Abstract
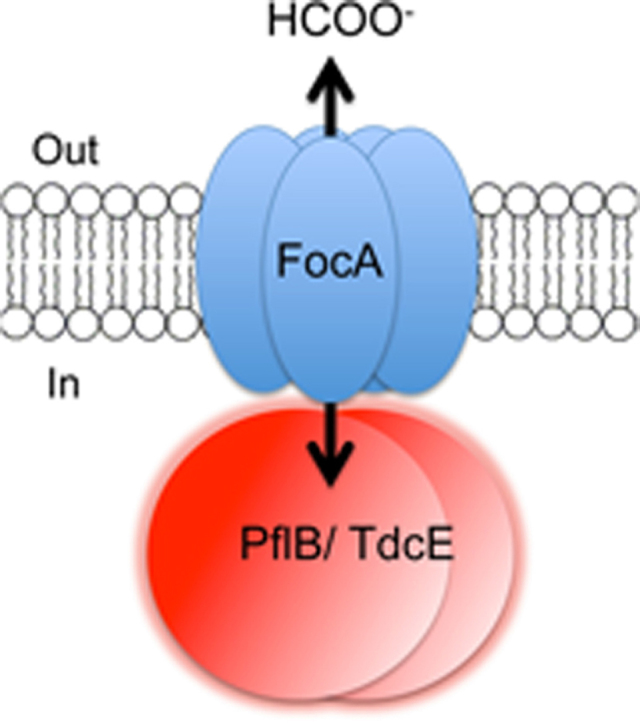
Formate is a major product of mixed-acid fermentation in Escherichia coli. Because formate can act as an uncoupler at high concentration it must be excreted from the cell. The FNT (formate-nitrite transporter) membrane channel FocA ensures formate is translocated across the cytoplasmic membrane. Two glycyl-radical enzymes (GREs), pyruvate formate-lyase (PflB) and 2-ketobutyrate formate-lyase (TdcE), generate formate as a product of catalysis during anaerobic growth of Escherichia coli. We demonstrate in this study that TdcE, like PflB, interacts specifically with FocA. His-tagged variants of two other predicted GREs encoded in the genome of E. coli were over-produced and purified and were shown not to interact with FocA, indicating that interaction with FocA is not a general property of GREs per se. Together, these data show that only the GREs TdcE and PflB interact with the FNT channel protein and suggest that, like PflB, TdcE can control formate translocation by FocA.
Abbreviations: CBD, chitin-binding domain; FNT, formate-nitrite transporters; FocA, formate channel A; GRE, glycyl-radical enzyme; PflB, pyruvate formate-lyase; TdcE, 2 ketobutyrate formate-lyase
Keywords: 2-Ketobutyrate formate-lyase, Formate channel, Formate-nitrite transporter superfamily, Glycyl-radical enzymes, Protein complexes
Graphical abstract
Highlights
-
•
2-ketobutyrate formate-lyase, TdcE, was purified as a chitin-binding protein fusion.
-
•
TdcE was shown to interact specifically with the formate channel protein FocA.
-
•
The predicted glycyl radical enzymes PflD and PflF do not interact with FocA.
-
•
Only glycyl enzymes that generate formate during catalysis interact with FocA.
1. Introduction
The FNT (formate-nitrite transporter) superfamily of pentameric membrane channels is widely distributed amongst archaea and bacteria, and examples are also found in certain fungi and unicellular eukarya. Despite the first member being described more than 20 years ago [1], it is only relatively recently through the advent of genome sequencing that the extent of this superfamily has been recognized and described [2], [3]. The FNT superfamily comprises minimally three sub-classes and they are characterized by translocating monovalent anions, often bi-directionally, as specific substrate [2], [3].
Along with NirC, the channel specific for translocating nitrite, and a recently described lactate/H+ symporter from the malarial parasite Plasmodium falciparum [4], [5], FocA is biochemically the best characterized of the FNT proteins [1], [6], [7], [8], [9]. FocA is the archetype of this class of channels and it is an integral membrane protein that forms a pentamer. Each protomer of the FocA pentamer has a substrate channel and this translocates formate bi-directionally [6], [7], [8]. FocA has been characterized structurally from a number of bacterial sources [6], [7], [8], and the purified protein isolated from Salmonella enterica was shown to transport not only formate but also other organic acids produced during mixed-acid fermentation [10]. Surprisingly, however, in vivo studies with E. coli have demonstrated that only formate appears to be translocated by FocA [1], [11], raising the question as to how this restricted substrate-specificity is achieved in vivo.
Amino acid alignments of FocA proteins from different microbial sources reveal that several highly conserved amino acids line the substrate pore of each protomer. Substitution of these amino acid residues demonstrated that some are important for directionality of substrate translocation, while exchange of others abolished substrate translocation completely [12]. Remarkably, the amino acids that were identified to line the substrate channel are conserved among FNT proteins.
The focA gene is co-transcribed with pflB, the gene encoding the glycyl-radical enzyme (GRE) pyruvate formate-lyase (PflB), which catalyzes the coenzyme A (CoA)-dependent homolytic cleavage of pyruvate generating acetyl-CoA and formate [12], [13]. Recent studies have demonstrated that PflB interacts directly with FocA to control formate translocation, suggesting that interaction with a specific partner protein might play a role in determining substrate passage through the channel [14]. Chemical cross-linking studies revealed that the N-terminal alpha-helix of FocA is important for the interaction with PflB [14].
As well as PflB, five further GREs are encoded on the genome of E. coli [15]. These include the 2-ketobutyrate formate-lyase TdcE [16], anaerobic ribonucleotide reductase [17], PflD [15], PflF [15] and GrcA (formerly known as YfiD; [18]). The functions of PflD and PflF remain to be resolved, while GrcA has been suggested to function as a mobile glycyl-radical domain that repairs oxygen-damaged GREs [19]. TdcE has a similar activity to that of PflB and cleaves the keto-acids pyruvate and 2-ketobutyrate to formate and either acetyl-CoA or propionyl-CoA, respectively [16], [20]. Comparison of the amino acid sequences of PflB and TdcE reveals an amino acid identity of 79%, while the identities between PflB and PflD or PflF are only in the low 20% range (Table S1).
Many of the amino acid residues identified in PflB to be cross-linked with amino acids residues in FocA [14] are conserved in TdcE [16], [20]. Surprisingly, however, many are also conserved in PflF and in PflD (Fig. S1). The amino acid similarity between PflB and TdcE, together with their common capability of formate generation, suggested that TdcE might also interact with FocA. The findings of the studies presented here confirm this hypothesis and provide insight into which amino acids on TdcE and PflB might be most important for the interaction with FocA.
2. Materials and methods
2.1. Strains and growth conditions
The strains and plasmids used in this study are listed in Table 1. For standard DNA work, E. coli strains were routinely grown at 37 °C on LB-agar plates or with shaking in LB-broth [21]. BL21(DE3) strains containing expression plasmids for protein purification were grown in shaking (100 rpm) cultures of TB (terrific broth) or LB medium [21] at 37 °C. TB medium consisted of 1.2% (w/v) peptone, 2.4% (w/v) yeast extract, 0.8% (w/v) glucose, 0.4% (w/v) glycerol, 0.37% (w/v) aspartate, 100 mM potassium phosphate buffer, pH 7, 2 mM MgSO4, and 0.01% (v/v) trace element solution SL-A [22]. Ampicillin and chloramphenicol, when required, were used at the final concentrations of 100 and 15 μg per ml, respectively. When pASK-IBA vectors were used, cells were cultivated in LB medium and induction of gene expression was achieved by adding 0.2 μg ml−1 anhydrotetracycline (AHT) after the OD600 nm of the culture had reached approximately 0.8. After addition of AHT, incubation of the culture was continued for a further 16 h at 16 °C. When pCA24N-based plasmids were used cells were cultivated in TB and 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to the cultures to induce expression of plasmid-encoded genes. This was also done when the culture attained an OD600 nm of 0.8 and, subsequent to IPTG addition, incubation was continued for 2–3 h prior to harvest of the cells.
Table 1.
Strains and plasmids.
| Strains | Genotype | Reference |
|---|---|---|
| MC4100 | F-, araD139, Δ(argF-lac)U169, λ-, rpsL150, relA1 deoC1, flhD5301, Δ(fruK-yeiR)725(fruA25), rbsR22, Δ(fimB-fimE)632(::IS1) | [26] |
| BL21(DE3) | F-ompT lon hsdS (r-m-) dcm+Tetrgal λ(DE3) endA | Invitrogen, Carlsbad, USA |
| BL21(DE3) Δact | BL21(DE3) carrying a deletion oft he pflA gene (CmR) | This study |
| RM220 | MC4100 but ΔpflB ΔpflA | [1] |
| Plasmids | ||
| pASK-IBA5 | Expression vector, AmpR | IBA Biotagnologies,Göttingen |
| pASK-IBA5focA | pASK-IBA5 focA+ | [9] |
| pCYB1 | Expression vector, AmpR | New England Biolabs |
| pCPF-1 | pCYB1 tdcE+ AmpR | This study |
| pflB (JW0886) | pCA24N derivative encoding His-tagged PflB | [27]a |
| pflD (JW3923) | pCA24N derivative encoding His-tagged PflD | [27]a |
| pflF (JW0807) | pCA24N derivative encoding His-tagged PflF | [27]a |
ASKA E.coli ORF plasmid library, Japan (National Bioresource Project).
For overproduction of the (chitin-binding domain) CBD-intein-TdcE fusion protein, cultures of E. coli BL21 (Δact) containing the expression plasmid pCPF-1 were grown aerobically in 1 l LB medium supplemented with 25 µg/ml chloramphenicol at 30 °C with continuous shaking to an OD600 of approximately 0.5. Gene expression was induced by adding 0.4 mM IPTG to the culture and incubation was continued at 30 °C for a further 3 h.
Generally, cells were harvested by centrifugation at 3000g for 30 min and at 4 °C and were washed once with 100 mM Tris–HCl buffer pH 7.4. Cell pellets were used immediately or were stored at −20 °C until use.
2.2. Plasmid construction
The tdcE gene was amplified using the oligonucleotides P-1: 5′-GCGGGGCATATGAAGGTAGATATTG-3′ and P-2: 5′-GCGATGGCTCTTCCGCAGAGCGCCTGGGTAAAGG-3′ and Pfu DNA polymerase (Promega) employing standard reaction conditions. The resulting DNA fragment was digested with the restriction enzymes NdeI and SapI (both from Fermentas) and was cloned into vector pCYB1 (New England Biolabs) digested with the same enzymes. The DNA insert of the resulting plasmid, pCPF-1, was verified by DNA sequencing.
2.3. Preparation of crude cell extracts
Wet cell paste was re-suspended at a ratio of 1g per 3 ml in 100 mM Tris–HCl, pH 8, including 150 mM NaCl, 5 μg DNase ml−1 and 0.2 mM phenylmethylsulfonyl fluoride. Cells were disrupted aerobically by sonication (Sonoplus Bandelin, Berlin) using a KE76 tip, 30 W power and 3 cycles of 4 min treatment with 0.5 s intervals. Unbroken cells and cell debris were removed by centrifugation for 30 min at 15,000g at 4 °C. The resultant supernatant was termed the crude extract and was used as the starting material for all studies reported herein.
2.4. Purification of tagged proteins
Strep-tagged FocA derivatives were purified from the membrane fraction prepared from crude extracts exactly according to [9]. N-terminally His-tagged proteins were purified using cobalt-NTA according to [23].
The CBD-intein-TdcE fusion protein was overproduced and it was subsequently purified using the IMPACT system according to the manufacturer's instructions (New England Biolabs). Briefly, the cell pellet (1 g wet weight) was re-suspended in 7 ml of buffer containing 20 mM Tris, pH 8.5, 500 mM NaCl, 1 mM EDTA and cell disruption was carried out by sonication (30 W power, 3×4 min with 0.5 s pulses) followed by centrifugation of the disrupted cell material for 30 min at 20,000g and 4 °C to separate cell debris from the crude extract. The resulting crude extract contained the overproduced CBD-intein-TdcE fusion protein and this was purified according to the manufacturer's instructions. By using the self-splicing activity of the intein, which is activated by addition of the reducing agent dithiothreitol (DTT) at 50 mM final concentration, the overproduced TdcE could be quantitatively released from the CBD-tag. To remove DTT the elution fractions were passaged over a PD10-column (GE-Healthcare).
2.5. Pull-down assay with Streptactin-coated magnetic beads
To identify any possible interaction between N-terminally Strep-tagged FocA and purified GREs, we used a pull-down assay with a 5% (w/v) suspension of Streptactin-coated magnetic beads (Mag Strep “type I” beads, IBA Biotechnology) or with 40 µl Streptactin sepharose matrix, as described [9], [14]. Briefly, aliquots (10–20 µg) of solubilized and purified FocA with an N-terminal Strep-tag were added to 25 µl of the Streptactin-coated magnetic beads and washed several times with buffer W (100 mM Tris HCl, 150 mM NaCl, 1 mM EDTA, 1 mM n-dodecyl-ß-maltoside, pH 8.0). Afterwards the FocA-loaded beads were incubated by gentle agitation for 2 h at 4 °C with 10 µg of purified GRE protein. After extensive washing with 10 ml buffer W, bound proteins were eluted by using 50 µl of buffer W containing 2.5 mM α-desthiobiotin. Unless stated otherwise 20 µl of the elution fraction were analyzed by SDS-PAGE and western blotting and interacting proteins were detected using either anti-PflB antiserum or anti-His-tag antibodies (Santa Cruz Biotechnology).
2.6. Polyacrylamide gel electrophoresis and immunoblotting
Aliquots of purified proteins (between 1 and 5 µg) or the indicated fractions from the pull-downs were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) using either 7.5% (w/v) or 10% (w/v) polyacrylamide gels [24]. Gels were either stained with Coomassie Brilliant Blue or the separated polypeptides were transferred to nitrocellulose membranes as described [25]. Antibodies raised against PflB (1: 3000) [16] or commercially available antibodies that detected the His-tag (1: 20,000; Santa Cruz Biotechnology) were used. Secondary antibody conjugated to horseradish peroxidase was obtained from Bio-Rad. Visualisation was done by the enhanced chemiluminescent reaction (Agilent Technologies).
3. Results and discussion
3.1. Purification of PflB, PflD, and TdcE and enrichment of PflF
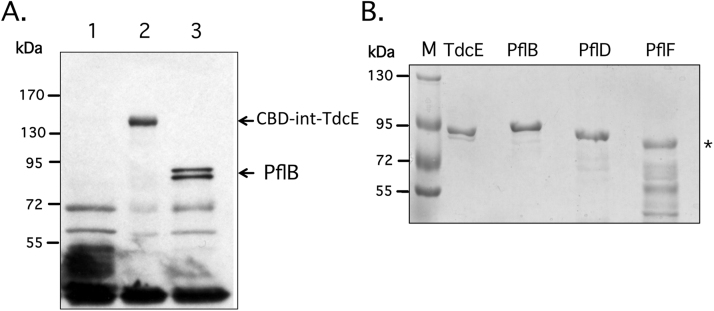
To demonstrate production of the CBD-intein-TdcE fusion, plasmid pCFP-1 (Table 1) was transformed into RM220, which lacks the genes coding for both PflB and its activating enzyme PflA [1] and after growth in LB medium, a polypeptide with a predicted molecular mass of approximately 140 kDa (TdcE, 86 kDa+intein-CBD, 55 kDa) was observed after western blotting of samples of crude extract and challenge with anti-PflB antiserum (Fig. 1A). This antiserum has been shown previously to react with epitopes on TdcE [16], [20]. As a control, strain RM220 without plasmid pCPF-1 showed no corresponding polypeptide while the crude extract derived from anaerobically grown wild type strain MC4100 revealed two polypeptides migrating at 85 and 82 kDa, which correspond to the full-length and the oxygen-fragmented species of PflB; the latter species formerly carried the glycyl radical [19], [20].
Fig. 1.
Purification/enrichment of E. coli glycyl radical enzymes. (A) A western blot is shown in which aliquots (50 μg protein) of crude extracts derived from anaerobically grown E. coli strains RM220 (ΔpflB ΔpflA) (lane 1), RM220/ pCPF-1 (lane 2), and MC4100 (lane 3) were separated by SDS-PAGE (7.5% w/v polyacrylamide). After transfer to a nitrocellulose membrane the polypeptides were challenged with anti-PflB antiserum. Molecular mass size-markers (Fermentas) are shown on the left and the migration positions of the chitin-binding domain-intein-TdcE (CBD-int-TdcE) fusion and the full-length and oxygen-fragmented pyruvate formate-lyase (PflB) are indicated on the right of the panel. (B) Aliquots of purified TdcE, His-PflB and His-PflD (2 μg protein) and enriched His-PflF (5 μg protein) were separated by SDS-PAGE (7.5% w/v polyacrylamide) and the gel was subsequently stained with Coomassie Brilliant Blue. M, represents molecular mass markers and the asterisk denotes the migration position of His-PflF.
For overproduction of the CBD-intein-TdcE fusion protein and purification of TdcE, plasmid pCPF-1 was transformed into a derivative of BL21(DE3), BL21(DE3)Δact, which is unable to synthesize PflA, and which is also required to introduce the radical into TdcE [20]. After successful overproduction, the CBD-intein-TdcE fusion was bound to chitin sepharose and the full-length TdcE protein was released by incubation with dithiothreitol, delivering nearly homogeneous protein (Fig. 1B). Approximately 1 mg of purified protein was obtained from 1 g wet-weight of E. coli cells.
Plasmids encoding the N-terminally His-tagged GREs PflB, PflD and PflF were introduced into E. coli BL21(DE3) and overproduction and purification of each protein was carried out as described in Materials and Methods. All three proteins were soluble and PflB and PflD could be purified to near-homogeneity, while PflF was enriched to about 40% purity (Fig. 1B).
3.2. TdcE interacts with the formate channel FocA
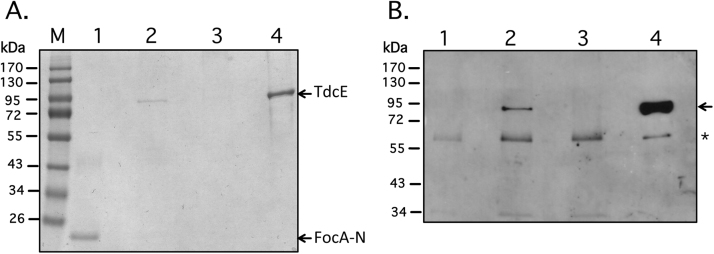
Previous studies had shown that TdcE can functionally replace PflB in the fermentative metabolism of E. coli [20]. This suggests that, like PflB [14], TdcE might be able to interact specifically with FocA. To test this, we mixed 10 μg of purified Strep-tagged FocA bound to Streptactin beads with 10 μg of purified TdcE and after incubation as described in the Materials and Methods, complexes formed between the proteins were isolated by pull-down assay (Fig. 2A). TdcE interacted with FocA, as observed after both Coomassie Blue staining of the gel, as well as after immunoblotting and challenge with anti-PflB antiserum (lane 2 in Fig. 2A and B, respectively). TdcE identified in the pull-down experiment co-migrated with purified TdcE protein (Fig. 2, lane 4). As a control, it could be shown that TdcE did not interact with the Streptactin-coated magnetic beads in the absence of Strep-tagged FocA (Fig. 2, lane 3).
Fig. 2.
Strep-tagged FocA pulls down TdcE. (A) A Coomassie Brilliant Blue-stained SDS-PAGE (10% w/v polyacrylamide) is shown. Lane M, molecular mass markers (Fermentas); lane 1, purified N-terminally Strep-tagged FocA (FocA-N; 1 μg); lane 2, pull-down of a FocA-N and TdcE mixture (10 μg each); lane 3, pull-down of TdcE (10 μg) without FocA-N; lane 4, purified TdcE (1 μg). (B) The same experiment as shown in part A. but after western blot analysis with anti-PflB antiserum. The migration positions of TdcE and FocA-N are indicated on the right side of panel A., while an arrow on the right side of panel B. indicates the migration position of TdcE. The asterisk denotes an unidentified polypeptide that acted as a loading control.
3.3. The E. coli GREs PflD and PflF do not interact with FocA
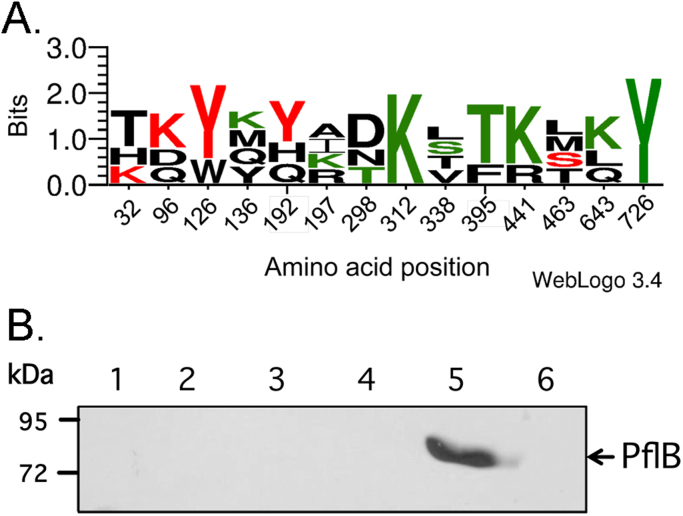
Of the 14 amino acid residues in PflB recently identified to form chemical cross-links with FocA [14], 8 of these residues are conserved in TdcE and include K96, Y126, Y192, K312, T395, K441, K643 and Y726 (Fig. 3A and S1). Residues K312 and Y726 are also conserved in the predicted GREs PflD and PflF, while Y126, T395 and K441 are shared between PflB, TdcE and PflD; none of the 14 amino acid residues is conserved in the catalytic subunit of anaerobic ribonucleotide reductase in E. coli [17]. It was therefore important to determine whether either PflD or PflF also interact with FocA. To test this, a pull-down experiment similar to that shown in Fig. 2 with purified, N-terminally His-tagged PflD or with the enriched fraction containing PflF was performed and cross-reacting polypeptides were detected using anti-His-tag antibodies (Fig. 3B). As a control a N-terminally His-tagged variant of PflB was also tested. The results clearly show that only His-tagged PflB was capable of interacting with Strep-tagged FocA and the interaction was FocA-dependent (Fig. 3B, compare lanes 5 and 6). Moreover, this experiment also demonstrated that the N-terminal His-tag did not prevent interaction of PflB with FocA. Together, these findings indicate that interaction with the formate channel FocA is not an inherent property of GREs but rather is highly specific, and interaction partners include PflB and its homologue TdcE.
Fig. 3.
FocA-N does not interact with His-PflD or His-PflF. (A) A comparison using the WebLogo 3.4 stacking algorithm [28] of the 14 amino acids (shown as red or green) identified in PflB by chemical cross-linking [14] to interact with amino acid residues in FocA and the corresponding residues in the GREs TdcE, PflD, and PflF is shown. Red amino acid residues are identical, or are highly conserved as in the case of S463 in PflB and T467 in TdcE, between PflB and TdcE. The amino acids residues neighboring these residues are also conserved between the two proteins (see Fig. S1). Other amino acid residues in PflB identified to cross-link with FocA but which are either common to PflD and/or PflF, or which occur only in PflB and are therefore not considered as strong candidates for being distinguishing interactors with FocA, are shown in green (see text for details). (B) A western blot with anti-His-tag antibodies is shown in which pull-down assays with His-PflD (lanes 1 and 2), His-PflF (lanes 3 and 4) and His-PflB (lanes 5 and 6) were carried out. Purified or enriched N-terminally His-tagged proteins (10 μg) were incubated with (lanes 1, 3 and 5) or without (lanes 2, 4 and 6) purified Strep-tagged FocA-N (10 μg). The migration position of His-PflB (PflB) is shown on the right and molecular mass markers are indicated in kDa on the left of the figure.
Examination of the amino acid residues in the immediate vicinity of the 14 residues in PflB identified by chemical cross-linking to interact with FocA reveals that at 6 locations (K312, S338, T395, K441, K643, and Y726) there is significant similarity between all four GREs, possibly excluding these as surfaces important in interacting strongly with FocA (Fig. S1). Five of the remaining 8 regions (centred around K32, K96, Y126, Y192, and S463; Fig. 3A) show a high conservation between only PflB and TdcE (Fig. S1), suggesting that these might be involved in the specific interaction with FocA and represent first targets for an amino acid exchange mutagenesis program to investigate the interactions between these proteins and the N-terminal helix of FocA [14]. The remaining three amino acids residues of the 14 identified to cross-link between PflB and FocA (K136, K197, and T298) and their corresponding adjacent amino acid residues show no strong similarity between PflB and TdcE (Fig. S1).
4. Conclusions
The findings of this study identify the glycyl-radical enzyme TdcE as a specific interaction partner of the formate-specific channel FocA. We rule out that FocA-binding is a general property of GREs. Rather, only TdcE and PflB interact with FocA and both have in common that they generate formate during catalysis, the substrate of FocA. Furthermore, this study has helped narrow down the key regions on TdcE/PflB important for high-affinity interaction with FocA and these will be analyzed in future mutagenesis programs.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (GRK 1026).
Footnotes
Transparency document associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2016.04.005.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Suppmann B., Sawers G. Isolation and characterization of hypophosphite-resistant mutants of Escherichia coli: identification of the FocA protein, encoded by the pfl operon, as a putative formate transporter. Mol. Microbiol. 1994;11:965–982. doi: 10.1111/j.1365-2958.1994.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 2.Waight A.B., Czyzewski B.K., Wang D.N. Ion selectivity and gating mechanisms of FNT channels. Curr. Opin. Struct. Biol. 2013;23:499–506. doi: 10.1016/j.sbi.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lü W., Du J., Schwarzer N.J., Wacker T., Andrade S.L., Einsle O. The formate/nitrite transporter family of anion channels. Biol. Chem. 2013;394:715–727. doi: 10.1515/hsz-2012-0339. [DOI] [PubMed] [Google Scholar]
- 4.Lü W., Schwarzer N.J., Du J., Gerbig-Smentek E., Andrade S.L., Einsle O. Structural and functional characterization of the nitrite channel NirC from Salmonella typhimurium. Proc. Natl. Acad. Sci. USA. 2012;109:18395–18400. doi: 10.1073/pnas.1210793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu B., Rambow J., Bock S., Holm-Bertelsen J., Weichert M., Blancke Soares A., Spielmann T., Beitz E. Identity of a Plasmodiium lactate/H+ symporter structurally unrelated to human transporters. Nat. Commun. 2015;6:6284. doi: 10.1038/ncomms7284. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., Huang Y., Wang J., Cheng C., Huang W., Lu P., Xu Y.N., Wang P., Yan N., Shi Y. Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel. Nature. 2009;462:467–472. doi: 10.1038/nature08610. [DOI] [PubMed] [Google Scholar]
- 7.Waight A.B., Love J., Wang D.N. Structure and mechanism of a pentameric formate channel. Nat. Struct. Mol. Biol. 2010;17:31–37. doi: 10.1038/nsmb.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lü W., Du J., Wacker T., Gerbig-Smentek E., Andrade S.L., Einsle O. pH-dependent gating in a FocA formate channel. Science. 2011;332:352–354. doi: 10.1126/science.1199098. [DOI] [PubMed] [Google Scholar]
- 9.Falke D., Schulz K., Doberenz C., Beyer L., Lilie H., Thiemer B., Sawers R.G. Unexpected oligomeric structure of the FocA formate channel of Escherichia coli: a paradigm for the formate-nitrite transporter family of integral membrane proteins. FEMS Microbiol. Lett. 2010;303:69–75. doi: 10.1111/j.1574-6968.2009.01862.x. [DOI] [PubMed] [Google Scholar]
- 10.Lü W., Du J., Schwarzer N.J., Gerbig-Smentek E., Einsle O., Andrade S.L. The formate channel FocA exports the products of mixed-acid fermentation. Proc. Natl. Acad. Sci. Usa. 2012;109:13254–13259. doi: 10.1073/pnas.1204201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beyer L., Doberenz C., Falke D., Hunger D., Suppmann B., Sawers R.G. Coordinating FocA and pyruvate formate-lyase synthesis in Escherichia coli: preferential translocation of formate over other mixed-acid fermentation products. J. Bacteriol. 2013;195:1428–1435. doi: 10.1128/JB.02166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunger D., Doberenz C., Sawers R.G. Identification of key residues in the formate channel FocA that control import and export of formate. Biol. Chem. 2014;395:813–825. doi: 10.1515/hsz-2014-0154. [DOI] [PubMed] [Google Scholar]
- 13.Sawers R.G., Clark D.P. Fermentative pyruvate and acetyl CoA metabolism. In: Curtiss R. III, editor. Ecosal--Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, D.C: 2004. http://www.ecosal.org [Google Scholar]
- 14.Doberenz C., Zorn M., Falke D., Nannemann D., Hunger D., Beyer L., Ihling C.H., Meiler J., Sinz A., Sawers R.G. Pyruvate formate-lyase interacts directly with the formate channel FocA to regulate formate translocation. J. Mol. Biol. 2014;426:2827–2839. doi: 10.1016/j.jmb.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reizer J., Reizer A., Saier M.H., Jr Novel phosphotransferase system genes revealed by bacterial genome analysis – a gene cluster encoding unique Enzyme I and the proteins of a fructose-like permease system. Microbiology. 1995;141:961–971. doi: 10.1099/13500872-141-4-961. [DOI] [PubMed] [Google Scholar]
- 16.Heßlinger C., Fairhurst S.A., Sawers G. Novel keto acid formate-lyase and propionate kinase activities are components of an anaerobic pathway in Escherichia coli that degrades L-threonine to propionate. Mol. Microbiol. 1998;27:477–492. doi: 10.1046/j.1365-2958.1998.00696.x. [DOI] [PubMed] [Google Scholar]
- 17.Mulliez E., Fontecave M., Gaillard J., Reichard P. An iron-sulfur center and a free radical in the active anaerobic ribonucleotide reductase of Escherichia coli. J. Biol. Chem. 1993;268:2296–2299. [PubMed] [Google Scholar]
- 18.Wyborn N.R., Messenger S.L., Sawers G., Roberts R.E., Attwood M.M., Green J. Expression of the Escherichia coli yfiD gene responds to intracellular pH, and reduces the accumulation of acidic metabolic end-products. Microbiology. 2002;148:1015–1026. doi: 10.1099/00221287-148-4-1015. [DOI] [PubMed] [Google Scholar]
- 19.Wagner A.F.V., Schultz S., Bomke J., Pils T., Lehmann W.D., Knappe J. YfiD of Escherichia coli and Y06I of bacteriophage T4 as autonomous glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem. Biophys. Res. Commun. 2001;285:456–462. doi: 10.1006/bbrc.2001.5186. [DOI] [PubMed] [Google Scholar]
- 20.Sawers G., Heßlinger C., Muller N., Kaiser M. The glycyl radical enzyme TdcE can replace pyruvate formate-lyase in glucose fermentation. J. Bacteriol. 1998;180:3509–3516. doi: 10.1128/jb.180.14.3509-3516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J. Cold Spring Harbor Laboratories; United States: 1972. Experiments in Molecular Genetics. [Google Scholar]
- 22.Hormann K., Andreesen J. Reductive cleavage of sarcosine and betaine by Eubacterium acidaminophilum via enzyme systems different from glycine reductase. Arch. Microbiol. 1989;153:50–59. [Google Scholar]
- 23.Stripp S.T., Soboh B., Lindenstrauss U., Braussemann M., Herzberg M., Nies D.H., Sawers R.G., Heberle J. HypD is the scaffold protein for Fe–(CN)2CO cofactor assembly in [NiFe]–hydrogenase maturation. Biochemistry. 2013;52:3289–3296. doi: 10.1021/bi400302v. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Towbin J., Staehlin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA. 1979;76:239–248. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casadaban M.J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 27.Kitagawa M. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 28.Crooks G.E., Hon G., Chandonia J.M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material