Abstract
Inhibitor of DNA binding protein 3 (ID3) has long been characterized as an oncogene that implicates its functional role through its Helix–Loop–Helix (HLH) domain upon protein–protein interaction. An insight into the dimerization brought by this domain helps in identifying the key residues that favor the mechanism behind it. Molecular dynamics (MD) simulations were performed for the HLH proteins ID3 and Transcription factor E2-alpha (E2A/E12) and their ensemble complex (ID3-E2A/E12) to gather information about the HLH domain region and its role in the interaction process. Further evaluation of the results by Principal Component Analysis (PCA) and Free Energy Landscape (FEL) helped in revealing residues of E2A/E12: Lys570, Ala595, Val598, and Ile599 and ID3: Glu53, Gln63, and Gln66 buried in their HLH motifs imparting key roles in dimerization process. Furthermore the T-pad analysis results helped in identifying the key fluctuations and conformational transitions using the intrinsic properties of the residues present in the domain region of the proteins thus specifying their crucial role towards molecular recognition. The study provides an insight into the interacting mechanism of the ID3-E2A/E12 complex and maps the structural transitions arising in the essential conformational space indicating the key structural changes within the helical regions of the motif. It thereby describes how the internal dynamics of the proteins might regulate their intrinsic structural features and its subsequent functionality.
Abbreviations: ID, Inhibitors of DNA binding protein; ID3, Inhibitor of DNA binding protein 3; E2A, Transcription factor E2-alpha; BHLH, Basic Helix–Loop–Helix; MD, Molecular dynamics; PCA, Principal Component Analysis; FEL, Free Energy Landscape
Keywords: Inhibitor of DNA binding protein 3, Transcription factor E2-alpha, Basic Helix–Loop–Helix, Molecular dynamics, Principal Component Analysis, Free Energy Landscape
Highlights
-
•
ID3 acts as a negative regulator of E2A class bHLH transcription factors.
-
•
ID3 and E2A transcription factor (Isoform E12) proteins were modeled.
-
•
Molecular docking and dynamic simulations were performed on the docked complex.
-
•
The activity of their HLH domain region was mapped through PCA and FEL analyses.
-
•
Seven key interacting residues favoring the dimerization were identified.
1. Introduction
The Inhibitors of DNA-binding or ID proteins are essential dominant negative regulators belonging to the family of basic helix–loop–helix protein (bHLH) transcription factors. The four ID proteins (ID 1–4) are well known to interact with and transform the activity of other family members of bHLH transcription factors thereby asserting a regulatory role in the cell cycle and differentiation [1], [2], [3], [4]. The members of bHLH family, except for ID proteins, contain a DNA binding region adjacent to the bHLH domain region that promotes protein dimerization [5], [6], [7]. The dimerization is favored by parallel arrangement of four helix bundles each of which is located towards the N-terminal region to the alpha helices [8], [9], [10], [11], [12]. The members also include E12/E47, E2-2, and HEB (grouped under class A members and also known as E proteins) that either dimerize to form homodimers functioning as transcription activators or as heterodimers that play the role of key transcription activators or suppressors [13], [14], [15].
ID proteins dimerize with the bHLH transcriptional regulators and sequester their activity by preventing them to bind to DNA, thereby functioning as dominant negative regulators [16]. Besides being a negative regulator of cell differentiation, studies have shown ID proteins committed to having a positive role too in cell cycle progression during the early and mid-late G1 phase mediated by several key proteins (non bHLH proteins as well) thus initiating variation in the transcription of many genes [17], [18], [19]. Corresponding to the role of ID in cell cycle progression, studies have also ascertained its oncogenic activity in certain cell lines such as pancreatic cancer, astrocytic cancer, neural tumor, and invasive breast carcinoma to name a few [20], [21], [22], [23]. Inhibitors of differentiation 3 or simply ID3 is one such protein whose purpose serves to dimerize with other transcription factors belonging to bHLH family transcriptional factors bringing about its functional stability and maintaining its subcellular localization for emphasizing its biological effect [24], [25], [26]. Well within the cell cycle machinery the Cdk2-dependent ID3 phosphorylation accounts for the G1–S phase transition that in turn negates the cell cycle arrest at G1 phase, a mechanism that involves the sequestering of its dimeric partners – bHLH transcription factors thereby preventing them to bind to E box sequences [27]. ID3 implicates its functional role upon such protein–protein interaction mechanism well mediated through its HLH domain. The heterodimerization of ID3 with its known interacting partners is favored by the hydrophobic residues (Met44, Asn45, Tyr48, Leu51, Leu64, Val67, Ile69, Leu70, Val73, Ile74, Tyr76, Ile77, Leu80) buried within its bHLH domain. Residues such as Met44, Asn45, Cys47, Tyr48, Ser49, Leu51, Leu54, Val55, Pro56, Pro59, Ser65, Glu68, Ile69, Leu70, Gln71, Val73, Ile74, Asp75, Tyr76, Ile77, Asp79, Leu80, and Gln81 have also been found to be conserved among the ID family of proteins. The dimerization has also been substantiated by regions preceding and succeeding the HLH domain implying their plausible role in molding the structure of the entire protein [28]. It therefore becomes an imperative task of understanding the structural aspects of the domain region thereby getting a broader knowledge of the underlying mechanism behind the dimerization favored by the HLH domain.
Transcription factor E2-alpha (E2A) is known to bind to DNA at the canonical E-box sequence (CANNTG where in N signifies any nucleotide) and mediate various cellular processes involving cell growth and differentiation. Its alternative splice product – E12 and E47 are known to interact with ID3. A few of the bHLH proteins are known to bind DNA as homodimers and few as heterodimers along with certain proteins. E12 homodimers bind poorly to DNA than E47, though any such binding activity is antagonized by the induction of ID3 protein [29]. The regulation of the E2A class bHLH proteins by increased or decreased levels of ID3 substantiates its activity in deciding major cell fates thus making such interactions a hallmark in understanding early stages of development of major diseases.
The imperative role of molecular dynamics (MD) simulation in unraveling the structural complexities of a biological molecule has steadily evolved with gradual increase in the precision time steps incurred in running any complex simulation. Though it seem to have added increased computational time to the chagrin of researchers, they sure have endowed researchers with prolific outcome in terms of understanding the periodicity of secondary structures, or the hydrogen bond dynamics that encompass within a biomolecular process [30], [31]. Analyses such as Principal Component Analysis (PCA) are efficiently used to assess the internal atomic motions of a molecule within the conformational space, thereby leading to its biological activity [32], [33]. This method samples the molecular conformations from a larger ensemble of structures using Cartesian coordinates and depicts the correlated motions of the particles by projecting the trajectory onto eigenvectors and their respective eigenvalues [34], [35], [36]. These chosen principal components are then used to construct free-energy landscape, a three dimensional representation of energy basins that comprises mapping of all possible conformations a protein can possibly adopt (closer to its native state) along with their corresponding energies [37]. This study deals with employment of molecular dynamics (MD) simulations, Principal Component Analysis (PCA) and free-energy landscape (FEL) to understand the protein interface of ID3 with E2A/E12. The study elucidates and thus identifies the key residues that provide the necessary framework for the dimerization of the proteins mediated by their HLH domains thereby providing insight on how well the antagonist effect might disturb its dimerization partner.
2. Materials and methods
2.1. Tertiary structure prediction of ID3 and E2A/E12
The protein sequence for ID3 and E2A was retrieved from the Uniprot (http://www.uniprot.org/) (119 a.a, Uniprot accession no – Q02535; 654 a.a, Uniprot accession no-P15923, Isoform E12 respectively) to predict their three-dimensional structure. To understand the structural dynamics and the apt conformation adaptation of the proteins, a full length model comprising of the N and C terminal flanking the bHLH domain was a pre-requisite. Hence, to obtain a complete modeled structure, the full length sequences of the proteins were submitted to and modeled using the Robetta server's (http://robetta.bakerlab.org/) structure prediction program. The server implements ROSETTA de novo structure prediction algorithm that performs Monte Carlo search through a space of conformations to obtain a minimal energy conformation [38]. Rosetta investigates structure by swapping torsion angles between a fragment in an obtained model and known structure fragments. The method constructed five models for each protein, each of which were then subjected to validation using the Structural Analysis and Verification Server (SAVES), with tools including PROCHECK and ERRAT [39], [40].
2.2. Molecular dynamics simulation for ID3 and E2A/E12
Molecular Dynamics simulations were carried out using the GROMACS 4.5.3 molecular dynamics package [41], [42] using Amber99sb force field [43] for understanding the molecular stability of the modeled three dimensional tertiary structure of ID3 and E2A/E12. The proteins were soaked in a cubic box with a dimension extending to 1.50 nm. Periodic boundary system was applied in all the direction to both the systems. The solvated systems were neutralized by replacing the water molecules with sodium ions. The ID3 system was neutralized by adding three sodium ions whereas the E2A/E12 system was neutralized through addition of twelve ions of sodium. Each of the systems was then subjected to two rounds of energy minimization of 50,000 steps using steepest descent algorithm followed by minimization through conjugate gradient method. The minimized systems were then equilibrated for 100 ps (ps) for position restraining at both NVT (constant number of particles, volume and temperature) and NPT (constant number of particles, pressure and temperature). The LINCS algorithm [44] was used to constrain all the bond angles whereas the geometry of water molecules was constrained by SETTLE algorithm [45]. V-rescale weak coupling method and Parrinello–Rahman method [46] were used to regulate temperature (at 310 K) and set the pressure (at 1 atm) respectively of the systems. The equilibrated systems were then set up for a 50 ns (ns) of simulation run with a time step of 2 femtosecond (fs). Structural coordinates for every 2 ps were saved and analyzed using suitable tools available in the GROMACS package. Certain steps in the running protocol were referred from our previous work with minor modifications in the input files to suit our need [47], [48].
2.3. Principal component and free energy landscape analysis
Principal Component Analysis (PCA) is a covariance-matrix based analytical technique that computes correlated fluctuations in concerted atomic motions within the MD trajectories and maps the chosen principal components (PCs) in an energy landscape or basins to define the free energy minima corresponding to their respective conformations [32], [49]. To begin with, a covariance matrix was calculated using protein alpha carbons (Cα) as reference structure for the rotational fit. Following the covariance matrix generation, a set of eigenvectors and eigenvalues was computed to define the dimensionality of the essential subspace. A majority of the motions (>90%) are represented by less than ten eigenvectors that illustrate the relevant concerted motions within an atomic system.
The cosine content (Ci) of the principal components (PCs) was evaluated as a measure of the characteristic features to define the trajectory motion of the protein is enough for the generation of FEL. In general the value ranges from 0 (no cosine) to 1 (perfect cosine) for a time period of T:
| (1) |
Cosine content closer to 1 is usually attributed to large scale motions within a protein and hence cannot be employed to generate an energy landscape [50]. However, PCs having cosine measures closer to 0.2 and sometime up to 0.5 are reliable and produce qualitatively smooth results with minimum energy cluster. The first 20 PCs (projection eigenvectors) of ID3 and E2A/E12 protein were retrieved and analyzed for their cosine distribution. After retrieving the suitable reaction coordinates (PCs), the free energy landscapes (FELs) were defined for each protein to investigate their near native conformations.
2.4. Molecular docking and simulation of ID3 and E2A/E12 complex
For understanding the molecular mechanism behind the dimerization of ID3 and E2A/E12, the predicted three dimensional structures of ID3 and E2A/E12 were docked with each other. The protein–protein docking was carried out using HADDOCK or High Ambiguity Driven protein-protein Docking approach [51], [52] to reveal the interacting residues involved in the dimerization. The approach implements ambiguous interaction restraints (AIRs) on the interacting residues to perform docking of the proteins. The docked structures are then ranked on the basis of sum of electrostatics, van der Waals, and AIR energy terms. Docking in HADDOCK is performed in three major steps that involve rigid body docking through energy minimization, followed by refinement through simulated annealing of the best complexes, and finally refinement of the best docked complexes using the 8 Å explicit solvent layer (TIP3P). Here, the docking was performed using the ensemble conformations of ID1 and E2A/E12 obtained from the FEL analysis. The HLH (42–85) motif of ID1 and HLH (547–607) motif of E2A were defined as active site residues for the ID3–E2A/E12 complex formation. The neighboring residues of the structures were taken as passive residues for the above-mentioned docking protocol. Based on HADDOCK score and an RMSD cut-off of 7.5 Å, the top cluster was selected and chosen for surface interface analysis using PISA (Protein Interfaces, Surfaces and Assemblies) web interface [53]. The lowest energy complex was then subjected to a 50 ns MD run as mentioned previously. The simulated complex was then further analyzed to get an insight into their complex stability.
2.5. T-pad analysis
The intrinsic plasticity of protein residues modulates various cellular processes via molecular recognition [54], [55], [56], [57]. The plasticity, previously known as the Angular Dispersion Index, of a protein's backbone was based on the circular spread of the Ramachandran angles Φ and Ψ denoted as CΦ and CΨ respectively. Circular spread, however, does not completely account for the backbone conformation of the protein. T-Pad analysis identifies the plasticity of the protein residue-by-residue along with the rigid sites in the protein. It also reveals the backbone transition concerning two conformations of the Ramachandran plot and identifies the hinge points that regulate the conformational changes favoring the proteins molecular activity [58]. It computes a protein angular dispersion of the angle ω or the PADω to quantify the plasticity of the protein backbone residues for the given ensemble structure. The PADω is computed as a function of ω=Φ+Ψ and is formulated in the ranges between 0° and 180°. For our study, the T-pad analysis was carried out to map the fluctuations as well as the conformational transitions of ID3 protein residues upon binding with E2A/E12 protein at the bHLH domain region. Accordingly, the 50 ns MD trajectory of the complex was prepared and analyzed by removing the water molecules and ions with the help of trjconv tool available under the GROMACS package.
3. Results and discussions
3.1. Tertiary structure prediction of ID3 and E2A/E12
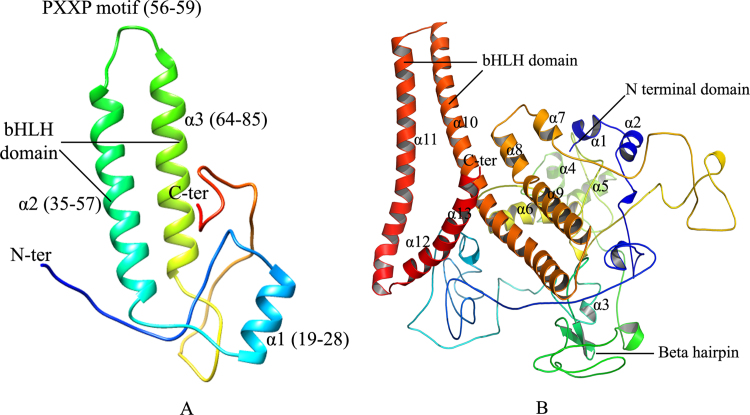
The ID3 protein was predicted using the structural information of the chain A of the solution NMR structure of the HLH domain of ID3 through ROBETTA full chain protein structure prediction server. The predicted modeled structure of ID3 (Fig. 1A) reveals the presence of three helices α1 (19–28), α2 (35–57), and α3 (64–85) connected by two loops (29–34) and (58–63), having a helical length of 15.17 Å, 33.03 Å, and 31.64 Å respectively. Apart from these three helices, the structure was found to encompass nine beta turns, four gamma turns, and one helix–helix interaction. The alpha helix α2 has a highly conserved motif PXXP (P56-G57-V58-P59) that lies at the N terminal end of the loop and is known to be crucial towards the activity of ID proteins [59]. Likewise residues that lie before a proline are more likely to be responsible for prevention of helix formation that in turn makes the region less affinitive towards DNA binding and thereby hinder its binding activity towards DNA [16]. The loop region within the HLH motif consists of residues Gly57, Arg60, Gly61, Thr62, and Gln63 which through previous experimental analysis have been regarded as less conserved [28]. An NCBI CD search showed the presence of HLH domain in the 42–85 lying in the region of α2 and α3 helix [60].
Fig. 1.
Predicted structure of ID3 (A) and E2A/E12 (B). (A) shows three helices α1–3, which contain the HLH motif by the residues 42–85 and also the PXXP motif (56–59). (B) shows 13 helices α1-13, containing the HLH motif in the residues 547–607 and a β hairpin loop.
The predicted structure of E2A/E12 protein is seen to encompass a bHLH domain, an N terminal domain, and 13 alpha helices (Fig. 1B). The N terminal domain is known for the transcriptional activation of E2A gene products while the bHLH domain is involved in homo/hetero dimerization to form a transcriptional unit or complex. The predicted modeled structure of the protein shows 13 helices i.e. α1 (3–7), α2 (13–19), α3 (190–193), α4 (313–319), α5 (329–337), α6 (385–404), α7 (477–482), α8 (490–498), α9 (501–519), α10 (525–581), α11 (588–627), α12 (632–638), and α13 (641–652). The structure has four long helices α6, α9 to α11 of length 27.89 Å, 28.62 Å, 79.91 Å, and 59.47 Å along with nine short helices α1 to α5, α7, α8, α12, α13 having a length of 7.88 Å, 10.93 Å, 6.29 Å, 10.06 Å, 12.77 Å, 9.51 Å, 13.74 Å, 11.14 Å, and 18.78 Å respectively. The predicted structure also includes one beta hairpin, nine helix–helix interactions, 75 beta turns, and four gamma turns. The HLH domain was seen to lie in the 547–607 regions within the α10 and α11 helix as revealed from the NCBI CD search.
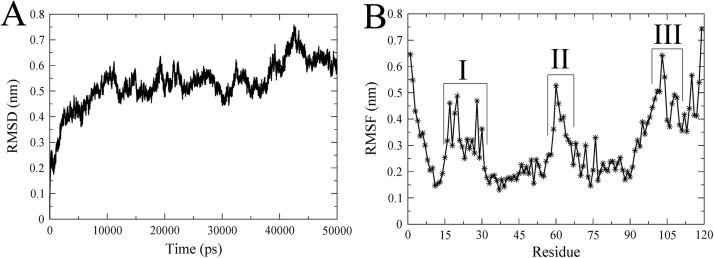
3.2. Stability analysis of ID3 through molecular dynamics
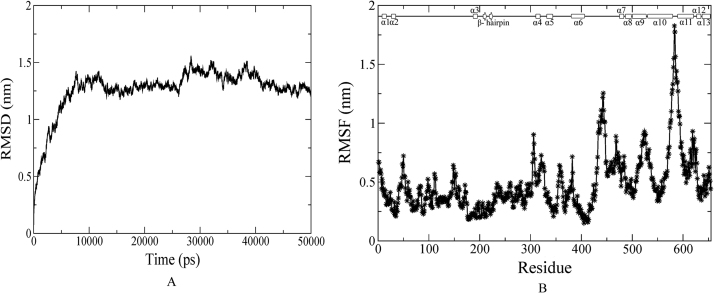
MD simulation for a time scale of 50 ns was performed to understand and verify the stability of the modeled structure of ID3. The backbone RMSD profile of ID3 shows that the protein reaches equilibrium after approximately 10 ns with a maximum RMS deviation to be 0.6 nm and is able to maintain it until the final production MD (Fig. 2A). The RMSF profile of the protein was also evaluated to account for the flexible regions within the structure (Fig. 2B). Region I falls well within the helical range of α1 (16–29) having a fluctuation ranging between 0.25–0.5 nm. For the second region II (61–65) a fluctuation of 0.4–0.55 nm is observed. For both regions I and II, the presence of Gly27, Gly29 and Gly61 can be attributed for the observed fluctuations owing to glycine's high conformational flexibility. The third flexible region III (103–108), which is a loop extending from the helix α3 towards the C-terminal, has an RMS fluctuation of 0.45–0.65 nm.
Fig. 2.
Stability analysis of ID3. (A) Backbone RMSD of ID3 for 50 ns MD run. (B) Backbone RMSF of ID3 showing three flexible regions within.
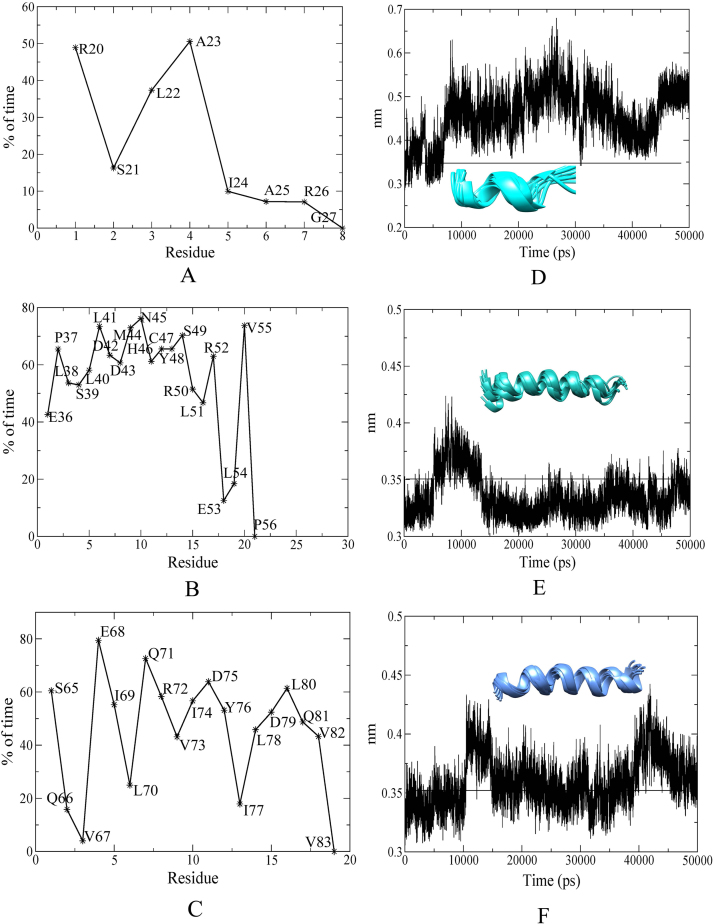
The dihedral angles phi (Φ) and psi (Ψ) of each helix were retrieved from the production MD simulation run of 50 ns to confirm their helical nature. The backbone dihedral angles describe the rotation of a polypeptide backbone around the bonds formed between N-Cα (Φ) and Cα-C (Ψ) thereby indicating an absolute measure of the quality of a modeled protein structure. For protein residues to lie within the constricted ranges of helix in a Ramachandran Plot, the backbone dihedral angles Φ and Ψ should lie within −95° to −35° and −15° to −70° respectively. Further classification to helix type (alpha, π, and 310) is done by summing up of the phi and psi angles on adjacent residues; a helical sum of ~−105° signifies the backbone residues adopt an alpha-helical conformation while a helical sum of −75 or −130 indicates the residues assume a π or 310 helical conformation [61]. Ramachandran plots for helix α1, α2, and α3 were generated to check whether the concerned residues of each helix lied within the normal ranges of alpha helix region (Fig. S1). It is evident that the helical (Φ) and (Ψ) of α2 and α3 lie well within the constricted regions of an alpha helix (Fig. S1B, C). However, the helix α1 shows a more dispersed pattern with its residues unevenly scattered along the entire plot suggesting that the underlying residues switch between different secondary structural elements (Fig. S1A). The average sum for helices α1, α2, and α3 are −86.50°, −105.16°, and −103.68° respectively. To further analyze the helical properties, percentage propensity of the residues of each helix to maintain its helicity through 50 ns simulation run was calculated as shown for helices α1, α2, and α3 respectively (Fig. 3A–C). Notably, for an alpha helix to maintain its helicity the backbone acceptor C=O and H–N donor pairing distance should be at a minimum distance of 3.5 Å. Considering this fact the classical backbone hydrogen bond profile (n−n+4) was also calculated for helices α1, α2, and α3 respectively (Fig. 3D–F). To gather more information about structural fluctuations, helical conformations of α1, α2, and α3 for every 5 ns MD simulation was obtained from conformations starting at 5–50 ns. A total of ten conformations for each helical region was generated and superimposed to check for variations among the structural elements of α1, α2, and α3 (inset of Fig. 3D–F). Combining all of the facts stated above, the helical properties of α1, α2, and α3 has been substantiated. Residues Arg20, Leu22, and Ala23 of α1 are able to attenuate their helical nature as observed from the helical propensity in spite of the hydrogen bond profile showing fluctuations ranging between 3.5 Å and 4.5 Å (Fig. 3A). This fluctuation can be reasoned with the presence of Gly27 that is well known to be a helix breaker owing to its high conformational flexibility thereby disrupting the regularity of an alpha helical backbone conformation. The presence of Glu19, Leu22, Ala23, and Ala25 however steadies the alpha helicity of α1 owing to their high helix forming propensities. The helical nature of α2 and α3 is noted to be smoothly maintained as shown in (Fig. 3B and C) with residues in individual helices being able to retain their helical propensity almost the entire period of simulation run. Thus the above mentioned results illustrate the reliability of the modeled structure of ID3.
Fig. 3.
Helical Propensity percentage and average n−n+4 hydrogen bond length of the alpha helical regions of ID3. Helical propensity is shown for (A) alpha helix α1, (B) alpha helix α2, and (C) alpha helix α3. Average n−n+4 hydrogen bond length is shown for (D) alpha helix α1, (E) alpha helix α2, and (F) alpha helix α3 with inset pictures showing superimposition of individual helices at each 5–50 ns MD simulation.
3.3. Principal component and FEL analysis of ID3
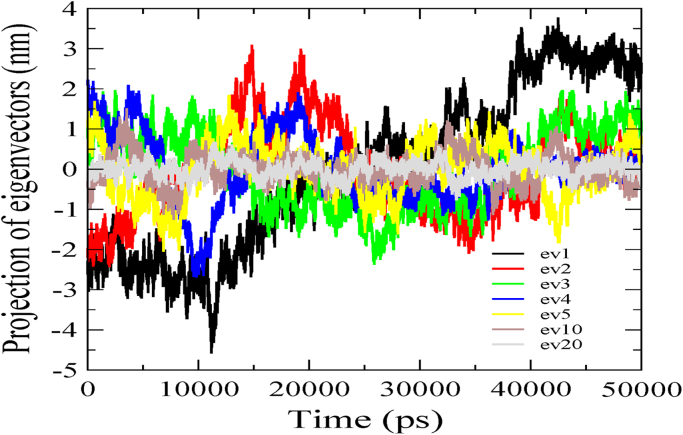
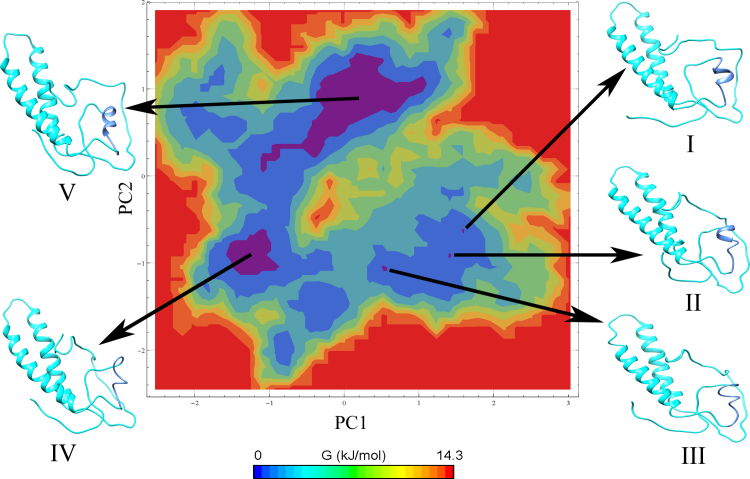
Principal component (PC) and Free Energy landscape (FEL) analyses were performed for ID3 to understand its structural and conformational stability during the simulation run. To begin with, a covariance matrix was generated on protein alpha carbon (Cα) atoms with a sum of eigenvalues to be 10.235 nm2. As mentioned earlier first few eigenvectors capture 90% of the internal motions of the protein thereby indicating that a large amount of the internal motion of the protein is restricted to a subspace with very small dimensions. Hence, for our study, we retrieved the first five, the tenth, and the twentieth projections from the protein trajectory and projected them onto the eigenvectors as obtained from the covariance matrix (Fig. 4). The subspace defined by the eigenvectors chosen from the cosine content analysis were picked up as representatives of the essential conformational subspace and used for constructing a free energy landscape. The contour map shows five different energy clusters namely I, II, III, IV, and V depicting the structural transition to distinct active conformational states (Fig. 5). The low energy representative structures of ID3 were retrieved at 18 ns, 21 ns, 23 ns, 30 ns, and 44 ns respectively. It is observed that low energy cluster V of the FEL shows much deviation from the initial predicted structure. The structural transition for cluster V occurs at 44 ns, suggesting that major fluctuations are observed during the first half of MD run and can be attributed for transition between clusters lying in the essential conformational subspace. Likewise, cluster III and IV whose transition occur at 23 ns and 30 ns respectively show almost similar structure deviation from the initial structure. The principal transition in these clusters within the configurational space is observed at the alpha helical region of α1 switching between two different secondary structural elements. The HLH motif however is seen to be in a metastable state with minimal transitions thus maintaining its structural integrity almost similar to that of the initial structure throughout the simulation period. In addition, the free energy representative structures were validated and are found to be within the permissible range of Ramachandran plot and the overall quality factor also defines the reliability of the simulated structures (Table S1).
Fig. 4.
PCA analyses of ID3. The projection of the trajectory by essential dynamics is shown depicting the motions along the first five, the tenth, and the twentieth eigenvectors during the 50 ns simulation run.
Fig. 5.
FEL analyses of ID3 depicting five low energy basins namely Region I, II, III, IV, and V along with their representative structures retrieved.
3.4. Stability analysis of E2A/E12 through molecular dynamics
The backbone RMSD profile of E2A was acquired to assess the convergence of the structure towards an equilibrium state. The plot elucidates the stability of the protein from the 11th ns to 50th ns with an RMS deviation to be around 0.1–0.2 nm (Fig. 6A). The RMSF profile was evaluated to look out for regions within the structure that are flexible and offer more fluctuations to the structure (Fig. 6B). The profile depicts higher fluctuations within residues lying towards the C-terminal of the protein. Evidently the loop residues buried within the bHLH domain as well as the helical regions (α10 and α11) are noticed to exhibit more fluctuations at a scale of 0.2 nm implying that these regions account for major structural transition during the production MD run. Besides these regions, loop regions that connect α6 and α7, and the β hairpin and α4 helix also depict low fluctuations up to 0.2 nm.
Fig. 6.
Stability Analysis of E2A/E12. (A) Backbone RMSD of E2A/E12 for 50 ns MD run. (B) Backbone RMSF of E2A/E12 showing the fluctuating regions within.
Helicity analysis was performed using the Φ and Ψ values for α10 and α11 extracted from the 50 ns production run. The average helical sum of Φ and Ψ for α10 and α11 helix was found to be −100.09 and −106.18 respectively which lies closer to the ranges for an alpha-helix. The helical percentage propensity of the residues of α10 and α11 illustrates the nature of helicity maintained by the two helices (Fig. S2). The region of α10 encompassing the bHLH region (547–581) shows propensity values of residues Gln547-Asn555, Glu558, and Glu571 are well maintained for more than 50% of the simulation time. The hydrogen bonding profile of the particular region show variations ranging from 3.5 to 4.5 Å which remains well stabilized until final production MD. The helical nature of the bHLH region (588–607) in α11 is, however, found to be well stabilized as observed in the per residue helical propensity values for more than half period of the simulation time.
3.5. Principal component and FEL analysis of E2A/E12
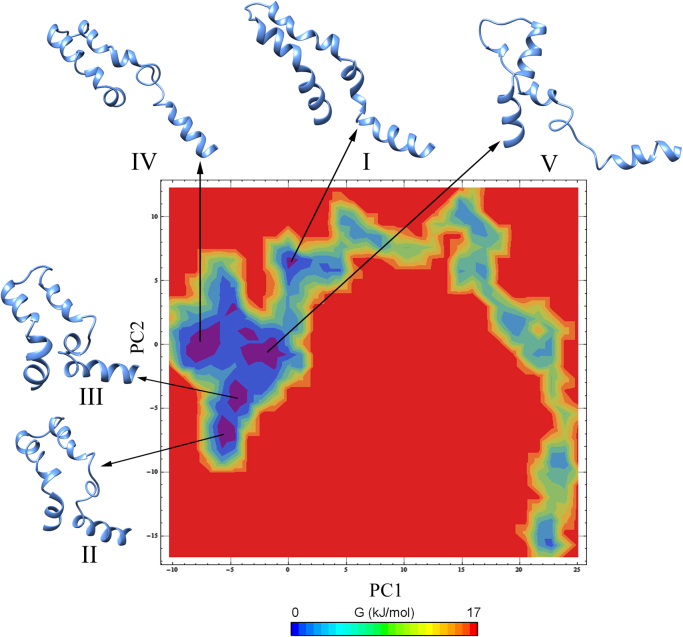
In a manner similar to the previous, essential dynamics and FEL analysis was performed to understand the structural and conformational stability of the bHLH region of E2A/E12 during the simulation run of 50 ns. Subsequently an energy landscape was generated to map the structural transition occurring within the domain region of the protein (Fig. 7). Five different populated minimum clusters retrieved at 18 ns, 21 ns, 23 ns, 30 ns, and 44 ns timescale were chosen as representatives portraying the structural transition occurring within the protein. The validated results of each free energy representative structures are found to be within the acceptable range of Ramachandran plot and the ERRAT values also define the reliability of the simulated structures (Table S1).
Fig. 7.
FEL Analysis of E2A/E12 depicting five low energy basins for the HLH domain namely Region I, II, III, IV, and V along with their representative structures obtained as function of principal components having cosine content less than 0.2 using projection eigenvectors.
3.6. Molecular Docking of ID3 and E2A/E12
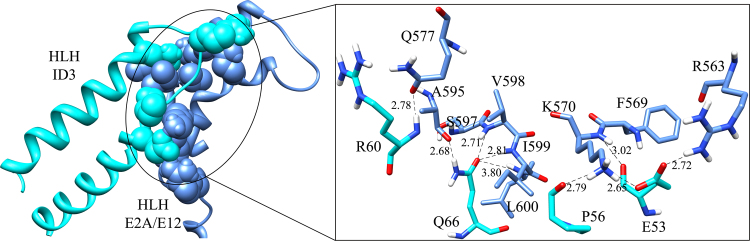
The best cluster of ID3 and E2A/E12 complex was selected in terms of the best binding energy (−61007.3 kcal mol−1) along with a buried surface area of 2302.56 Å2 (Table 1). The ID3–E2A/E12 complex has an interface area of 1183.7 Å2, encompassing a total of 26 residues of ID3 and 34 residues of E2A with a −15.0 kcal mol−1 solvation free energy gain upon interface formation (Table 2). A framework of nine hydrogen bonds and four salt bridges were maintained to stabilize the interface complex (Table 3). The bHLH domain of ID3 effectively makes strong interaction with the bHLH domain of E2A/E12. The α2, the loop, and α3 residues (Glu53, Pro56, Arg60, and Gln66) connect the bHLH domain of ID3 with the α10 (Lys570 and Gln577) and α11 residues (Arg563, Ala595, Val598, Ile599, and Leu600) of E2A/E12 (Fig. 8) through strong hydrogen and hydrophobic interactions. Such an interaction between the HLH domains of the two proteins is known to negatively regulate cell differentiation and promote anti-tumor suppressor properties. Through our study, we describe the interaction levels of the two bHLH proteins providing an insight into the key hotspot residues that might be responsible in the regulatory activity of the proteins as mediated by ID3.
Table 1.
HADDOCK energies for the docked complex of ID3 and E2A/E12.
| Structure | Binding energy (kcal mol−1) | Van der Waals energy (kcal mol−1) | Electrostatic energy (kcal mol−1) | AIR energy (kcal mol−1) | Buried surface area (Å2) |
|---|---|---|---|---|---|
| ID3–E2A | −61007.3 | −83.8859 | −224.483 | 1030.03 | 2302.56 |
Table 2.
Interface description for the docked complex of ID3-E2A/E12.1
| No. | ID3 |
E2A |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chain | NA | NR | Chain | NA | NR | Interface area (Å2) | ΔG (kcal/mol) | ΔG P-value | NHB | NSB | |
| 1 | A | 121 | 26 | B | 116 | 34 | 1183.7 | −15.0 | 0.336 | 9 | 4 |
Table 3.
Hydrogen bonds and salt bridges observed between ID3 and E2A/E12.
| S. no. | ID3 | E2A | Distance (Å) |
|---|---|---|---|
| Hydrogen bonds | |||
| 1 | A:ARG 60[N] | B:GLN 577[OE1] | 2.78 |
| 2 | A:GLN 66[HE22] | B:ALA 595[O] | 2.68 |
| 3 | A:GLU 53[OE1] | B:LYS 570[HZ3] | 2.65 |
| 4 | A:GLU 53[OE2] | B:ARG 563[HH22] | 2.72 |
| 5 | A:GLU 53[O] | B:LYS 570[N] | 3.02 |
| 6 | A:PRO 56[O] | B:LYS 570[HZ1] | 2.79 |
| 7 | A:GLN 66[OE1] | B:LEU 600[N] | 3.80 |
| 8 | A:GLN 66[OE1] | B:VAL 598[N] | 2.71 |
| 9 | A:GLN 66[OE1] | B:ILE 599[N] | 2.81 |
| Salt bridges | |||
| 1 | A:GLU 53[OE1] | B:LYS 570[NZ] | 2.65 |
| 2 | A:GLU 53[OE2] | B:LYS 570[NZ] | 3.76 |
| 3 | A:GLU 53[OE2] | B:ARG 563[NH1] | 3.80 |
| 4 | A:GLU 53[OE2] | B:ARG 563[NH2] | 2.72 |
Fig. 8.
HADDOCK bio-molecular complex of ID3–E2A/E12. The zoomed image shows the interacting residues between the HLH domains of ID3 (color code: cyan) and of E2A/E12 (color code: cornflower blue) forming the hydrogen bonds and salt bridges.
3.7. Stability analysis of ID3–E2A/E12 complex
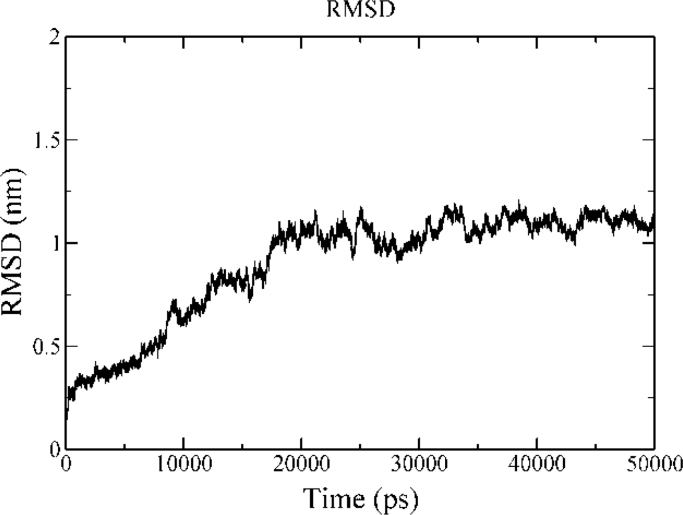
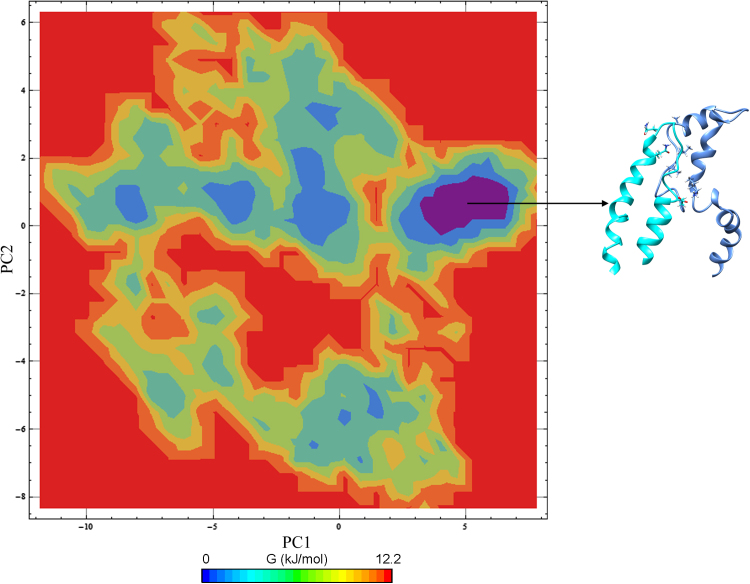
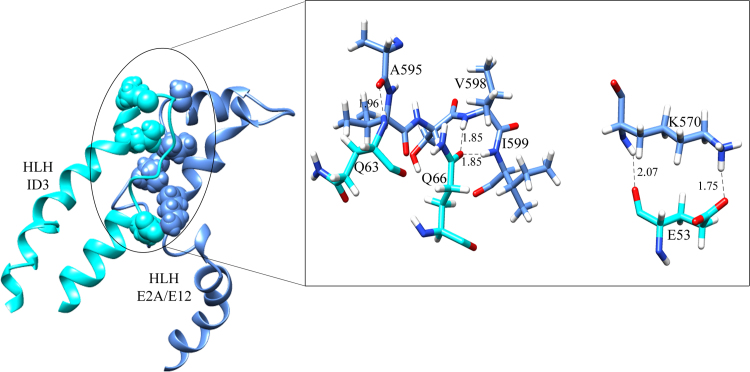
MD protocols for a 50 ns timescale of ID3–E2A/E12 complex was monitored by the analysis of their backbone RMSD, which show the complex was stabilized after 18 ns and maintained till the end of the production MD run with a maximum RMSD fluctuation 0.25 nm (Fig. 9). The H-bonds stability was also analyzed throughout the MD simulation and shown seven hydrogen bonds in the region of HLH motif are well maintained and stabilized (Fig. S3). Though, the simulated complex has seven hydrogen bonds throughout the simulation period in contrast to docked complex, regions beyond the HLH domain of ID3 and E2A/E12 significantly play a role for the complex stability. This difference in the hydrogen bond pattern was observed due to the intrinsic nature of E2A/E12 and its subsequent conformational changes observed upon complex formation. The free energy landscape generated using Cartesian coordinate's showed a single populated free energy cluster of the complex which was further evaluated for the selection of representative structure (Fig. 10). The interface analysis of the complex revealed the lowest energy representative structure to have a solvation free energy of −8.5 kcal mol−1 during the MD simulation (Table 4). The simulated free energy representative structure shows significant stability towards complex formation favored by a framework of five hydrogen bonds and one salt bridge, and also by a good interface area if compared with the native ID3–E2A/E12 complex (Fig. 11). These hydrogen bonds are mediated by residues of E2A/E12 (Lys570, Ala595, Val598, and Ile599) and ID3 (Glu53, Gln63, and Gln66), some of which were previously noted to maintain interactions in the native docked complex as well. This indicates the stronger contribution of these residues towards maintenance as a strong complex for promulgating their molecular function.(Table 5)
Fig. 9.
The RMSD graph of ID3–E2A/E12 complex throughout the simulation depicting its stability for 50 ns MD run.
Fig. 10.
The Free energy landscape of ID3–E2A/E12 complex showing the interacting HLH domain region. A single representative complex structure encompassing the domains is shown from the most populated free energy minimum cluster. Color code: Cyan – ID3 and cornflower blue – E2A/E12.
Table 4.
Interface description of the free energy representative structure derived from 50 ns MD simulation of ID3–E2A/E12 complex.2
| No. | ID3 |
E2A |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chain | NA | NR | Chain | NA | NR | Interface area (Å2) | ΔG (kcal/mol) | ΔG P-value | NHB | NSB | |
| 1 | A | 116 | 29 | B | 119 | 32 | 1112.5 | −8.5 | 0.533 | 5 | 1 |
Fig. 11.
The interacting conformation of free energy representative ID3–E2A/E12 complex. The zoomed image shows the interacting residues between the HLH domains of ID3 (color code: cyan) and of E2A/E12 (color code: cornflower blue) forming the hydrogen bonds and salt bridges.
Table 5.
Hydrogen bonds and salt bridges of free energy representative ID3–E2A/E12 complex.
| S. no. | ID3 | E2A | Distance (Å) |
|---|---|---|---|
| Hydrogen bonds | |||
| 1 | A:GLU 53[OE1] | B:LYS 570[HZ2] | 1.78 |
| 2 | A:GLU 53[O] | B:LYS 570[H] | 2.07 |
| 3 | A:GLN 66[OE1] | B:ILE 599[H] | 1.85 |
| 4 | A:GLN 66[OE1] | B:VAL 598[H] | 1.85 |
| 5 | A:GLN 63[H] | B:ALA 595[O] | 1.97 |
| Salt bridges | |||
| 1 | A:GLU 53[OE1] | B:LYS 570[NZ] | 2.76 |
3.8. Structural transition analysis
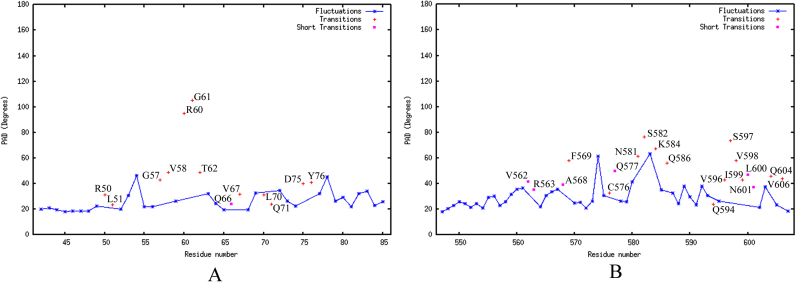
Structural integrity of the docked complex was further evaluated using the T-pad analysis to identify the regions (or amino acids) that undergo conformational transitions. Understanding the intrinsic properties, such as plasticity, of a protein helps in identifying the flexible and rigid residues within a protein that may have a plausible role in regulating various molecular recognition processes. The T-pad analysis of ID3–E2A/E12 gave a quantitative description of the residual fluctuations and transitions occurring within the interacting regions of the ID3 protein upon complex formation with E2A/E12 from its 50 ns MD simulation data. The angular dispersion plot depicts the residual fluctuation, transition and short transition of the HLH domains of proteins upon molecular association as shown in (Fig. 12). For the HLH region of ID3 protein, the pad analysis identified the regions which are highly flexible and significantly play a role for its molecular recognition towards E2A/E12. The plot reveals that the N-terminal α2 helix residues (Asp42-Ser49 and Arg52-Pro56), loop connecting α2 and α3 (Pro59 and Gln63), and α3 helix residues (Leu64, Ser65, Glu68, Ile69, Arg72-Ile74, and Ile77-Ala85) attaining fluctuations with PAD degrees ranging from 20–50° during the 50 ns MD time scale. Residues belonging to α2 helix (Arg50, Leu51, and Gly57), the loop connecting α2 and α3 (Val58, Arg60, Gly61, and Thr62), and the α3 helix (Val67, Leu70, Gln71, Asp75, and Tyr76) exhibit long transitions while residue Gln66 experiences short transition as observed from their PAD degrees (Fig. 12A). For the HLH region of E2A/E12 protein, residues belonging to the α10 helix (Phe569, Cys576, and Asn581), loop connecting α10 and α11 (Ser582, Lys584, and Gln586), and α11 helix (Gln594, Val596-Ile599, Gln604, and Val606) exhibit long transitions (from 20–79°) based upon the PAD angles (Fig. 12B). Residues namely Val562, Arg563, Ala568, Gln577, Leu600, and Asn601 are noticed to achieve shorter fluctuations. All these fluctuation, short transition and transition of both HLH motif of ID3 and E2A/E12 are significantly involved for its effective complex formation and anticipated for their different hydrogen bond pattern in the complex stability.
Fig. 12.
The residue Fluctuations, Transitions and Short Transitions of bHLH domain region of (A) ID3 and (B) E2A/E12 upon complex formation were calculated using T-pad tool and plotted based on the residue and their PAD degree from 50 ns MD simulation. The residue fluctuations are represented as starred connected lines (blue) and full transition and short transitions as plus (red) and rectangle (magenta) symbols respectively.
4. Conclusion
The dimerization of bHLH proteins has been well documented to modify diverse cellular processes, including negatively regulating the cell differentiation and promoting anti-tumor suppressor properties which is evident by the dimerization of ID3 with other bHLH proteins. The interaction mimics of the antagonist effect of ID3 with E2A/E12 provides a structural insight into how the helix–loop–helix domain might mediate the structural mechanism and vice versa. Accordingly, the structure of ID3 and E2A/E12 was predicted and analyzed for their stability through 50 ns MD simulation. Ensemble conformations of ID3 and E2A/E12 were obtained through PCA and FEL analysis and subjected to molecular interaction studies using HADDOCK docking protocol. The key residues that favor the molecular interaction between the HLH motif of ID3 and E2A/E12 were identified and confirmed further through 50 ns MD simulation and cross verified by the results of PCA and FEL analysis. Our findings reveal the key residues of E2A/E12 (Lys570, Ala595, Val598, and Ile599) and ID3 (Glu53, Gln63, and Gln66) residing in the HLH motif, observed to be involved for the significant complex stabilization are also maintained in the lowest free energy representative structure ID3–E2A/E12 complex. Furthermore, structural transition analysis performed on the bHLH domain regions of ID3–E2A/E12 complex identified the key residues which undergo the transitions and fluctuations in order to regulate the molecular recognition and its subsequent functionality. The aforementioned interactions significantly induce considerable structural changes in the helical regions of HLH domain which are proven to regulate the antagonist effect of ID3. However, the binding mechanism of ID3 towards the homo/hetero-dimeric interaction of E2A with DNA remains to be further investigated.
Acknowledgment
RK thanks the UGC Research Award (No. F.30-1/2013 (SA-II)/RA-2012-14-OB-TAM-1399) for providing financial assistance. NST (No. F.14-2(ST)/2010(SA-III)) and MK (No. F.14–2(SC)/2009(SA-III)) thank Rajiv Gandhi National Fellowship for pursuing their PhD and Centre for Excellence in Bioinformatics, Pondicherry University for providing the facilities to carry out the work.
Footnotes
Chain A represents ID3 and Chain B represents E2A. NA and NR represents number of atoms and number of residues respectively in the interface area. ΔG denotes solvation free energy gained upon interface formation, ΔG P-value denotes nature of hydrophobicity upon interface. NHB indicates number of hydrogen bonds and NSB indicates number of salt bridges across the interface.
Chain A represents ID3 and Chain B represents E2A. NA and NR represent number of atoms and number of residues respectively in the interface area. ΔG denotes solvation free energy gained upon interface formation, ΔG P-value denotes nature of hydrophobicity upon interface. NHB indicates number of hydrogen bonds and NSB indicates number of salt bridges across the interface.
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2015.12.002..
Contributor Information
Nishith Saurav Topno, Email: ntopno09@gmail.com.
Muthu Kannan, Email: mkannan52@gmail.com.
Ramadas Krishna, Email: ramadaskr@gmail.com.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Yokota Y., Mori S. Role of Id family proteins in growth control. J. Cell. Physiol. 2002;190:21–28. doi: 10.1002/jcp.10042. [DOI] [PubMed] [Google Scholar]
- 2.Ruzinova M.B., Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 3.Fong S., Debs R.J., Desprez P.Y. Id genes and proteins as promising targets in cancer therapy. Trends Mol. Med. 2004;10:387–392. doi: 10.1016/j.molmed.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Perk J., Iavarone A., Benezra R. Id family of helix–loop–helix proteins in cancer. Nat. Rev. Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 5.Engel I., Murre C. The function of E- and Id proteins in lymphocyte development. Nat. Rev. Immunol. 2001;1:193–199. doi: 10.1038/35105060. [DOI] [PubMed] [Google Scholar]
- 6.Davis R.L., Cheng P.F., Lassar A.B., Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- 7.Voronova A., Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix–loop–helix protein map to distinct domains. Proc. Natl. Acad. Sci. USA. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anthony-Cahill S.J., Benfield P.A., Fairman R., Wasserman Z.R., Brenner S.L., Stafford W.F., 3rd Molecular characterization of helix–loop–helix peptides. Science. 1992;255:979–983. doi: 10.1126/science.1312255. [DOI] [PubMed] [Google Scholar]
- 9.Ferre-D’Amare A.R., Prendergast G.C., Ziff E.B., Burley S.K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 10.Ellenberger T., Fass D., Arnaud M., Harrison S.C. Crystal structure of transcription factor E47: E-box recognition by a basic region helix–loop–helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 11.Nair S.K., Burley S.K. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 12.Phillips S.E. Built by association: structure and function of helix–loop–helix DNA-binding proteins. Structure. 1994;2:1–4. doi: 10.1016/s0969-2126(00)00002-2. [DOI] [PubMed] [Google Scholar]
- 13.Kee B.L. E and ID proteins branch out. Nat. Rev. Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 14.Wang D., Claus C.L., Vaccarelli G., Braunstein M., Schmitt T.M., Zuniga-Pflucker J.C. The basic helix–loop–helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J. Immunol. 2006;177:109–119. doi: 10.4049/jimmunol.177.1.109. [DOI] [PubMed] [Google Scholar]
- 15.Massari M.E., Murre C. Helix–loop–helix proteins: regulators of transcription in eukaryotic organisms. Mol. Cell. Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benezra R., Davis R.L., Lockshon D., Turner D.L., Weintraub H. The protein Id: a negative regulator of helix–loop–helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 17.Norton J.D., Atherton G.T. Coupling of cell growth control and apoptosis functions of Id proteins. Mol. Cell. Biol. 1998;18:2371–2381. doi: 10.1128/mcb.18.4.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norton J.D., Deed R.W., Craggs G., Sablitzky F. Id helix–loop–helix proteins in cell growth and differentiation. Trends Cell Biol. 1998;8:58–65. [PubMed] [Google Scholar]
- 19.Hara E., Yamaguchi T., Nojima H., Ide T., Campisi J., Okayama H. Id-related genes encoding helix–loop–helix proteins are required for G1 progression and are repressed in senescent human fibroblasts. J. Biol. Chem. 1994;269:2139–2145. [PubMed] [Google Scholar]
- 20.Norton J.D. ID helix–loop–helix proteins in cell growth, differentiation and tumorigenesis. J. Cell Sci. 2000;113:3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- 21.Andres-Barquin P.J., Hernandez M.C., Hayes T.E., McKay R.D., Israel M.A. Id genes encoding inhibitors of transcription are expressed during in vitro astrocyte differentiation and in cell lines derived from astrocytic tumors. Cancer Res. 1997;57:215–220. [PubMed] [Google Scholar]
- 22.Lin C.Q., Singh J., Murata K., Itahana Y., Parrinello S., Liang S.H. A role for Id-1 in the aggressive phenotype and steroid hormone response of human breast cancer cells. Cancer Res. 2000;60:1332–1340. [PubMed] [Google Scholar]
- 23.Lyden D., Young A.Z., Zagzag D., Yan W., Gerald W., O’Reilly R. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401:670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 24.Benezra R., Rafii S., Lyden D. The Id proteins and angiogenesis. Oncogene. 2001;20:8334–8341. doi: 10.1038/sj.onc.1205160. [DOI] [PubMed] [Google Scholar]
- 25.Sikder H.A., Devlin M.K., Dunlap S., Ryu B., Alani R.M. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3:525–530. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 26.Zebedee Z., Hara E. Id proteins in cell cycle control and cellular senescence. Oncogene. 2001;20:8317–8325. doi: 10.1038/sj.onc.1205092. [DOI] [PubMed] [Google Scholar]
- 27.Deed R.W., Hara E., Atherton G.T., Peters G., Norton J.D. Regulation of Id3 cell cycle function by Cdk-2-dependent phosphorylation. Mol. Cell. Biol. 1997;17:6815–6821. doi: 10.1128/mcb.17.12.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiewitz S.D., Cabrele C. Synthesis and conformational properties of protein fragments based on the Id family of DNA-binding and cell-differentiation inhibitors. Biopolymers. 2005;80:762–774. doi: 10.1002/bip.20287. [DOI] [PubMed] [Google Scholar]
- 29.Loveys D.A., Streiff M.B., Kato G.J. E2A basic-helix-loop-helix transcription factors are negatively regulated by serum growth factors and by the Id3 protein. Nucleic Acids Res. 1996;24:2813–2820. doi: 10.1093/nar/24.14.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 31.Brooks C.L., 3rd Protein and peptide folding explored with molecular simulations. Acc. Chem. Res. 2002;35:447–454. doi: 10.1021/ar0100172. [DOI] [PubMed] [Google Scholar]
- 32.Amadei A., Linssen A.B., Berendsen H.J. Essential dynamics of proteins. Proteins. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 33.Tavernelli I., Cotesta S., Di Iorio E.E. Protein dynamics, thermal stability, and free-energy landscapes: a molecular dynamics investigation. Biophys. J. 2003;85:2641–2649. doi: 10.1016/s0006-3495(03)74687-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mu Y., Nguyen P.H., Stock G. Energy landscape of a small peptide revealed by dihedral angle principal component analysis. Proteins. 2005;58:45–52. doi: 10.1002/prot.20310. [DOI] [PubMed] [Google Scholar]
- 35.Ichiye T., Karplus M. Collective motions in proteins: a covariance analysis of atomic fluctuations in molecular dynamics and normal mode simulations. Proteins. 1991;11:205–217. doi: 10.1002/prot.340110305. [DOI] [PubMed] [Google Scholar]
- 36.Tournier A.L., Smith J.C. Principal components of the protein dynamical transition. Phys. Rev. Lett. 2003;91:208106. doi: 10.1103/PhysRevLett.91.208106. [DOI] [PubMed] [Google Scholar]
- 37.Gruebele M. Protein folding: the free energy surface. Curr. Opin. Struct. Biol. 2002;12:161–168. doi: 10.1016/s0959-440x(02)00304-4. [DOI] [PubMed] [Google Scholar]
- 38.Raman S., Vernon R., Thompson J., Tyka M., Sadreyev R., Pei J. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77(Suppl 9):S89–S99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 40.Colovos C., Yeates T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 42.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 43.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 45.Miyamoto S., Kollman P.A. Settle: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992;13:952–962. [Google Scholar]
- 46.Martonak R., Laio A., Parrinello M. Predicting crystal structures: the Parrinello–Rahman method revisited. Phys. Rev. Lett. 2003;90:075503. doi: 10.1103/PhysRevLett.90.075503. [DOI] [PubMed] [Google Scholar]
- 47.Muthu K., Panneerselvam M., Jayaraman M., Topno N.S., Das A.A., Ramadas K. Structural insights into interacting mechanism of ID1 protein with an antagonist ID1/3-PA7 and agonist ETS-1 in treatment of ovarian cancer: molecular docking and dynamics studies. J. Mol. Model. 2012;18:4865–4884. doi: 10.1007/s00894-012-1489-x. [DOI] [PubMed] [Google Scholar]
- 48.Muthu K., Panneerselvam M., Topno N.S., Jayaraman M., Ramadas K. Structural perspective of ARHI mediated inhibition of STAT3 signaling: an insight into the inactive to active transition of ARHI and its interaction with STAT3 and importinbeta. Cell. Signal. 2015;27:739–755. doi: 10.1016/j.cellsig.2014.11.036. [DOI] [PubMed] [Google Scholar]
- 49.G. Stock, A. Jain, L. Riccardi, P.H. Nguyen, Exploring the Energy Landscape of Small Peptides and Proteins by Molecular Dynamics Simulations, 2012, pp. 55–77.
- 50.Maisuradze G.G., Leitner D.M. Free energy landscape of a biomolecule in dihedral principal component space: sampling convergence and correspondence between structures and minima. Proteins. 2007;67:569–578. doi: 10.1002/prot.21344. [DOI] [PubMed] [Google Scholar]
- 51.de Vries S.J., van Dijk A.D., Krzeminski M., van Dijk M., Thureau A., Hsu V. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 52.de Vries S.J., van Dijk M., Bonvin A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 53.Krissinel E. Crystal contacts as nature’s docking solutions. J. Comput. Chem. 2010;31:133–143. doi: 10.1002/jcc.21303. [DOI] [PubMed] [Google Scholar]
- 54.Huber R. Conformational flexibility in protein molecules. Nature. 1979;280:538–539. doi: 10.1038/280538a0. [DOI] [PubMed] [Google Scholar]
- 55.Teilum K., Olsen J.G., Kragelund B.B. Protein stability, flexibility and function. Biochim. Et. Biophys. Acta. 2011;1814:969–976. doi: 10.1016/j.bbapap.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 56.Lin J.H. Accommodating protein flexibility for structure-based drug design. Curr. Top. Med. Chem. 2011;11:171–178. doi: 10.2174/156802611794863580. [DOI] [PubMed] [Google Scholar]
- 57.Nocker M., Cozzini P. Induced fit simulations on nuclear receptors. Curr. Top. Med. Chem. 2011;11:133–147. doi: 10.2174/156802611794863526. [DOI] [PubMed] [Google Scholar]
- 58.Caliandro R., Rossetti G., Carloni P. Local fluctuations and conformational transitions in proteins. J. Chem. Theory Comput. 2012;8:4775–4785. doi: 10.1021/ct300610y. [DOI] [PubMed] [Google Scholar]
- 59.Pesce S., Benezra R. The loop region of the helix–loop–helix protein Id1 is critical for its dominant negative activity. Mol. Cell. Biol. 1993;13:7874–7880. doi: 10.1128/mcb.13.12.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bounpheng M.A., Melnikova I.N., Dimas J.J., Christy B.A. Identification of a novel transcriptional activity of mammalian Id proteins. Nucleic Acids Res. 1999;27:1740–1746. doi: 10.1093/nar/27.7.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo Z., Mohanty U., Noehre J., Sawyer T.K., Sherman W., Krilov G. Probing the alpha-helical structural stability of stapled p53 peptides: molecular dynamics simulations and analysis. Chem. Biol. Drug Des. 2010;75:348–359. doi: 10.1111/j.1747-0285.2010.00951.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material