Abstract
Structural polymorphism of DNA has constantly been evolving from the time of illustration of the double helical model of DNA by Watson and Crick. A variety of non-canonical DNA structures have constantly been documented across the globe. DNA attracted worldwide attention as a carrier of genetic information. In addition to the classical Watson–Crick duplex, DNA can actually adopt diverse structures during its active participation in cellular processes like replication, transcription, recombination and repair. Structures like hairpin, cruciform, triplex, G-triplex, quadruplex, i-motif and other alternative non-canonical DNA structures have been studied at length and have also shown their in vivo occurrence. This review mainly focuses on non-canonical structures adopted by DNA oligonucleotides which have certain prerequisites for their formation in terms of sequence, its length, number and orientation of strands along with varied solution conditions. This conformational polymorphism of DNA might be the basis of different functional properties of a specific set of DNA sequences, further giving some insights for various extremely complicated biological phenomena. Many of these structures have already shown their linkages with diseases like cancer and genetic disorders, hence making them an extremely striking target for structure-specific drug designing and therapeutic applications.
Keywords: Hairpin, Triplex, Quadruplex, i-motif, Structural polymorphism, Alternate DNA structures
Graphical abstract
Highlights
-
•
DNA can adopt diverse range of structures other than classical Watson–Crick duplex.
-
•
Discussion of alternate structures like hairpin, cruciform, triplex, quadruplex etc.
-
•
This review gives some insights for the biological relevance of DNA structures.
1. Introduction
In eukaryotic cells, deoxyribose nucleic acid (DNA) is present in its supercoiled form which is stabilized by ancillary proteins. However, during the biological processes such as replication and transcription, DNA gets unwind and may form structures that differ from the Watson–Crick B-form of DNA. This biological phenomenon of adopting various conformations by this marvelous biomolecule is known as structural polymorphism and it depends upon a number of factors like oligonucleotide sequence, solution condition, hydration, ions, proteins, ligands and superhelical stress. B-form of DNA double helix, proposed by Watson and Crick, generally accounts for most of the DNA behavior in the cell [1]. Widely studied DNA conformations are A, B and Z forms, while DNA structures like bulge, hairpin, cruciform, parallel-stranded DNA, triplex, quadruplex, and i-motif are also well documented. These alternative DNA structures might not only be important for interactions with proteins involved in replication, gene expression and recombination but these would also have an impact on DNA damage, repair and genetic stability [2]. They also play different roles in the formation of nucleosomes and other supramolecular structures involving DNA [3].
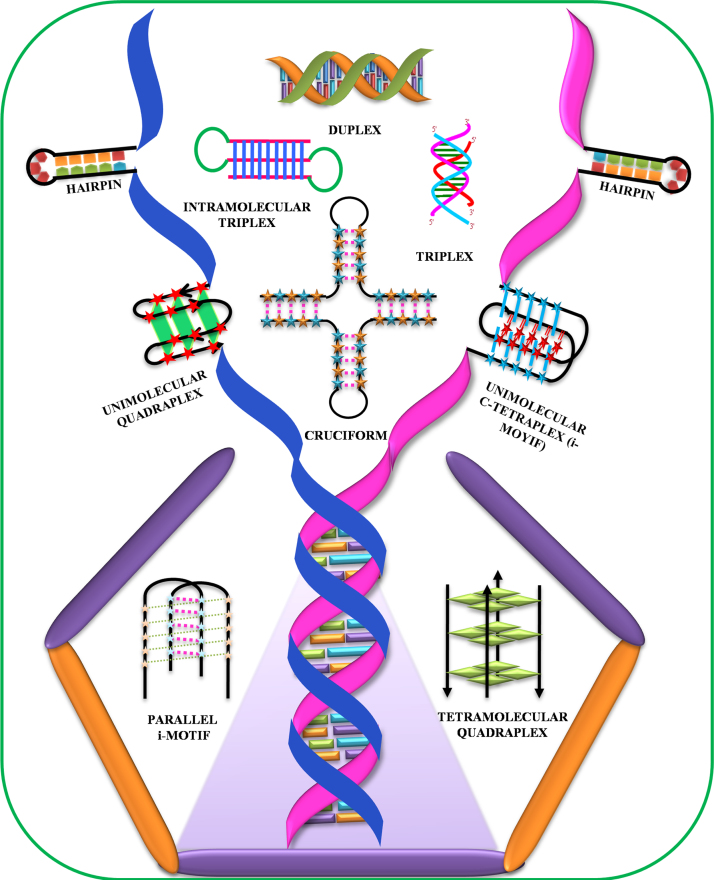
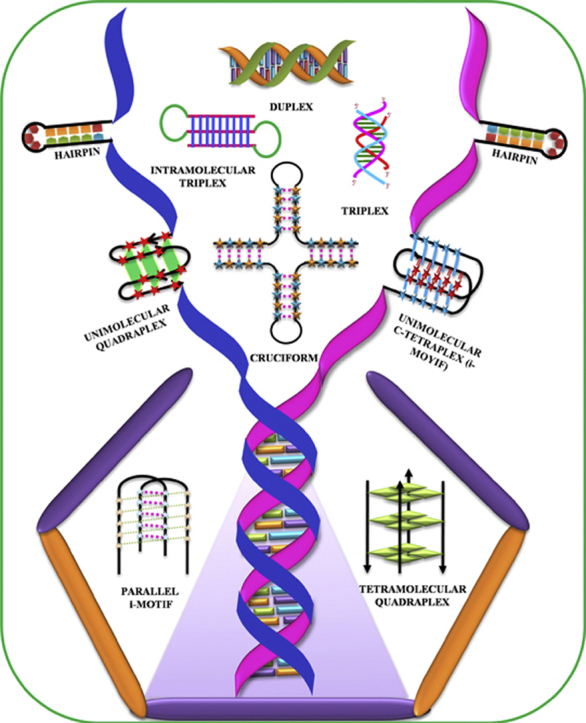
With the exception of the A-form, which is usually adopted by RNA duplexes and is capable of accommodating any sequence, all known DNA structures are sequence-dependent. The sequence requirement for Z-DNA is an alternating purine–pyrimidine/GC-rich sequence and such Z-DNA forming sequences have shown their occurrence near chromosomal breakpoints involving the c-MYC and BCL-2 genes [4]. DNA sequences containing (CGG)n repeats have been shown to form Z-DNA at high salt and millimolar concentrations of Ni2+ ions [5]. Further, their stability can be enhanced with methylation of CGG repeats. The conformational properties of DNA sequences containing (CCA)n and (TGG)n repeats were reported by Zemánek et al. using CD spectroscopy, polyacrylamide gel electrophoresis, and UV absorption spectroscopy. This study revealed that (CCA)n repeats associate to form i-motif structure at acidic pH, while it existed as single strand at alkaline or neutral pH. DNA sequences containing (TGG)n stretches form antiparallel homoduplex or hairpin structure at low salt concentrations, whereas these sequences adopt G-quadruplex structure in the presence of potassium ions at physiological pH [6]. The inverted repeat DNA sequences are capable of forming cruciform structures, while the G-quadruplex and i-motif formation require a contiguous stretch of guanines and cytosines respectively [7], [8]. Some of the sequence requirements for the formation of various non-canonical DNA structures are summarized in Table 1. Most of these non-canonical DNA structures have already shown their in vivo existence, making them biologically very significant. Some of these fascinating DNA structures (Fig. 1) are discussed in the following sections.
Table1.
Summarizing the sequence requirements for various DNA structures.
| S.No. | Structures | Sequence | Reference |
|---|---|---|---|
| 1. | Parallel-Stranded DNA | 5′-CCTATTAAATCC | [51] |
| 5′-AAAAAAAAAATAATTTTAAATATTT | [52] | ||
| 2. | Hairpin | (CAG)n/(CTG)n | [35] |
| 5′-TGGGGA/GCCCCA (Hairpin and duplex) | [11,12] | ||
| 3. | Cruciform | 5′-ATGGTCTTACCTA | [92] |
| 4. | Triplex | 5′-(AAG)5 (Intermolecular triplex) | [93] |
| 5′-C2TC5TC2T5G2AG5AG2T5G2AG5AG2 | [58] | ||
| 5. | i-motif | 5′-CCCTAACCCTAA (Bimolecular) | [65,66] |
| 5′-(CCCTAACCCTAA)2 (Unimolecular) | [66] | ||
| 6. | Quadruplex | 5′-AG6AG3AG3TG2 (Dimeric parallel-stranded) | [73] |
| 5′-GGTTGGTGTGGTTGG (Antiparallel unimolecular) | [94] | ||
| 5′-TTAGGGTTAGGG (Antiparallel tetramer) | [70] | ||
| 7. | Z-DNA | 5′-(CGCGCGCGCGCG)2 | [95] |
| 8. | A-DNA | 5′-(GCGGGCCCGC)2 | [96] |
Fig. 1.
Bouquet of non-canonical DNA structures.
2. Cruciform DNA
DNA supercoiling and base sequences are the two prime factors which are responsible for the structural complexity of DNA molecule. A cruciform structure is formed when interstrand base pairing in duplex DNA with inverted repeats, convert to intrastrand base pairing. It is well documented that an essential requirement for cruciform DNA formation is an inverted repeat DNA sequence which should be embedded in an A+T-rich region [9]. It is of great importance in many biological processes, including the nucleosome positioning, replication and regulation of gene expression. Two mechanisms are proposed for the formation of cruciform structure which differ in salt concentration, activation energy and temperature [10]. The thermodynamic stability of cruciform is very less in comparison to normal B-DNA and showed highly retarded mobility in polyacrylamide gel. Two classes of cruciform have been found till now, one with four-fold symmetry with all arms perpendicular to each other, and another with arms at an acute angle. Cruciform structures are generally a preferential target for many proteins such as HMG proteins, H1, H5 histones, and help to target many diseases [9].
3. Hairpin DNA
DNA sequences with inverted repeats (IRs) or palindromes lead to the formation of hairpin structure, which might also be in equilibrium with duplex DNA, depending upon salt or oligomer concentrations. A single nucleotide polymorphism (SNP) in an 11-mer DNA oligonucleotide (d-TGGGG(A/G)CCCCA) had been shown to exhibit equilibrium between duplex and hairpin [11], [12], while its RNA counterpart (UGGGG(G/A)CCCCA) adopted only hairpin structure [13]. This polymorphism of DNA sequences of Locus Control region (LCR) of beta-globin gene had been discussed from our laboratory in a review [14]. Hairpins have a base-paired stem and a small loop of unpaired bases, which are usually formed via two main mechanisms. In the first mechanism, hairpin is formed in the same way as cruciform structure is formed from double stranded DNA. In the second mechanism, single stranded DNA (ssDNA), produced during various cellular processes like replication on the template for lagging-strand synthesis, DNA repair, rolling-circle replication (RCR), and infection by some viruses leads to hairpin formation [15]. Single strand DNA binding protein (SSB) facilitate the binding of RecA to ssDNA without sequence specificity and leads to hairpin formation [16]. Basically, the possibility of hairpin or cruciform formation at the protein binding site can affect the coiling state of DNA which may either facilitate or prevent the DNA–protein interactions, and alter the gene expression. Although hairpins forming long palindromes are genetically unstable, yet the hairpin structure is known to play a key role in a number of cellular processes such as gene expression, recombination, and transcription.
Poly(dG)•Poly(dC) sequences exhibit A- as well B-forms depending on the solution conditions. These sequences adopt A-form of DNA at higher (molar) salt conditions, while at low (millimolar) salt concentration, they exist in B-form [17]. The transition from B- to A-form can be induced in the presence of methanol or hexamine cobalt or spermine or with the use of high oligomer concentration [18], [19], [20], [21]. On the basis of CD spectroscopy, NMR spectroscopy and unrestricted molecular dynamics, d(CCCCGGGG) was shown to exhibit A- as well as B-like DNA characteristics. This study suggested that stacking between bases is accountable for A-form, while backbone framework is responsible for B-DNA signatures [22].
On the contrary, the sequence with inverted guanine and cytosine tracts i.e. d(GGGGCCCC) showed both B- as well as A-form characteristics which were the attributes of base stacking of cytosine and guanine bases respectively [23]. It had been reported that A-like base stacking geometry of guanine bases in B-DNA is recognized by transcription factors [24] and, therefore, such sequences may play an important role in cellular processes such as transcription and replication [25].
4. DNA bubble or bulge duplex
These DNA structures are formed in double-stranded DNA due to the presence of unpaired nucleotides on one strand and their structure determination along with the dynamics have been reviewed by Turner [26]. Bulges can have one or more nucleotides and are classified in different types depending on their location: on one strand, on both strands (internal loop) or at a junction. These bulges may resemble the replication or transcription bubble thereby affecting the DNA–protein interaction during such events [27]. In addition, DNA bulges are capable of releasing the bending energy of highly bent duplex DNA, thereby enhancing the process of wrapping of DNA around histones and thus acting as a pivotal element for DNA condensation [28].
5. Slipped DNA (S-DNA)
S-DNA is formed by sequences containing stretches of direct repeats and is involved in frame shift mutagenesis during replication. Slippage of several repeat units produces a three way junction in the form of a bulge or a hairpin which is thermodynamically unfavorable, relative to the duplex DNA [29]. Adding or deleting a single base shifts the reading frame, thereby encoding the amino acids different from those present in wild-type protein. Moreover, S-DNA involved in GC-rich triplet repeat mutagenesis (CTG)n•(CAG)n, and (CGG)n•(CCG)n have been associated with genetic neurodegenerative diseases or fragile sites [30], [31]. Originally, expansions were limited to trinucleotide repeats only. Later, it was found that tetrameric (CCTG)n•(CAGG)n [32], pentameric (TGGAA)n•(TTCCA)n [33] and dodecameric (C4GC4GCG)n•(CGCG4CG4)n [34] repeats are also associated with a number of human genetic diseases [35].
6. Bent/Curved DNA
The phenomenon of sequence-directed DNA bending was discovered more than 30 years ago; when restriction fragments from the kinetoplast body of Leishmania tarentolae appeared to migrate anomalously slow on polyacrylamide gels [36]. A subsequent study comparing the electrophoretic migration of circularly permuted DNA molecules of the same length, with the bend positioned differently in each, localized the phenomenon to short stretches of 4–6 adenines, which were repeated in phase with the helical repeat [37].
Bending is important in DNA packaging and in regulating diverse cellular processes. A nucleosome core particle consists of 147 base pairs of DNA wrapped around a histone octamer. The phenomenon of preferential binding of histone octamer at particular positions on a long piece of DNA is called nucleosome positioning. It plays an important role in gene regulation since nucleosome binding to specific DNA regions impede transcription factor binding at these sites, thereby influencing transcription both negatively as well as positively [38]. DNA bending is also believed to facilitate the initial recognition of the mismatched base for repair mechanisms in the cell [39].
7. Parallel-stranded DNA
Apart from the usual antiparallel duplex, DNA also has the ability to form parallel-stranded duplex (ps-duplex), which is stabilized by reverse Watson–Crick or Hoogsteen hydrogen bonding. They may play an important role in regulation of transcription, mutational processes and chromosomal folding.
Purine-rich sequences (dG•dA)n are over-represented in the eukaryotic genome [40] including centromeres and are involved in various biological processes. Such sequences are capable of forming peculiar structures like antiparallel homoduplex, parallel stranded homoduplex (ps-homoduplex), hairpin, and DNA triplex depending on the solution conditions [41], [42]. The sequences with GA repeats, (GA)n associate together to adapt ps-homoduplex at physiological conditions [43], while a single-stranded helical structure [44], [45], [46] was formed at low pH, where adenine is protonated. Such single stranded helical structure is also formed in the presence of ethanol, where adenine is not protonated [47]. A similar sequence (AG)20,30 had been shown to exist into ps-homoduplex having A+•A+ and G•G pairs [46].
DNA, RNA triplex and quadruplex could also be formed using parallel-stranded stretches as a template [48], [49]. Ps-duplexes were shown to be less stable than the antiparallel duplexes [50]. Parvathy et al. demonstrated that C*C+ bonding on both sides of a sequence containing complementary A-T base pairs is responsible for the formation of ps-duplex at acidic pH, which is further confirmed by NMR [51]. Recently, modified nucleosides (dG.isoCd) and (dC.isoGd) have been introduced in order to stabilize ps-duplexes containing G:C pairs [52].
8. Triplex DNA
Triplexes are formed by recognition of oligopurine•oligopyrimidine duplex by single-stranded triplex-forming oligonucleotides (TFOs) in a sequence-specific manner via Hoogsteen or reverse Hoogsteen hydrogen bonds. The existence of triplexes was first shown in 1957, by Felsenfeld et al. [53] which initiated the idea that double helices containing only purines in one chain could bind a third strand polynucleotide containing either pyrimidines [54] or purines [55]. A potential biological function of triplex was discovered in 1968, when Morgan and Wells reported that triplex formation was able to inhibit transcription [54]. The biological relevance of triplexes were not paid any attention for almost 20 years, until two research groups led by Peter Dervan [56] and Claude Hélène [57] in 1987 simultaneously published that short oligonucleotides could be used to induce a DNA cleavage at a specific site on DNA through triplex formation.
Triplexes can be formed either from RNA or DNA chains or their combinations, giving rise to intramolecular or intermolecular triplex, which can occur in both the purine and pyrimidine motifs. Formation of triplex DNA depends on several factors like oligonucleotide length, base composition, pH, divalent cation, and temperature. In a report from our own laboratory, the formation of an intramolecular purine–motif triplex containing human c-jun proto-oncogene target was reported [58]. Specificity and selectivity of the third strand in duplex recognition has led to a variety of potential applications of triplexes in molecular biology, diagnostics, and therapeutics [59], [60], [61]. It was also shown that the triple helix-forming oligonucleotides may inhibit transcription of a specific gene [62]. The research on triplex has become very significant after the detection of triplex within human cells using psoralens [63].
9. i-motif DNA
The complementary strand of G-rich sequences, creates a very interesting structure by making C*C+ base pair, in which two parallel cytosine-rich strands forming duplex are intercalated in antiparallel orientation, resulting into the formation of “intercalated-motif or i-motif DNA” [64], [65], [66]. These structures are known to be formed by one, two or four strands leading to the formation of uni-, bi- and tetramolecular structures, which differ in the strand orientation, sequence length, number of C*C+ base pairs etc. [65], [66]. These structures are stabilized by acidic pH and hence are quite extensively being used as pH switches for a large number of nanotechnology applications [67]. It is assumed that these pH switches or molecular motors will have the ability to function as the natural proteins motors, which are present inside the cells. Also, various ligands have now been designed to stabilize i-motif structures so that they can be explored for more biological applications [64].
10. Guanine quadruplex
Based on the cyclic arrangement of four guanines forming G-tetrads through Hoogsteen hydrogen bonding, a new family of nucleic acid structures was discovered in 1962 by Gellert et al., which was called as G-quadruplex [68]. Guanine-rich DNA sequences have shown their occurrence in most extensively characterized telomeric region of eukaryotic chromosomes [69]. Bioinformatics had been extensively used for investigating the prevalence of G-rich sequences forming quadruplexes in the genomes of various organisms, especially in regions like promoter, 5′-UTR, and oncogenes.
G-quadruplexes are highly polymorphic in nature and may form intramolecular/intermolecular, and parallel/antiparallel structures. Our parent laboratory had also reported an antiparallel tetrameric quadruplex formed by the double repeat (d-TTAGGGTTAGGG) of human telomeric sequence [70]. The observed high stability of quadruplex is due to hydrogen bonding occurring within each quartet, stacking of hydrophobic quartets upon one another and coordination of monovalent counterions (Na+, K+). G-quadruplexes can be classified in terms of strand stoichiometry (uni-, bi- and tetramolecular), orientation (parallel, anti-parallel and mixed), shapes (chair or basket) and loops (lateral, propeller, diagonal, V-shaped) [71]. Some advanced structures like (3+1) G-quadruplex, G-triplex and G-quadruplex with bulges have also now been well documented [8].
Till recently, reports had been limited to uni-, bi- or tetramolecular G-quadruplexes [72], but an interesting dimeric quadruplex formed by human CEB1 Minisatellite had now been reported, which had two stacked subunits involving a parallel snapback arrangement. This scaffold comprises of three double-chain reversal loops, a V-shaped loop and three G-quartet layers. These two subunits are stacked by facing their 5′-end away from each other giving rise to multiple stacking rotamers [73]. So far, only right-handed quadruplexes have been studied, but recently, a left-handed G-quadruplex has also been reported [74]. NMR and X-ray studies of a G-rich sequence from AGRO100 exhibited anti-proliferative activity against cancer cells and had been shown to adopt a parallel-stranded quadruplex motif. The CD spectra showed an inverted profile from that of right-handed topology, confirming its left-handed conformation [74]. Several intriguing structures expand the repertoire of G-quadruplex for the better understanding of this highly polymorphic structure [75], [76].
11. Biological applications of alternative DNA structures
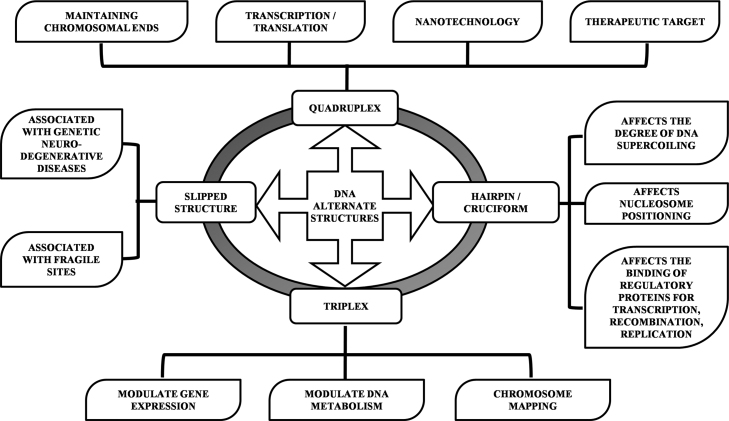
In the last few decades, DNA has been granted the status of ‘Book of life’ with the complete set of instructions to decipher it. As researchers uncover the veil over the existence of wealth of unusual DNA structures, they were baffled over their biological relevance and considered them as transient structures. Scientists have characterized almost more than a dozen of non-canonical DNA structures, among which quadruplex, triplex, cruciform and slipped structures are most common [77]. However, many researchers advocate that these structures represent hotspots for mutations, such as deletions or expansions and also participate in various biological processes governed by the interaction of proteins such as recombination, repair, replication and translocation etc. [78], [79]. Biological applications of various alternative DNA structures are systematically summarized and illustrated in Fig. 2.
Fig. 2.
Pictorial representation of biological applications of non-canonical DNA structures.
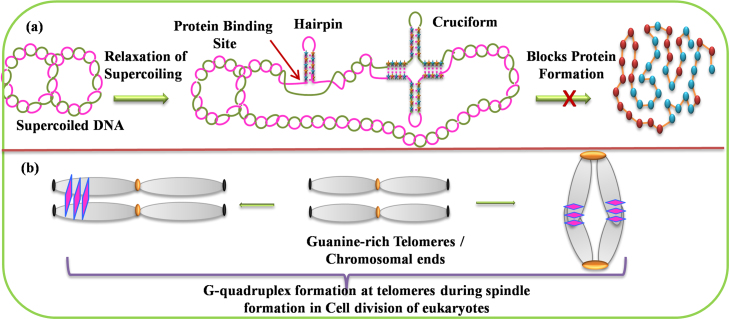
Structured forms of DNA, with intrastrand pairing with inverted repeat sequences form hairpin or cruciform and their thermodynamic stability comes from the relaxation of negative supercoiling and thus affects the degree of DNA supercoiling as well as the positioning of nucleosomes in vivo. There are three ways in which such stem-loop systems can interact with proteins and impact cell physiology: (i) cruciform formation modifies the coiling state of DNA, which is known to affect the binding of regulatory proteins; (ii) the DNA–protein interaction can be inhibited, if a hairpin overlaps a protein recognition site; [80] and (iii) proteins can directly recognize and bind DNA hairpins (Fig. 3a) [9], [15], [30].
Fig. 3.
Biologically relevant structures. (a) Hairpin and Cruciform at protein binding site. (b)Formation of Guanine-quadruplex at chromosomal ends/telomeres.
Likewise, triplex forming sequences are located predominantly in non-coding regions and may play a significant role in regulation, recombination and evolution [81]. The intramolecular DNA triplexes may exist in vivo and may act as molecular switches to modulate gene expression and other DNA metabolism events in a structure-dependent manner, in addition to the well-established sequence-specific regulation [82]. Moser and Dervan demonstrated specific cleavage of plasmid DNA with a short oligonucleotide conjugated to EDTA-Fe and suggested its potential use in chromosome mapping [56]. Effect of pharmaceutical compounds on triplexes [83], [84] and transcription directed therapeutics might extend their applications in future.
It is well known that G-quartet structures play crucial roles in various cellular processes like in maintaining chromosomal ends, transcription, translation etc. [72], along with their wide application in nanotechnology because of their self-assembly formation and stability [85], [86], [87]. The role of G-quadruplex structure in neurodegenerative diseases and non-coding transcriptome has also recently been reported [88].
An illustration showing G-quadruplex formation at chromosomal telomeres suggests its clear biological relevance during the spindle formation in cell division of eukaryotes (Fig. 3b). Recently, G-quadruplex formation has been visualized in the mammalian living system [89] and in the promoter region of oncogene using cross-linking strategy [90]. A quadruplex-forming aptamer with an ability to bind to certain cellular proteins have been found to inhibit proliferation in various cancer cells. It has also been reported that G-quadruplex formation may provide a mean for long-range sensing and communication between distal genomic locations to coordinate regulatory transactions in genomic DNA [91].
12. Outlook and future directions
Understanding about non-canonical DNA structures further opens up a new arena of exploring their biological applications which might be relevant for understanding the mechanism of regulations of the cellular processes. As most of these DNA structures have already shown their in vivo existence in recent past, the mechanistic models of their functioning should now be the focus of the scientific community. Many of these structures like guanine quadruplex have now been linked with the diseases such as cancer and genetic disorders, providing the clues for the future correlation of the same with customized designing of various therapeutic targets. This review reinforces the role of alternative DNA structures in gene regulation along with emphasizing upon the need of understanding their formation and control mechanisms at various genomic locations. Profound insights into these mechanisms of non-canonical DNA structures could also help us devise efficient strategies to combat diseases by altering gene expression.
Acknowledgments
Authors would like to thank Prof. M.M. Chaturvedi, (Director, Cluster Innovation Centre, University of Delhi) for his support and encouragement. Authors would also like to express their gratitude to the University of Delhi (Letter No RC/2014/6820), Delhi for its R&D grants.
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.bbrep.2016.01.013.
Appendix A. Transparency Document
Supplementary material
References
- 1.Watson J.D., Crick F.H. Molecular structure of nucleic acids. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Wang G., Vasquez K.M. Impact of alternative DNA structures on DNA damage, DNA repair, and genetic instability. DNA Repair. 2014;19:143–151. doi: 10.1016/j.dnarep.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinden R.R. Elsevier; California, United States: 2012. DNA Structure and Function. [Google Scholar]
- 4.Wang G., Vasquez K.M. Non-B DNA structure-induced genetic instability. Mutat. Res. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Renčiuk D., Kypr J., Vorlíčková M. CGG repeats associated with fragile X chromosome form left‐handed Z‐DNA structure. Biopolymers. 2011;95:174–181. doi: 10.1002/bip.21555. [DOI] [PubMed] [Google Scholar]
- 6.Zemánek M., Kypr J., Vorlíčková M. Conformational properties of DNA containing (CCA)n and (TGG)n trinucleotide repeats. Int. J. Biol. Macromol. 2005;36:23–32. doi: 10.1016/j.ijbiomac.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Kaushik M., Kaushik S., Bansal A. Structural diversity and specific recognition of four stranded G-quadruplex DNA. Curr. Mol. Med. 2011;11:744–769. doi: 10.2174/156652411798062421. [DOI] [PubMed] [Google Scholar]
- 8.Kaushik M., Kaushik S., Kukreti S. Advancement in the structural polymorphism of G-quadruplexes. Int. Rev. Biophys. Chem. 2014;5 [Google Scholar]
- 9.Brázda V., Laister R.C., Jagelská E.B., Arrowsmith C. Cruciform structures are a common DNA feature important for regulating biological processes. BMC Mol. Biol. 2011;12:33. doi: 10.1186/1471-2199-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murchie A.I., Lilley D.M. The mechanism of cruciform formation in supercoiled DNA: initial opening of central base pairs in salt-dependent extrusion. Nucleic Acids Res. 1987;15:9641–9654. doi: 10.1093/nar/15.23.9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaushik M., Kukreti R., Grover D. Hairpin–duplex equilibrium reflected in the A→B transition in an undecamer quasi‐palindrome present in the locus control region of the human β‐globin gene cluster. Nucleic Acids Res. 2003;31:6904–6915. doi: 10.1093/nar/gkg887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaushik M., Kukreti S. Structural polymorphism exhibited by a quasipalindrome present in the locus control region (LCR) of the human β-globin gene cluster. Nucleic Acids Res. 2006;34:3511–3522. doi: 10.1093/nar/gkl456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaushik M., Kukreti S. Differential structural status of the RNA counterpart of an undecamer quasi-palindromic DNA sequence present in LCR of human β-globin gene cluster. J. Biomol. Struct. Dyn. 2015;33:244–252. doi: 10.1080/07391102.2013.877402. [DOI] [PubMed] [Google Scholar]
- 14.Kukreti S., Kaur H., Kaushik M. Structural polymorphism at LCR and its role in beta-globin gene regulation. Biochimie. 2010;92:1199–1206. doi: 10.1016/j.biochi.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 15.Bikard D., Loot C., Baharoglu Z., Mazel D. Folded DNA in action: hairpin formation and biological functions in prokaryotes. Microbiol. Mol. Biol. Rev. 2010;74:570–588. doi: 10.1128/MMBR.00026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy M.S., Vaze M.B., Madhusudan K., Muniyappa K. Binding of SSB and RecA protein to DNA-containing stem loop structures: SSB ensures the polarity of RecA polymerization on single-stranded DNA. Biochemistry. 2000;39:14250–14262. doi: 10.1021/bi001187+. [DOI] [PubMed] [Google Scholar]
- 17.Nishimura Y., Torigoe C., Tsuboi M. An A-form poly(dG) poly(dC) in H2O solution. Biopolymers. 1985;24:1841–1844. doi: 10.1002/bip.360240913. [DOI] [PubMed] [Google Scholar]
- 18.Vorlícková M., Minyat E.E., Kypr J. Cooperative changes in the chiroptical properties of DNA induced by methanol. Biopolymers. 1984;23:1–4. doi: 10.1002/bip.360230102. [DOI] [PubMed] [Google Scholar]
- 19.Nishimura Y., Torigoe C., Tsuboi M. Salt induced B-A transition of poly(dG).poly(dC) and the stabilization of A-form by its methylation. Nucleic Acids Res. 1986;14:2737–2749. doi: 10.1093/nar/14.6.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benevides J., Wang A.H.J., Rich A., Kyogoku Y., van der Marel G.A., van Boom J.H., Thomas G.J., Jr Raman spectra of single crystals of r(GCG) d(CGC) and d(CCCCGGGG) as models for a DNA, their structure transitions in aqueous solution, and comparison with double-helical poly(dG).poly(dC) Biochemistry. 1986;25:41–50. doi: 10.1021/bi00349a007. [DOI] [PubMed] [Google Scholar]
- 21.Robinson H., Wang A.H.J. Neomycin, spermine and hexaamminecobalt (III) share common structural motifs in converting B-to A-DNA. Nucleic Acids Res. 1996;4:676–682. doi: 10.1093/nar/24.4.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trantírek L., Stefl R., Vorlíckova M., Koca J., Sklenár V., Kypr J. An A-type double-helix of DNA having B-type puckering of the deoxyribose rings. J. Mol. Biol. 2000;297:907–922. doi: 10.1006/jmbi.2000.3592. [DOI] [PubMed] [Google Scholar]
- 23.Stefl R., Trantírek L., Vorlícková M., Koca J., Sklenár V., Kypr J. A-like guanine-guanine stacking in the aqueous DNA duplex of d(GGGGCCCC) J. Mol. Biol. 2001;307:513–524. doi: 10.1006/jmbi.2001.4484. [DOI] [PubMed] [Google Scholar]
- 24.Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986;46:123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- 25.Timsit Y. DNA structure and polymerase fidelity. J. Mol. Biol. 1999;293:835–853. doi: 10.1006/jmbi.1999.3199. [DOI] [PubMed] [Google Scholar]
- 26.Turner D.H. Bulges in nucleic acids. Curr. Opin. Struct. Biol. 1992;2:334–337. [Google Scholar]
- 27.Shin J., Lee O.C., Sung W. How a short double-stranded DNA bends. J. Chem. Phy. 2015;142:155101. doi: 10.1063/1.4916379. [DOI] [PubMed] [Google Scholar]
- 28.Grinevich A.A., Ryasik A.A., Yakushevich L.V. 130 Modeling the DNA bubbles dynamics. J. Biomol. Struct. Dyn. 2015;33:84. [Google Scholar]
- 29.Harvey S.C. Slipped structures in DNA triplet repeat sequences: entropic contributions to genetic instabilities. Biochemistry. 1997;36:3047–3049. doi: 10.1021/bi962771e. [DOI] [PubMed] [Google Scholar]
- 30.V.N. Potaman, R.R. Sinden, DNA: alternative conformations and biology. In: Proceedings of the Madame Curie Bioscience Database [Internet], Landes Bioscience, Austin TX, 2000. Available from: 〈http://www.ncbi.nlm.nih.gov/books/NBK6545/〉
- 31.Sinden R.R., Potaman V.N., Oussatcheva E.A. Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA. J. Biosci. 2002;27:53–65. doi: 10.1007/BF02703683. [DOI] [PubMed] [Google Scholar]
- 32.Liquori C.L., Ricker K., Moseley M.L. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa K., Dürr A., Klopstock T. Pentanucleotide repeats at the spinocerebellar ataxia type 31 (SCA31) locus in caucasians. Neurology. 2001;77:1853–1855. doi: 10.1212/WNL.0b013e3182377e3a. [DOI] [PubMed] [Google Scholar]
- 34.Michelucci R., Pasini E., Riguzzi P. Genetics of epilepsy and relevance to current practice. Curr. Neurol. Neurosci. Rep. 2012;12:445–455. doi: 10.1007/s11910-012-0281-8. [DOI] [PubMed] [Google Scholar]
- 35.Mirkin S.M. DNA structures, repeat expansions and human hereditary disorders. Curr. Opin. Struct. Biol. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Marini J.C., Levene S.D., Crothers D.M., Englund P.T. Bent helical structure in kinetoplast DNA. Proc. Natl. Acad. Sci. USA. 1982;79:7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu H.M., Crothers D.M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984;308:509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- 38.Haran T.E., Mohanty U. The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 2009;42:41–81. doi: 10.1017/S0033583509004752. [DOI] [PubMed] [Google Scholar]
- 39.Sharma M., Predeus A.V., Mukherjee S., Feig M. DNA bending propensity in the presence of base mismatches: implications for DNA repair. J. Phys. Chem. B. 2013;117:6194–6205. doi: 10.1021/jp403127a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manor H., Sridhara Rao B., Martin R.G. Abundance and degree of dispersion of genomic d(GA)n.d(TC)n sequences. J. Mol. Evol. 1988;27(1988):96–101. doi: 10.1007/BF02138367. [DOI] [PubMed] [Google Scholar]
- 41.Kejnovská I., Kypr J., Vondrusková J., Vorlícková M. Towards a better understanding of the unusual conformations of the alternating guanine-adenine repeat strands of DNA. Biopolymers. 2007;85:19–27. doi: 10.1002/bip.20597. [DOI] [PubMed] [Google Scholar]
- 42.Vorlíčková M., Kejnovská I., Bednářová K., Renčiuk D., Kypr J. Circular dichroism spectroscopy of DNA: from duplexes to quadruplexes. Chirality. 2012;24:691–698. doi: 10.1002/chir.22064. [DOI] [PubMed] [Google Scholar]
- 43.Rippe K., Fritsch V., Westhof E., Jovin T.M. Alternating d(G-A) sequences form a parallel-stranded DNA homoduplex. EMBO J. 1992;11:3777–3786. doi: 10.1002/j.1460-2075.1992.tb05463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dolinnaya N.G., Fresco J.R. Single-stranded nucleic acid helical secondary structure stabilized by ionic bonds: d(A(+)-G)10. Proc. Natl. Acad. Sci. USA. 1992;89:9242–9246. doi: 10.1073/pnas.89.19.9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dolinnaya N.G., Braswell E.H., Fossella J.A., Klump H., Fresco J.R. Molecular and thermodynamic properties of d(A(+)-G)10, a single-stranded nucleic acid helix without paired or stacked bases. Biochemistry. 1993;32:10263–10270. doi: 10.1021/bi00089a049. [DOI] [PubMed] [Google Scholar]
- 46.Dolinnaya N.G., Ulku A., Fresco J.R. Parallel-stranded linear homoduplexes of d(A+-G)n>10 and d(A-G)n>10 manifesting the contrasting ionic strength sensitivities of poly(A+•A+) and DNA. Nucleic Acids Res. 1997;25:1100–1107. doi: 10.1093/nar/25.6.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vorlícková M., Kejnovská I., Kovanda J., Kypr J. Dimerization of the guanine-adenine repeat strands of DNA. Nucleic Acids Res. 1997;27:581–586. doi: 10.1093/nar/27.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van De Sande J.H., Ramsing N.B., Germann M.W. Parallel stranded DNA. Science. 1988;241:551–557. doi: 10.1126/science.3399890. [DOI] [PubMed] [Google Scholar]
- 49.Soyfer V.N., Potaman V.N. Springer Science & Business Media; New York, United States: 2012. Triple-Helical Nucleic Acids. [Google Scholar]
- 50.Barsky D., Colvin M.E. Guanine-cytosine base pairs in parallel-stranded DNA: an ab initio study of the keto-amino wobble pair versus the enol-imino minor tautomer pair. J. Phys. Chem. A. 2000;104:8570–8576. [Google Scholar]
- 51.Parvathy V.R., Bhaumik S.R., Chary K.V. NMR structure of a parallel-stranded DNA duplex at atomic resolution. Nucleic Acids Res. 2002;30:1500–1511. doi: 10.1093/nar/30.7.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ming X., Ding P., Leonard P. Parallel-stranded DNA: enhancing duplex stability by the ‘G-clamp’and a pyrrolo-dC derivative. Org. Biomol. Chem. 2012;10:1861–1869. doi: 10.1039/c2ob06606h. [DOI] [PubMed] [Google Scholar]
- 53.Felsenfeld G., Davies D.R., Rich A. Formation of a three-stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023–2024. [Google Scholar]
- 54.Morgan A., Wells R. Specificity of the three-stranded complex formation between double-stranded DNA and single-stranded RNA containing repeating nucleotide sequences. J. Mol. Biol. 1968;37:63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]
- 55.Beal P.A., Dervan P.B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991;251:1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- 56.Moser H.E., Dervan P.B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 57.Le Doan T., Perrouault L., Praseuth D. Sequence-specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo-(α]-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987;15:7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaushik S., Kaushik M., Svinarchuk F. Presence of divalent cation is not mandatory for the formation of intramolecular purine-motif triplex containing human c-jun protooncogene target. Biochemistry. 2011;50:4132–4142. doi: 10.1021/bi1012589. [DOI] [PubMed] [Google Scholar]
- 59.Knauert M.P., Glazer P.M. Triplex forming oligonucleotides: sequence-specific tools for gene targeting. Hum. Mol. Genet. 2001;10:2243–2251. doi: 10.1093/hmg/10.20.2243. [DOI] [PubMed] [Google Scholar]
- 60.Jain A., Wang G., Vasquez K.M. DNA triple helices: biological consequences and therapeutic potential. Biochimie. 2008;90:1117–1130. doi: 10.1016/j.biochi.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buske F.A., Mattick J.S., Bailey T.L. Potential in vivo roles of nucleic acid triple-helices. RNA Biol. 2011;8:427–439. doi: 10.4161/rna.8.3.14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duval-Valentin G., Thuong N.T., Hélène C. Specific inhibition of transcription by triple helix-forming oligonucleotides. Proc. Natl. Acad. Sci. USA. 1992;89:504–508. doi: 10.1073/pnas.89.2.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ping Y.-H., Rana T.M. Mechanism of site-specific psoralen photoadducts formation in triplex DNA directed by psoralen-conjugated oligonucleotides. Biochemistry. 2005;44:2501–2509. doi: 10.1021/bi0488707. [DOI] [PubMed] [Google Scholar]
- 64.Day H.A., Pavlou P., Waller Z.A. i-Motif DNA: Structure, stability and targeting with ligands. Bioorg. Med. Chem. 2014;22:4407–4418. doi: 10.1016/j.bmc.2014.05.047. [DOI] [PubMed] [Google Scholar]
- 65.Kaushik M., Prasad M., Kaushik S. Structural transition from dimeric to tetrameric i‐motif, caused by the presence of TAA at the 3′‐end of human telomeric C‐rich sequence. Biopolymers. 2010;93:150–160. doi: 10.1002/bip.21313. [DOI] [PubMed] [Google Scholar]
- 66.Kaushik M., Suehl N., Marky L.A. Calorimetric unfolding of the bimolecular and i-motif complexes of the human telomere complementary strand, d(C3TA2)4. Biophys. Chem. 2007;126:154–164. doi: 10.1016/j.bpc.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 67.Dong Y., Yang Z., Liu D. DNA nanotechnology based on i-motif structures. Acc. Chem. Res. 2014;47:1853–1860. doi: 10.1021/ar500073a. [DOI] [PubMed] [Google Scholar]
- 68.Gellert M., Lipsett M.N., Davies D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaushik M., Bansal A., Saxena S., Kukreti S. Possibility of an antiparallel (tetramer) quadruplex exhibited by the double repeat of the human telomere. Biochemistry. 2007;46:7119–7131. doi: 10.1021/bi0621009. [DOI] [PubMed] [Google Scholar]
- 71.Karsisiotis A.I., O’Kane C., da Silva M.W. DNA quadruplex folding formalism – a tutorial on quadruplex topologies. Methods. 2013;64:28–35. doi: 10.1016/j.ymeth.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 72.Huppert J.L. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 73.Adrian M., Ang D.J., Lech C.J. Structure and conformational dynamics of a stacked dimeric G-quadruplex formed by the human CEB1 minisatellite. J. Am. Chem. Soc. 2014;136:6297–6305. doi: 10.1021/ja4125274. [DOI] [PubMed] [Google Scholar]
- 74.Chung W.J., Heddi B., Schmitt E. Structure of a left-handed DNA G-quadruplex. Proc. Natl. Acad. Sci. USA. 2015;112:2729–2733. doi: 10.1073/pnas.1418718112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petraccone L. Higher-order quadruplex structures. Top. Curr. Chem. 2013;330:23–46. doi: 10.1007/128_2012_350. [DOI] [PubMed] [Google Scholar]
- 76.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bacolla A., Cooper D.N., Vasquez K.M. DNA structure matters. Genome Med. 2013;5:51. doi: 10.1186/gm455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pearson C.E., Zorbas H., Price G.B., Zannis‐Hadjopoulos M. Inverted repeats, stem‐loops, and cruciforms: significance for initiation of DNA replication. J. Cell. Biochem. 1996;63:1–22. doi: 10.1002/(SICI)1097-4644(199610)63:1%3C1::AID-JCB1%3E3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 79.Zhao J., Bacolla A., Wang G., Vasquez K.M. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Horwitz M., Loeb L.A. An E. coli promoter that regulates transcription by DNA superhelix-induced cruciform extrusion. Science. 1988;241:703–705. doi: 10.1126/science.2456617. [DOI] [PubMed] [Google Scholar]
- 81.Behe M.J. An overabundance of long oligopurine tracts occurs in the genome of simple and complex eukaryotes. Nucleic Acids Res. 1995;23:689–695. doi: 10.1093/nar/23.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zain R., Sun J. Do natural DNA triple-helical structures occur and function in vivo? Cell. Mol. Life Sci. 2003;60:862–870. doi: 10.1007/s00018-003-3046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Besch R., Giovannangeli C., Degitz K. Triplex-forming oligonucleotides-sequence-specific DNA ligands as tools for gene inhibition and for modulation of DNA-associated functions. Curr. Drug Targets. 2004;5:691–703. doi: 10.2174/1389450043345100. [DOI] [PubMed] [Google Scholar]
- 84.Mergny J., Duval-Valentin G., Nguyen C. Triple helix-specific ligands. Science. 1992;256:1681–1684. doi: 10.1126/science.256.5064.1681. [DOI] [PubMed] [Google Scholar]
- 85.Cree S.L., Kennedy M.A. Relevance of G-quadruplex structures to pharmacogenetics. Front. Pharmacol. 2014;5:160. doi: 10.3389/fphar.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mendoza O., Porrini M., Salgado G.F. Orienting tetramolecular G‐quadruplex formation: the quest for the elusive RNA antiparallel quadruplex. Chem. Eur. J. 2015;21:6732–6739. doi: 10.1002/chem.201500358. [DOI] [PubMed] [Google Scholar]
- 87.Yatsunyk L.A., Pietrement O., Albrecht D., Tran P.L.T. Guided assembly of tetramolecular G-quadruplexes. ACS Nano. 2013;7:5701–5710. doi: 10.1021/nn402321g. [DOI] [PubMed] [Google Scholar]
- 88.Simone R., Pietro-Fratta P., Neidle S., Parkinson G.N., Isaacs A.M. G-quadruplexes: emerging roles in neurodegenerative diseases and the non-coding transcriptome. FEBS Lett. 2015;589:1653–1668. doi: 10.1016/j.febslet.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 89.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yuan L., Tian T., Chen Y. Existence of G-quadruplex structures in promoter region of oncogenes confirmed by G-quadruplex DNA cross-linking strategy. Sci. Rep. 2013;3:1–9. doi: 10.1038/srep01811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang C., Liu H.-H, Zheng K.-W. DNA G-quadruplex formation in response to remote downstream transcription activity: long-range sensing and signal transducing in DNA double helix. Nucleic Acids Res. 2013;41:7144–7152. doi: 10.1093/nar/gkt443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pelham C., Jimenez T., Rodova M., Rudolph A., Chipps E., Islam M.R. Regulation of HFE expression by poly (ADP-ribose) polymerase-1 (PARP1) through an inverted repeat DNA sequence in the distal promoter. Biochim. Biophys. Acta Gene Regul. Mech. 1829;2013:1257–1265. doi: 10.1016/j.bbagrm.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rajeswari M.R. DNA triplex structures in neurodegenerative disorder, Friedreich’s ataxia. J. Biosci. 2012;37:519–532. doi: 10.1007/s12038-012-9219-1. [DOI] [PubMed] [Google Scholar]
- 94.Chiorcea-Paquim A.M., Oliveira-Brett A.M. Redox behaviour of G-quadruplexes. Electrochim. Acta. 2014;126:162–170. [Google Scholar]
- 95.Pan F., Roland C., Sagui C. Ion distributions around left-and right-handed DNA and RNA duplexes: a comparative study. Nucleic Acids Res. 2014;42:13981–13996. doi: 10.1093/nar/gku1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu X.J., Shakked Z., Olson W.K. A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material