Abstract
Human AlkB homolog 3 (ALKBH3), a homolog of the Escherichia coli protein AlkB, demethylates 1-methyladenine and 3-methylcytosine (3-meC) in single-stranded DNA and RNA by oxidative demethylation. Immunohistochemical analyses on clinical cancer specimens and knockdown experiments using RNA interference in vitro and in vivo indicate that ALKBH3 is a promising molecular target for the treatment of prostate, pancreatic, and non-small cell lung cancer. Therefore, an inhibitor for ALKBH3 demethylase is expected to be a first-in-class molecular-targeted drug for cancer treatment. Here, we report the development of a novel, quantitative real-time PCR-based assay for ALKBH3 demethylase activity against 3-meC by highly active recombinant ALKBH3 protein using a silkworm expression system. This assay enables us to screen for inhibitors of ALKBH3 demethylase, which may result in the development of a novel molecular-targeted drug for cancer therapy.
Abbreviations: ALKBH, AlkB homolog; 1-meA, 1-methyladenine; 3-meC, 3-methylcytosine; 2OG, 2-oxoglutarate; FTO, fat mass and obesity-associated; ss, single-stranded; ds, double-stranded; CRPC, castrate resistant prostate cancer; NACLC, non-small cell lung cancer; LC-MS/MS, liquid chromatography-tandem mass spectrometry
Keywords: AlkB, ALKBH3, Demethylation, 3-methylcytosine, RT-PCR
Highlights
-
•
Highly active recombinant ALKBH3 protein was produced from silkworms.
-
•
A novel, high-throughput assay for ALKBH3 demethylase activity was developed.
-
•
This assay enables us to screen for inhibitors of ALKBH3 demethylase.
-
•
A prominent ALKBH3-targeted drug may be developed for the treatment of cancers.
1. Introduction
AlkB is an Escherichia coli enzyme that demethylates 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) in single-stranded (ss)DNA and double-stranded (ds)DNA through its 2-oxoglutarate- (2OG) and Fe(II)-dependent oxygenase domain [1]. In humans, there are nine AlkB homologs (ALKBH) with the 2OG and Fe(II)-dependent oxygenase domain, which include ALKBH1 to ALKBH8 [2], [3] and fat mass and obesity-associated (FTO) [4], also known as ALKBH9. Of the ALKBH family proteins, ALKBH3 preferentially demethylates 1-meA and 3-meC in ssDNA and RNA in a 2OG and Fe(II)-dependent manner [5], [6].
ALKBH3, originally cloned as prostate cancer antigen-1 (PCA-1), is overexpressed in prostate cancer tissues compared with noncancerous tissues [7]. Higher expression of PCA-1 was immunohistochemically confirmed in human prostate cancer tissues but not in normal adjacent prostate tissues or benign prostatic hyperplasia. Prostate cancer is the second leading cause of cancer-related deaths in Western countries. Hormone therapy is effective for patients with prostate cancer; however, many patients develop castrate resistant prostate cancer (CRPC) within a few years. At present, effective therapies are limited for prostate cancers resistant to hormone therapy. Higher expression of ALKBH3 in prostate cancer specimens is correlated with the development of CRPC. Higher expression of ALKBH3 is also observed in pancreatic cancer [8], non-small cell lung cancer (NSCLC) [9], as well as renal cancer [10] specimens. In patients with these intractable cancers and high ALKBH3 expression also have a poorer prognosis.
In CRPC, pancreatic cancer, and NSCLC, the development of effective molecular-targeted drugs is highly desired. The knockdown of ALKBH3 by RNA interference induced apoptotic cell death in vitro and suppressed tumor formation in in vivo xenograft models [8], [11], [12]. These results strongly indicate that ALKBH3 is a promising molecular target for cancer therapy. However, to develop a molecular-targeted drug for ALKBH3, it is necessary to establish an assay to measure enzymatic activity, which could be used to screen for low molecular weight compound inhibitors. Few assays to measure the demethylase activity of ALKBH3 have been described [13], [14], [15]. These assays, although sensitive and fast, require special equipment or reagents such as capillary electrophoresis with laser-based fluorescence detection or mass spectrometer and radiolabeled substrates. Therefore, they are unsuitable for a high-throughput screening of low molecular weight compounds. In the present study, we obtained highly active recombinant ALKBH3 protein using a silkworm expression system. The production of recombinant ALKBH3 from silkworms enabled us to develop a real-time PCR-based quantitative assay of its demethylation activity against 3-meC in ssDNA.
2. Materials and methods
2.1. Expression and purification of recombinant ALKBH3 from silkworms
A silkworm vector, pMONFT21, for expression of human ALKBH3 (NM_139178.3) cDNA encoding the FLAG-His-tagged ALKBH3 (FLAG-His-ALKBH3) recombinant protein was constructed by Link Genomics (Tokyo, Japan). The expression vector was cotransfected with ABv baculovirus DNA into BmN cells to obtain the recombinant virus (Sysmex, Hyogo, Japan). The silkworm pupae were infected with the recombinant virus on the first day after pupation. Six days after infection, ten silkworm pupae expressing FLAG-His-ALKBH3 were homogenized at 10,000 rpm for 5 min in lysis buffer containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM ethylene glycol tetraacetic acid (EGTA), 10% glycerol, 10 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol. The homogenates were then solubilized by incubating with 1% Triton X-100 at 4 °C for 1 h. The lysate was centrifuged at 4 °C for 1 h at 100,000g. The protein purification was performed in two steps in order to achieve higher purity. First, the supernatant was purified by a 5-mL, 1.6×2.5-cm HisTrap HP affinity column (GE Healthcare) at 4 °C, using AKTA Prime Plus (GE Healthcare). The fractions containing recombinant FLAG-His-ALKBH3 were eluted with a linear gradient of 60–280 mM imidazole in 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 10% glycerol, and 0.1% Triton X-100. Subsequently, the eluate fractions containing recombinant ALKBH3 were diluted five-fold with 20 mM Tris-HCl (pH 8.0), 10% glycerol, and 0.1% Triton X-100 (binding buffer), and applied to a 0.5×5.0-cm Mono Q 5/50 GL anion ion exchange column (GE Healthcare) at 4 °C, using AKTA Prime Plus (GE Healthcare). After extensive washing with binding buffer containing 50 mM NaCl, recombinant FLAG-His-ALKBH3 was eluted with a linear gradient of 500 mM NaCl in the binding buffer, followed by dialysis in 20 mM Tris-HCl (pH 8.0), 50 mM NaCl, 50% glycerol, and 0.1% Triton X-100. Purity was confirmed on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel (Bio-Rad) stained with SimplyBlue SafeStain (Life Technologies).
2.2. Western blot analysis with anti-ALKBH3 antibody
Purified recombinant FLAG-His-ALKBH3 was resolved on a 10% denaturing acrylamide gel (Bio-Rad) and transferred to a polyvinylidene difluoride membrane (Millipore). The membrane was blocked with 3% bovine serum albumin at room temperature for 1 h. The membrane was incubated with anti-ALKBH3 antibody (1:5000 dilution, Millipore #09-882) overnight at 4 °C and then incubated with horseradish peroxidase-conjugated anti-rabbit IgG (Santa Cruz Biotechnology). Bound horseradish peroxidase conjugates were visualized using enhanced chemiluminescence reagent (GE Healthcare) under ImageQuant LAS 4000 (GE Healthcare).
2.3. Quantitative assay of ALKBH3 3-meC demethylase activity by real-time PCR
The reaction solution of the ALKBH3 demethylation assay comprised purified recombinant FLAG-His-ALKBH3 from silkworms (4 ng), 2 mM ascorbic acid, 100 μM 2-oxoglutarate, 40 μM Fe(SO)4·7H2O, and 80 fmol oligo ssDNA containing 3-meC (GGAAACAGCTATGACCATGATTACTAGACATTGCCATTCTCGATAGGATCCGGTCAAACCTAGACGAATTCCGGT-3-meC-GTGACTGGGAAAACCCTGGCG, Gene Design) as a substrate in 50 mM Tris-HCl (pH 8.0). The reaction mixture was incubated for 1 h at 37 °C, and then diluted 20-fold with distilled water for quantitative real-time PCR (qRT-PCR) analysis. The non-methylated ssDNA was used as a standard template. The qRT-PCR analysis was performed using a CFX96 real-time PCR system (Bio-Rad) in a total reaction volume of 20 µL containing 2 µL of the diluted reaction mixture, and 18 µL of Bio-Rad iQ SYBR Green Supermix with 200 nmol of each primer (sense: GGAAACAGCTATGACCATGATTAC, antisense: CGCCAGGGTTTTCCCAGTCGTG). Thermal cycling conditions were as follows: an initial incubation at 95 °C for 10 s to activate the polymerase followed by 40 cycles of 95 °C for 5 s, 61 °C for 30 s and 72 °C for 15 s. The level of the amplified products was calculated from a standard curve as the demethylation activity of ALKBH3 and ALKBH2. Recombinant His-tagged ALKBH2 (His-ALKBH2) and His-tagged ALKBH3 (His-ALKBH3) from E. coli were purchased from Abcam.
2.4. Digestion of ssDNA oligo containing 3-meC demethylated by ALKBH3 to nucleosides for LC-MS/MS analysis
After incubation with or without silkworm recombinant ALKBH3, the ssDNA oligo containing 3-meC was purified by ethanol-precipitation. The precipitate was suspended in 45 µL of H2O, after which 5 µL of 0.1 m ammonium acetate (pH 5.3) and 0.5 units of nuclease P1 (Wako) were added. After incubating for 2 h at 45 °C, 5.5 µL of 1 m ammonium bicarbonate and 0.002 units of venom phosphodiesterase II (Wako) were added to the mixture, followed by additional incubation for 2 h at 25 °C. Thereafter, the mixture was incubated for 1 h at 37 °C with 0.5 units alkaline phosphatase (NEW ENGLAND BioLabs). HCl (1.3 µL, 0.1 N), H2O (50 µL) and chloroform (20 µL) were then added. The sample was vortexed and the resulting suspension was centrifuged for 5 min at 5000g. The aqueous layer was collected and evaporated to dryness. The nucleosides left behind were solubilized in Milli-Q water.
2.5. LC-MS/MS confirmation of 3-meC demethylation by ALKBH3
LC-ESI-MS/MS analyses were performed on a Waters ACQUITY UPLC system (Waters) coupled to a Quattro Premier XE triple quadrupole mass spectrometer (Waters). For DNA samples, LC separations were carried out at 50 °C with an ACQUITY UPLC BEH C18 column, 1.7 µm, 2.1×100 mm (Waters). Mobile-phase A was 5 mM ammonium formate and 0.2% (v/v) formic acid, and mobile-phase B was acetonitrile. The analyte was eluted with mixed mobile-phase A and B (98/2, v/v) at a flow rate of 0.3 mL/min. The data for one run was acquired for 4 min. The mass spectrometer was operated using an electrospray ionization (ESI) source in the positive mode. ESI-MS/MS was conducted in the negative ion mode. The ionization parameters were capillary voltage, 3.0 kV; extractor voltage, 2 V; source temperature, 120 °C; desolvation temperature, 350 °C; desolvation gas flow, 800 L/h; cone gas flow, 50 L/h. Selected reaction monitoring (SRM) transitions (m/z of precursor ion/m/z of product ion) and parameters (cone voltage and collision energy) for the deoxynucleosides are listed in Supplementary Table 1. Inter-channel delay and inter-scan delay were set at 0.01 and 0.05 s, respectively. The dwell time for each SRM was set at 50 ms.
Statistics. Differences between values were statistically analyzed using a Student’s t-test. A p-value<0.05 was considered statistically significant.
3. Results
3.1. Production and purification of recombinant ALKBH3 from silkworm pupae
To develop an in vitro assay of ALKBH3 demethylase activity, a reliable supply of soluble and enzymatically active ALKBH3 protein is required. In protein expression, E. coli strains are routinely used; however, since recombinant ALKBH3 protein was mainly contained in inclusion bodies as aggregates, we could not obtain a sufficient amount of soluble recombinant ALKBH3 protein. Western blot analysis did not yield any band corresponding to the human ALKBH3 protein, which indicated that the yield was either extremely low or there was no yield at all (data not shown). The baculovirus-silkworm larvae expression system has been used to efficiently produce large amounts of recombinant proteins with biological activity [16]. Therefore, we tested whether a silkworm expression system would enable the production and purification of soluble recombinant ALKBH3 protein.
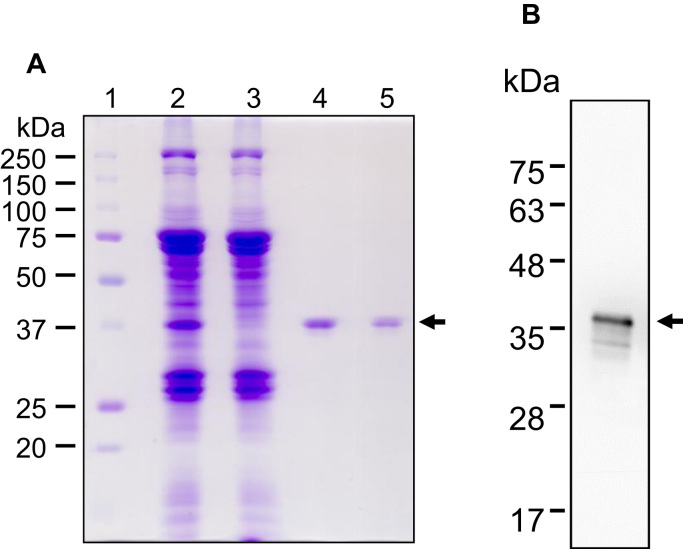
A silkworm expression vector containing FLAG-His-ALKBH3 cDNA was constructed and recombinant FLAG-His-ALKBH3 was expressed in 10 silkworm pupae. We discovered that the successful solubilization of recombinant FLAG-His-ALKBH3 can be achieved by the addition of 1% Triton X-100 to the homogenate of silkworm pupae (Fig. 1A). The supernatant containing soluble recombinant FLAG-His-ALKBH3 was obtained after ultracentrifugation of the homogenate. In order to achieve high purity, the recombinant FLAG-His-ALKBH3 was purified by a two-step affinity chromatography using a nickel affinity column and an anion exchange column (Supplementary Table 2). Purified recombinant FLAG-His-ALKBH3 was detected as a single, approximately 37-kDa band, which is its estimated molecular weight on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels. Immunoblot analysis with an anti-ALKBH3 antibody, anti-Histag antibody and anti-FLAG tag antibody indicated that these bands were the recombinant ALKBH3. (Fig. 1B, Supplementary Fig. 1A and B). The yield of purified recombinant FLAG-His-ALKBH3 was 0.6 mg from 10 silkworm pupae.
Fig. 1.
Purification of recombinant FLAG-His-ALKBH3 protein from silkworm pupae. (A) Silkworm pupae expressing FLAG-His-ALKBH3 were homogenized and purified by two-step affinity chromatography. Aliquots of the homogenate supernatant and purified fractions were resolved on a 10% SDS-PAGE gel and stained with Coomassie R-250. Lane 1, protein molecular weight markers; lane 2, input lysate; lane 3, flow-through fraction of nickel column; lane 4, eluant from the nickel column; lane 5, eluant purified by ion-exchange column. (B) Immunoblot analysis of purified recombinant FLAG-His-ALKBH3 protein (2 ng) using an anti-ALKBH3 antibody.
3.2. Potent 3-meC demethylation activity of recombinant ALKBH3 from silkworms
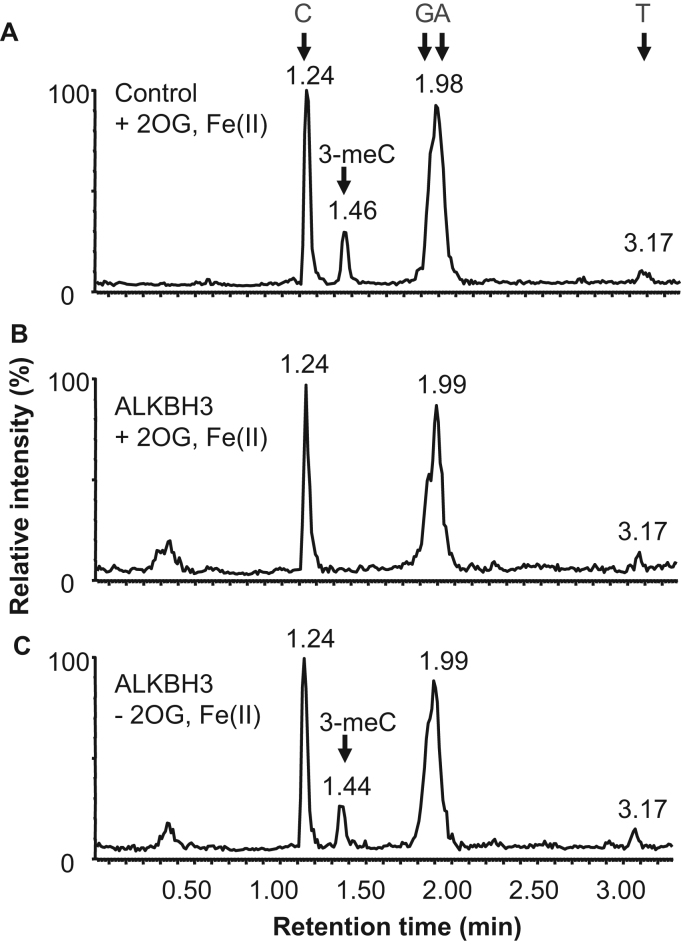
The enzymatic activity of purified recombinant FLAG-His-ALKBH3 was studied. ALKBH3 has potent demethylation activity against 3-meC in ssDNA in the presence of 2-OG and Fe(II). Therefore, as a substrate to study ALKBH3's enzymatic activity, we synthesized a unique 97 bp ssDNA oligonucleotide containing one 3-meC residue (referred to as 3-meC ssDNA). The sequence of the oligonucleotide was derived from a sequence found in a conventional cloning vector and its length was determined by taking into consideration the detection sensitivity of the following qRT-PCR analysis. 3-meC ssDNA was incubated with recombinant FLAG-His-ALKBH3 in the presence of 2-OG and Fe(II) at 37 °C for 1 h. After 1 h incubation, the 3-meC ssDNA in the reaction mixture was precipitated by ethanol and then resuspended in distilled water. Then the 3-meC ssDNA was incubated with nuclease P1 and venom phosphodiesterase II to digest phosphodiester bonds of the nucleotides, followed by incubation with alkaline phosphatase. The obtained nucleoside solution was treated with chloroform to remove proteins and then dried. The levels of nucleic acid bases and 3-meC in the samples were analyzed with LC-MS/MS. A peak representing 3-meC, which was detected in the control sample (Fig. 2A), could not be detected in the sample incubated with recombinant ALKBH3 in the presence of 2-OG and Fe(II) (Fig. 2B). However, recombinant ALKBH3 did not exhibit 3-meC demethylase activity in the absence of 2-OG and Fe(II) (Fig. 2C). The peak intensities of the bases (C, G, A, and T) were comparable in the samples incubated with or without recombinant ALKBH3 in the presence or absence of 2-OG and Fe(II). These results indicate that recombinant FLAG-His-ALKBH3 was successfully purified from silkworms and retained its demethylase activity.
Fig. 2.
Recombinant ALKBH3 from silkworm demethylates 3-meC in ssDNA oligonucleotides in a 2-OG and Fe(II)-dependent manner. Single-stranded DNA oligonucleotides containing 3-meC was incubated without (A) or with FLAG-His-ALKBH3 in the presence (B) or absence (C) of 2-OG/Fe(II). The ssDNA oligonucleotides were digested to nucleosides and analyzed by LC-MS/MS. Retention times are shown above each peak. A peak representing 3-meC was detected at a retention time of approximately 1.4 min.
3.3. The development of a quantitative assay to measure ALKBH3 demethylase activity by qRT-PCR
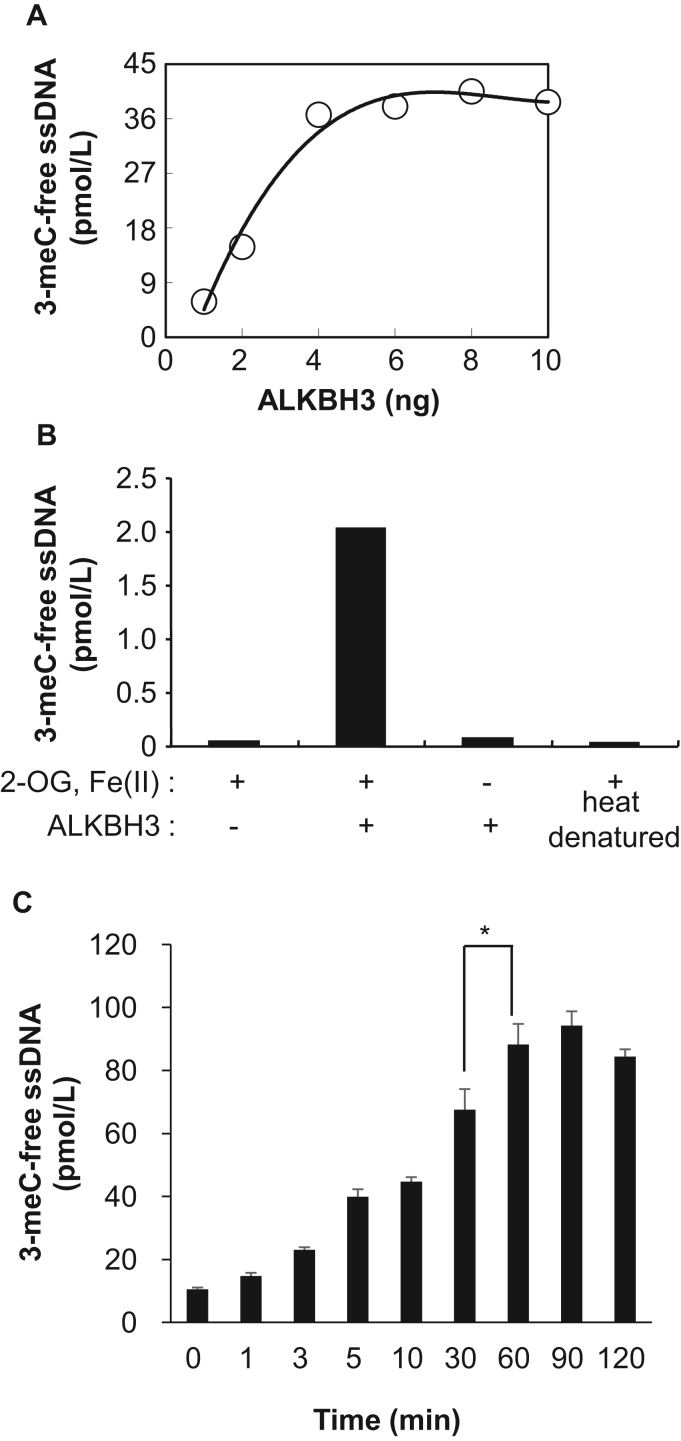
Previous evidence indicated that ALKBH3 might be a promising molecular target for CRPC, pancreatic cancer and NSCLC. To develop a molecular-targeted drug that targets ALKBH3, the establishment of an inhibitor screen for ALKBH3 demethylase is necessary. Since the N3-position of cytosine is involved in Watson-Crick base paring in dsDNA through hydrogen bonds, we assumed that a methylated residue at that position would disturb the annealing of a PCR primer. Moreover, qRT-PCR might provide a more quantitative and higher-throughput assay for screening small molecular weight compounds that inhibit ALKBH3 demethylase activity. Thus, for our ALKBH3 demethylation assay, we decided to perform qRT-PCR using a reverse primer with a mismatch at the second and third bases before the 3′-end (Supplementary Table 3) and found that the qRT-PCR detected the DNA demethylation in a methylated DNA content-dependent manner (Supplementary Fig. 2). First, we examined the concentration-dependence of recombinant ALKBH3 in the demethylation reaction. In a 10 μL reaction, the substrate, an oligo ssDNA containing one 3-meC, was incubated with various amounts of recombinant ALKBH3 in the presence of 2-OG and Fe(II) for 1 h. The reaction was then diluted 20-fold with distilled water. This dilution step was found to be suitable for the following reasons: further recombinant ALKBH3 demethylase activity was effectively quenched, and dilution did not interfere with the following qRT-PCR reaction in a 96-well plate. The diluted samples were then directly added to a qRT-PCR reaction solution. qRT-PCR analysis indicated that demethylation activity against 3-meC ssDNA was linear in a range of 0–6 ng of recombinant ALKBH3 (Fig. 3A). Therefore, we decided to use 4 ng of recombinant ALKBH3, which had approximately 90% the maximum demethylation activity observed in our assay.
Fig. 3.
qRT-PCR analysis of recombinant ALKBH3 demethylation activity against 3-meC ssDNA. Demethylation activity of recombinant ALKBH3 was analyzed by qRT-PCR. (A) The amount of demethylated product was estimated from a standard curve using ssDNA oligonucleotides without 3-meC. (B) The demethylation of 3-meC ssDNA by FLAG-His-ALKBH3 was measured in the presence or absence of 2-OG and/or Fe (II). Heat denatured: recombinant FLAG-His-ALKBH3 was heat-inactivated at 95 °C for 10 min. (C) The level of demethylated 3-meC ssDNA converted by recombinant ALKBH3 was determined in a time-dependent manner. All data shown are representative of at least three independent experiments. *p<0.05.
It is accepted that 2-OG and Fe(II) are essential for ALKBH3 demethylation activity. Indeed, the dependence of ALKBH3 demethylase activity on 2-OG and Fe(II) was quantitatively confirmed in our assay. As shown in Fig. 3B, the demethylase activity of recombinant ALKBH3 against 3-meC in ssDNA was observed only when both 2-OG and Fe(II) were present. On the other hand, heat-denatured recombinant ALKBH3 exhibited no enzymatic activity even in the presence of 2-OG or Fe(II).
Next, the dependence of ALKBH3 demethylase activity on the time of incubation was determined in our qRT-PCR assay. Recombinant ALKBH3 was incubated with 3-meC ssDNA in the presence of 2-OG and Fe(II) for 1–120 min. As shown in Fig. 3C and Supplementary Fig. 3, 3-meC demethylation by recombinant ALKBH3 was rapid, and approximately 50% of 3-meC in ssDNA was demethylated within 10 min. To achieve the complete demethylation of 3-meC in ssDNA by recombinant ALKBH3, an incubation time of 1 h was required.
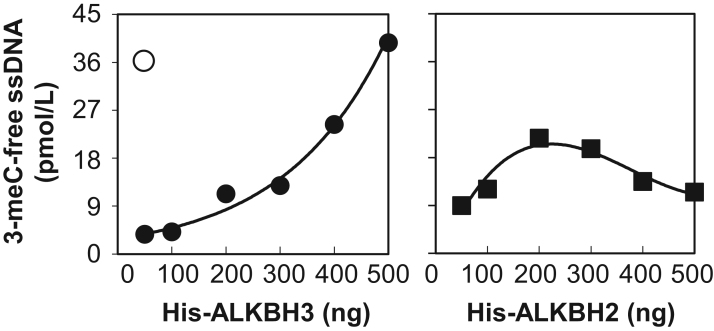
Finally, we compared the 3-meC demethylation activity of recombinant FLAG-His-ALKBH3 from silkworms with commercially available recombinant His-ALKBH2 and His-ALKBH3 from E. coli. ALKBH3 prefers as a substrate 3-meC in ssDNA to that in dsDNA. On the other hand, ALKBH2 preferentially demethylates 3-meC in dsDNA. Our qRT-PCR assay showed that recombinant FLAG-His-ALKBH3 from silkworms had demethylase activity against 3-meC ssDNA more than 400-fold greater than recombinant ALKBH3 from E. coli (Fig. 4). As expected, ALKBH2 showed weak demethylation activity against 3-meC in ssDNA. These results indicate that our qRT-PCR-based ALKBH3 demethylase assay can be useful for screening potential ALKBH3 inhibitors.
Fig. 4.
Comparison of 3-meC ssDNA demethylase activity of ALKBH2 and ALKBH3 from E. coli and FLAG-His-ALKBH3 from silkworms. Concentration-dependent demethylase activity of ALKBH2 and ALKBH3 from E. coli were determined in our qRT-PCR assay and compared to the activity of 4 ng of recombinant FLAG-His-ALKBH3 from silkworms (open circle). Data shown are representative of three independent experiments.
4. Discussion
We [7] and Liu et al. [17] have reported that ALKBH3, which was originally cloned as PCA-1, is highly expressed in prostate cancer tissues but not in normal prostate tissues or benign prostatic hyperplasia. The knockdown of ALKBH3 induces apoptosis of prostate cancer cells in vitro and suppresses tumor formation in an in vivo xenograft model [7], [12]. ALKBH3 is unique in its enzymatic activity by being able to demethylate 1-meA and 3-meC in ssDNA/RNA in a 2-OG/Fe(II)-dependent manner [5], [6]. Dango et al. have demonstrated that ALKBH3 enzymatic activity is critical for the growth of prostate cancer cells through the following mechanism [18]. ALKBH3 is associated with a helicase, activating signal cointegrator complex 3 (ASCC3). 3-meC produced by endogenous cellular metabolism or exogenous alkylating agents blocks Watson-Crick base pairing, leading to errors during DNA synthesis. ASCC3 unwinds dsDNA into ssDNA to expose 3-meC, making it accessible for demethylation by ALKBH3. On the other hand, RNA demethylation by ALKBH3 seems to increase the efficiency of protein synthesis in cancer cells (unpublished data). Recently, the high expression of ALKBH3 was also found in pancreatic cancers [8] and NSCLCs [9]. Therefore, ALKBH3 is a promising molecular target for the treatment of these cancers, and an inhibitor of its demethylase activity may represent a first-in-class cancer therapeutic agent. Here, we have established a useful demethylation assay for ALKBH3.
To develop a new molecular-targeted drug for cancer, a specific and sensitive assay to measure a target molecule’s activity must be established before screening for potential inhibitors from a library of low molecular weight compounds. A few assays to measure the demethylase activity of AlkB have been described [13], [14], [15]. These assays are sensitive and fast but require special equipment or reagents, such as radiolabeled substrates, capillary electrophoresis with laser-based fluorescence detection or mass spectrometry. Therefore, they are unsuitable for a high throughput screen of low molecular weight compounds. We established a qRT-PCR assay to measure ALKBH3 demethylase activity, which is based on the poor annealing of a primer that carries two mismatch bases to an ssDNA substrate containing 3-meC. A notable advantage of our assay is that many compounds (up to 384 samples depending on the qRT-PCR machine used) can be tested rapidly (within 2 h) and quantitatively in parallel.
Recently, by using our assay, Nakao et al. have reported that 1-(1H-5-methylbenzimidazol-2-yl)-4-benzyl-3-methyl-1H-pyrazol-5-ol has potent inhibitory activity against ALKBH3 demethylation [19]. Moreover, this compound inhibits prostate cancer cell growth in vitro and showed anti-tumor activity in an in vivo prostate cancer xenograft model. Optimization of the specificity and potency of this compound is the subject of future studies. However, these results demonstrate that our assay is useful in screening a large-scale chemical library.
Taken together, we have developed a quantitative, sensitive and high-throughput assay to screen for inhibitors of ALKBH3 demethylase. Since ALKBH3 represents a potential molecular target for CRPC, pancreatic cancer, NSCLC, and renal cancer [10], a high throughput screen using our assay might contribute to the development of a novel molecular-targeted agent.
Acknowledgements
This study was supported in part by a Grant-in-Aid for Scientific Research (26293015), by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics and Structural Life Science) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and Scientific Research provided by the Program for Promotion of Fundamental Sciences in Health Sciences of the National Institute of Biomedical Innovation (NIBIO).
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.02.007.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Trewick S.C., Henshaw T.F., Hausinger R.P., Lindahl T., Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 2.Drablos F., Feyzi E., Aas P.A., Vaagbo C.B., Kavli B., Bratlie M.S., Pena-Diaz J., Otterlei M., Slupphaug G., Krokan H.E. Alkylation damage in DNA and RNA—repair mechanisms and medical significance. DNA Repair. 2004;3:1389–1407. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Tsujikawa K., Koike K., Kitae K., Shinkawa A., Arima H., Suzuki T., Tsuchiya M., Makino Y., Furukawa T., Konishi N., Yamamoto H. Expression and sub-cellular localization of human ABH family molecules. J. Cell. Mol. Med. 2007;11:1105–1116. doi: 10.1111/j.1582-4934.2007.00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aas P.A., Otterlei M., Falnes P.O., Vagbo C.B., Skorpen F., Akbari M., Sundheim O., Bjoras M., Slupphaug G., Seeberg E., Krokan H.E. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859–863. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 6.Duncan T., Trewick S.C., Koivisto P., Bates P.A., Lindahl T., Sedgwick B. Reversal of DNA alkylation damage by two human dioxygenases. Proc. Natl. Acad. Sci. USA. 2002;99:16660–16665. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konishi N., Nakamura M., Ishida E., Shimada K., Mitsui E., Yoshikawa R., Yamamoto H., Tsujikawa K. High expression of a new marker PCA-1 in human prostate carcinoma. Clin. Cancer Res. 2005;11:5090–5097. doi: 10.1158/1078-0432.CCR-05-0195. [DOI] [PubMed] [Google Scholar]
- 8.Yamato I., Sho M., Shimada K., Hotta K., Ueda Y., Yasuda S., Shigi N., Konishi N., Tsujikawa K., Nakajima Y. PCA-1/ALKBH3 contributes to pancreatic cancer by supporting apoptotic resistance and angiogenesis. Cancer Res. 2012;72:4829–4839. doi: 10.1158/0008-5472.CAN-12-0328. [DOI] [PubMed] [Google Scholar]
- 9.Tasaki M., Shimada K., Kimura H., Tsujikawa K., Konishi N. ALKBH3, a human AlkB homologue, contributes to cell survival in Human non-small-cell lung cancer. Br. J. Cancer. 2011;104:700–706. doi: 10.1038/sj.bjc.6606012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hotta K., Sho M., Fujimoto K., Shimada K., Yamato I., Anai S., Harada H., Tsujikawa K., Konishi N., Shinohara N., Nakajima Y. Clinical significance and therapeutic potential of prostate cancer antigen-1/ALKBH3 in human renal cell carcinoma. Oncol. Rep. 2015;34:648–654. doi: 10.3892/or.2015.4017. [DOI] [PubMed] [Google Scholar]
- 11.Shimada K., Nakamura M., Ishida E., Higuchi T., Yamamoto H., Tsujikawa K., Konishi N. Prostate cancer antigen-1 contributes to cell survival and invasion though discoidin receptor 1 in human prostate cancer. Cancer Sci. 2008;99:39–45. doi: 10.1111/j.1349-7006.2007.00655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koike K., Ueda Y., Hase H., Kitae K., Fusamae Y., Masai S., Inagaki T., Saigo Y., Hirasawa S., Nakajima K., Ohshio I., Makino Y., Konishi N., Yamamoto H., Tsujikawa K. Anti-tumor effect of AlkB homolog 3 knockdown in hormone- independent prostate cancer cells. Curr. Cancer Drug Targets. 2012;12:847–856. doi: 10.2174/156800912802429283. [DOI] [PubMed] [Google Scholar]
- 13.Karkhanina A.A., Mecinovic J., Musheev M.U., Krylova S.M., Petrov A.P., Hewitson K.S., Flashman E., Schofield C.J., Krylov S.N. Direct analysis of enzyme-catalyzed DNA demethylation. Anal. Chem. 2009;81:5871–5875. doi: 10.1021/ac9010556. [DOI] [PubMed] [Google Scholar]
- 14.Welford R.W., Schlemminger I., McNeill L.A., Hewitson K.S., Schofield C.J. The selectivity and inhibition of AlkB. J. Biol. Chem. 2003;278:10157–10161. doi: 10.1074/jbc.M211058200. [DOI] [PubMed] [Google Scholar]
- 15.Woon E.C., Demetriades M., Bagg E.A., Aik W., Krylova S.M., Ma J.H., Chan M., Walport L.J., Wegman D.W., Dack K.N., McDonough M.A., Krylov S.N., Schofield C.J. Dynamic combinatorial mass spectrometry leads to inhibitors of a 2-oxoglutarate-dependent nucleic acid demethylase. J. Med. Chem. 2012;55:2173–2184. doi: 10.1021/jm201417e. [DOI] [PubMed] [Google Scholar]
- 16.Honjo E., Shoyama Y., Tamada T., Shigematsu H., Hatanaka T., Kanaji S., Arima K., Ito Y., Izuhara K., Kuroki R. Expression of the extracellular region of the human interleukin-4 receptor alpha chain and interleukin-13 receptor alpha1 chain by a silkworm-baculovirus system. Protein Expr. Purif. 2008;60:25–30. doi: 10.1016/j.pep.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 17.Liu B.Q., Wu Y.D., Li P.H., Wei J.X., Zhang T., Liu R.L. Prostate cancer antigen-1 as a potential novel marker for prostate cancer. Asian J. Androl. 2007;9:821–826. doi: 10.1111/j.1745-7262.2007.00279.x. [DOI] [PubMed] [Google Scholar]
- 18.Dango S., Mosammaparast N., Sowa M.E., Xiong L.J., Wu F., Park K., Rubin M., Gygi S., Harper J.W., Shi Y. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol. cell. 2011;44:373–384. doi: 10.1016/j.molcel.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakao S., Mabuchi M., Shimizu T., Itoh Y., Takeuchi Y., Ueda M., Mizuno H., Shigi N., Ohshio I., Jinguji K., Ueda Y., Yamamoto M., Furukawa T., Aoki S., Tsujikawa K., Tanaka A. Design and synthesis of prostate cancer antigen-1 (PCA-1/ALKBH3) inhibitors as anti-prostate cancer drugs. Bioorg. Med. Chem. Lett. 2014;24:1071–1074. doi: 10.1016/j.bmcl.2014.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material