myotubes infected with lentivirus containing either empty shRNA (pControl) or shRNAs (KD1). (B-C) Nitrocellulose membranes were also probed with anti-GAPDH antibodies to show equal loading. The graphs were obtained using Image J software to quantify MyHC and p21 normalized with GAPDH signals of three different experiments. Statistical significance was determined using a t-test analysis. *:p<0.05; **: p<0.01; ***: p<0.005.
3.2. In silico analysis of potential glycosylation sites of murine GASP-2
In silico analysis of GASP-2 with the NetNGlyc Server revealed one consensus N-glycosylation site at asparagine N497. The NetOGlyc Server allowed us to highlight 6 serine or threonine, S170, S171, T177, T178, T182, T206 with a mucin-type O-glycosylation potential higher than the threshold. All were located between the Follistatin/Kazal and the Igc2 domains except for T206 located in the Igc2 domains (Fig. S1).
To examine the conservation of these glycosylation sites, an alignment of GASP-2 primary sequences from 30 vertebrates was performed and represented as a logo. The logo indicated that the potential N-glycosylation site, N497, was highly conserved, correlating with the NetNGlyc result. In addition, all the potential O-glycosylation sites seemed conserved among vertebrates (Fig. S1).
3.3. In vitro validation of hGASP-2 state of glycosylation
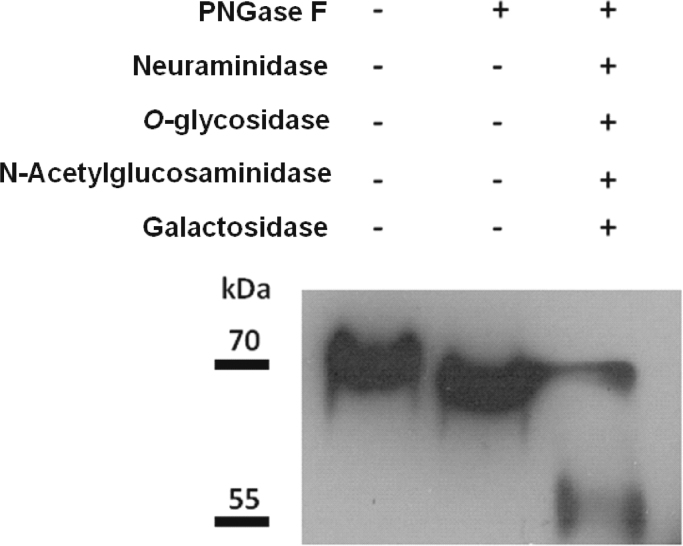
We first investigated the glycosylation state of the human recombinant protein GASP-2 by enzymatic digestions. Western analyses after N-deglycosylation by PNGase F revealed that hGASP-2 migrated faster than the untreated protein but the observed molecular weight was still higher than the predicted one (Fig. 5). PNGase F treatment was completed by four other enzymes (O-Glycosidase, α-(2→3,6,8,9)-Neuraminidase, β-N-Acetylglucosaminidase,β-(1→4)-Galactosidase) to remove the mucin-type O-glycans. This treatment allowed hGASP-2 to migrate at approximately 55 kDa, corresponding to its predicted molecular weight without post-translational modifications. This result confirmed that GASP-2 is N- and O-glycosylated and suggested that these glycosylations are the main post-translational modifications of GASP-2.
Fig. 5.
Characterization of the glycosylation state of hGASP-2 protein. Glycosylated (lane 1), N-deglycosylated (lane 3); N- and O-deglycosylated hGASP-2 protein were detected by Western blot with specific anti-GASP-2 antibody (1:1000).
3.4. Fully deglycosylated GASP-2 protein can inhibit myostatin
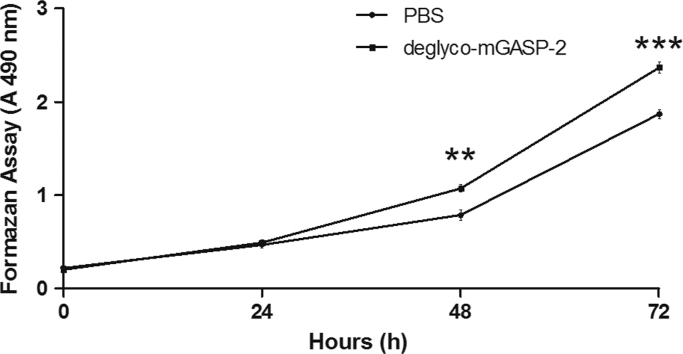
To determine whether the N- and O-glycosylation is required for GASP-2 to inhibit myostatin, we produced and purified the murine GASP-2 in a prokaryotic system to obtain a recombinant protein without glycosylation. We then tested the activity of the deglycosylated GASP-2 on C2C12 myoblasts. C2C12 proliferation is increased in the presence of 1 µg ml−1 deglyco-mGASP-2 (Fig. 6). This increase of the proliferation rate was comparable with the one observed after the treatment of the fully glycosylated commercial hGASP-2 (Fig. 6).
Fig. 6.
Effects of deglycosylated mGASP-2 on C2C12 myoblast proliferation. Proliferation analysis of C2C12 cells in the absence (PBS) or presence of the murine deglycosylated GASP-2 protein (deglyco-mGASP-2, produced in E.coli) cultured for 72 h as measured by formazan assay. Each point corresponds to the mean ± S.D. of three independent experiments. Statistical significance was determined using a t-test analysis. **:p<0.01; ***: p<0.005.
4. Discussion
In this work, we investigated the function of GASP-2 during myogenesis and the importance of glycosylation on its protein activity. Although GASP-2 was shown to interact in vitro with myostatin [8], [9], no data are available concerning the effect of this interaction in a muscular context. A recent study showed that knock-out Gasp-1-/- or Gasp2-/- mice, present a muscular atrophy from 8 months of age and defects in muscle regeneration [17], strongly suggesting an involvement of GASP-2 in myostatin regulation during muscle development. To get a better understanding on GASP-2 functions at cellular and molecular levels, we treated C2C12 cells with either GASP-2 as recombinant proteins or viral particles containing several specific shRNAs against Gasp-2. We showed here for the first time that GASP-2, like its paralogue GASP-1, promotes the proliferation and the differentiation of myoblasts by the inhibition of the myostatin canonical pathway [14], [18], [19]. No significant differences were observed on the development of myoblasts treated with GASP-1 or GASP-2 (data not shown). These data are consistent with the phenotypes described in Gasp-1 and Gasp-2 knock-out mice, even if the atrophy is less pronounced in Gasp-2-/-mice [17]. This could suggest that GASPs are equally involved in muscle development and have a functional redundancy. Therefore, it would be interesting to analyze Gasp-1 and Gasp-2 gene expression levels in Gasp2-/-and Gasp-1-/- mice respectively. In addition, double knock-out Gasp1-/-; Gasp2-/- mice analyses would confirm the involvement of these genes during muscle development. In addition to redundancy between GASP-1 and GASP-2, some studies have reported around myostatin and GDF11 redundancy by characterizing double knock-out Mstn-/-; Gdf11-/- [20], [21], [22]. These mice present only severe axial skeletal defects but no abnormality of skeletal muscle as Gdf-11-/- single mice [20]. All these data suggested no function of GDF-11 in muscle development. Although GASP-2 can inhibit both myostatin and GDF-11, the effects on myogenesis that we observed should be the result of myostatin regulation [17].
We further investigated the structure/function relationship of GASP-2 during its interaction with myostatin. Particularly we focused our study on how GASP-2 glycosylation may influence the interaction between the two proteins and the inhibition function of GASP-2. The two main forms of glycosylation found on secreted glycoproteins are the N-glycans and the mucin-type O-glycans. Thus, we first identified by in silico analyses the potential sites where GASP-2 could be glycosylated. We determined that GASP-2 sequence present a single consensus sequence for N-glycosylation localized in the Netrin domain, that we have later found occupied, and six potential sites of O-glycosylation between the Follistatin/Kazal and the IgC2 domains. Although we are able to prove the presence of mucin-type O-glycans on GASP-2, we cannot determine their exact number and position on the protein. In the aim of providing us a tool to analyze the effect of a fully deglycosylated protein, we decided to produce mGASP-2 in a bacterial system. C2C12 cells treated with deglyco-mGASP-2 did not present any difference in their proliferation compared to cells treated with the fully glycosylated hGASP-2. Nevertheless, we cannot exclude differences in effectiveness with lower protein concentration. Like its paralogue GASP-1, our result seems to prove that glycosylation is not necessary on GASP-2 inhibitor function of myostatin actions. Furthermore, the N-glycosylation could play a role in GASP-2 secretion like it was shown for GASP-1 [19].
We previously reported that the N-glycan site in the Netrin domain and the localization of the O-glycans sites are particularly well conserved among mammals and between the two GASPs [5]. This high degree of conservation tends to indicate that these glycans may play a role in other GASPs biological activities. As the Netrin domain is known to interact with myostatin propeptide, the N-glycosylation could modulate this interaction and help to maintain myostatin in its latent complex form [6], [10], [23]. An interaction test between the propeptide and the deglycosylated form of GASP-2 will help to confirm this hypothesis. It is well known that O-glycans participate in the tridimensional structure [24]. In a muscular context, their presence near the Follistatin/Kazal domain may modulate the interaction with myostatin. Moreover, mucin-type O-glycans were shown to protect proteins against proteolysis [25]. During the interaction between GASPs and myostatin latent complex, their presence may prevent the propeptide degradation and myostatin activation by hiding the cleavage site of the propeptide from BMP-1/Tolloid family [26].
In addition to skeletal muscle, GASPs proteins present a wide tissular expression pattern including kidney, ovary, testis, lung, brain and are highly expressed in pancreas [4]. This latter organ secrets large amount of digestive enzymes, such as trypsin, elastase and chymotrypsin, known to be targets of protease inhibitors. While only an antitrypsin activity of GASPs second Kunitz domain has been described, 3D structure studies indicate that trypsin is not the prime target of this domain [8], [23], [27], suggesting that most relevant proteases remain to be identified. The role of GASP glycosylation in this context should be explored.
5. Conclusions
This structure-function relationship study of GASP-2 demonstrates for the first time its effect on the myostatin signaling pathway during myogenesis. We also show that the lack of any type of glycosylation does not seem to affect its inhibitory role of myostatin actions. Nevertheless, these glycans structures could be involved in other GASP-2 biological activities and need to be investigated in the future.
Acknowledgments
This work was supported by the French National Institute of Agricultural Research (INRA) and by the Limousin Regional Council. LP was supported by a Ph.D. fellowship from INRA/Region Limousin. We thank Pr. Fabrice Lalloué for his help during some of the experiments. The authors thank Dr. Benoît Laporte for his critical reading of the manuscript.
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.03.001.
Contributor Information
Luce Pèrié, Email: luce.perie@etu.unilim.fr.
Alexis Parenté, Email: alexis.parente@etu.unilim.fr.
Caroline Brun, Email: cbrun@ohri.ca.
Laetitia Magnol, Email: laetitia.magnol@unilim.fr.
Patrick Pélissier, Email: patrick.pelissier@unilim.fr.
Véronique Blanquet, Email: veronique.blanquet@unilim.fr.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.McPherron A.C., Lawler A.M., Lee S.J. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 2.Lee S.-J. Regulation of muscle mass by myostatin. Annu. Rev. Cell Dev. Biol. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- 3.Trexler M., Bányai L., Patthy L. A human protein containing multiple types of protease-inhibitory modules. Proc. Natl. Acad. Sci. USA. 2001;98:3705–3709. doi: 10.1073/pnas.061028398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trexler M., Bányai L., Patthy L. Distinct expression pattern of two related human proteins containing multiple types of protease-inhibitory modules. Biol. Chem. 2002;383:223–228. doi: 10.1515/BC.2002.023. [DOI] [PubMed] [Google Scholar]
- 5.Monestier O., Brun C., Cocquempot O., Petit D., Blanquet V. GASP/WFIKKN proteins: evolutionary aspects of their functions. PLoS ONE. 2012;7:e43710. doi: 10.1371/journal.pone.0043710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill J.J. Regulation of myostatin in vivo by growth and differentiation factor-associated serum protein-1: a novel protein with protease inhibitor and follistatin domains. Mol. Endocrinol. 2003;17:1144–1154. doi: 10.1210/me.2002-0366. [DOI] [PubMed] [Google Scholar]
- 7.Phillips D.J., de Kretser D.M. Follistatin: a multifunctional regulatory protein. Front. Neuroendocrinol. 1998;19:287–322. doi: 10.1006/frne.1998.0169. [DOI] [PubMed] [Google Scholar]
- 8.Kondas K., Szlama G., Trexler M., Patthy L. Both WFIKKN1 and WFIKKN2 have high affinity for growth and differentiation factors 8 and 11. J. Biol. Chem. 2008;283:23677–23684. doi: 10.1074/jbc.M803025200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker R.G., Angerman E.B., Kattamuri C., Lee Y.-S., Lee S.-J., Thompson T.B. Alternative binding Modes identified for growth and differentiation factor-associated serum protein (GASP) family antagonism of myostatin. J. Biol. Chem. 2015;290:7506–7516. doi: 10.1074/jbc.M114.624130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas M. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J. Biol. Chem. 2000;275:40235–40243. doi: 10.1074/jbc.M004356200. [DOI] [PubMed] [Google Scholar]
- 11.Amthor H., Nicholas G., McKinnell I., Kemp C.F., Sharma M., Kambadur R. Follistatin complexes myostatin and antagonises myostatin-mediated inhibition of myogenesis. Dev. Biol. 2004;270:19–30. doi: 10.1016/j.ydbio.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 12.Lee S.J. Extracellular regulation of myostatin: a molecular rheostat for muscle mass. Immunol. Endocr. Metab. Agents Med. Chem. 2010;10:183. doi: 10.2174/187152210793663748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monestier O., Brun C., Heu K., Passet B., Malhouroux M., Magnol L. Ubiquitous Gasp1 overexpression in mice leads mainly to a hypermuscular phenotype. BMC Genom. 2012;13:541. doi: 10.1186/1471-2164-13-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brun C., Périé L., Baraige F., Vernus B., Bonnieu A., Blanquet V. Absence of hyperplasia in Gasp-1 overexpressing mice is dependent on myostatin up-regulation. Cell. Physiol. Biochem. 2014;34:1241–1259. doi: 10.1159/000366335. [DOI] [PubMed] [Google Scholar]
- 15.Yaffe D., Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 16.Julenius K., Mølgaard A., Gupta R., Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15:153–164. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y.-S., Lee S.-J. Regulation of GDF-11 and myostatin activity by GASP-1 and GASP-2. Proc. Natl. Acad. Sci. USA. 2013;110:E3713–E3722. doi: 10.1073/pnas.1309907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonala S., Lokireddy S., Arigela H., Teng S., Wahli W., Sharma M. Peroxisome proliferator-activated receptor β/δ induces myogenesis by modulating myostatin activity. J. Biol. Chem. 2012;287:12935–12951. doi: 10.1074/jbc.M111.319145. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Brun C., Monestier O., Legardinier S., Maftah A., Blanquet V. Murine GASP-1 N-glycosylation is not essential for its activity on C2C12 myogenic cells but alters its secretion. Cell. Physiol. Biochem. 2012;30:791–804. doi: 10.1159/000341458. [DOI] [PubMed] [Google Scholar]
- 20.McPherron A.C., Huynh T.V., Lee S.-J. Redundancy of Myostatin and growth/differentiation factor 11 function. BMC Dev. Biol. 2009;9:24. doi: 10.1186/1471-213X-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brun C.E., Rudnicki M.A. GDF11 and the mythical fountain of youth. Cell Metab. 2015;22:54–56. doi: 10.1016/j.cmet.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Egerman M.A., Cadena S.M., Gilbert J.A., Meyer A., Nelson H.N., Swalley S.E. GDF11 increases with age and inhibits skeletal muscle regeneration. Cell Metab. 2015;22:164–174. doi: 10.1016/j.cmet.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondás K., Szláma G., Nagy A., Trexler M., Patthy L. Biological functions of the WAP domain-containing multidomain proteins WFIKKN1 and WFIKKN2. Biochem. Soc. Trans. 2011;39:1416–1420. doi: 10.1042/BST0391416. [DOI] [PubMed] [Google Scholar]
- 24.Gerken T.A., Butenhof K.J., Shogren R. Effects of glycosylation on the conformation and dynamics of O-linked glycoproteins: carbon-13 NMR studies of ovine submaxillary mucin. Biochemistry. 1989;28:5536–5543. doi: 10.1021/bi00439a030. [DOI] [PubMed] [Google Scholar]
- 25.Semenov A.G., Postnikov A.B., Tamm N.N., Seferian K.R., Karpova N.S., Bloshchitsyna M.N. Processing of pro-brain Natriuretic peptide is suppressed by O-glycosylation in the region close to the cleavage site. Clin. Chem. 2009;55:489–498. doi: 10.1373/clinchem.2008.113373. [DOI] [PubMed] [Google Scholar]
- 26.Wolfman N.M., McPherron A.C., Pappano W.N., Davies M.V., Song K., Tomkinson K.N., Wright J.F., Zhao L., Sebald S.M., Greenspan D.S., Lee S.J. Activation of latent myostatin by the BMP-1/tolloid family of metalloproteinases. Proc. Natl. Acad. Sci. USA. 2003;100:15842–15846. doi: 10.1073/pnas.2534946100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liepinsh E., Nagy A., Trexler M., Patthy L., Otting G. Second kunitz-type protease inhibitor domain of the human WFIKKN1 Protein. J. Biomol. NMR. 2006;35:73–78. doi: 10.1007/s10858-006-9013-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material