Abstract
Skeletal muscle formation in vertebrates is derived from the paraxial mesoderm, which develops into myogenic precursor cells and finally differentiates into mature myofibers. This myogenic program involves temporal-spatial molecular events performed by transcription regulators (such as members of the Pax, MRFs and Six families) and signaling pathways (such as Wnts, BMP and Shh signaling). Epigenetic regulation, including histone post-translational modifications is crucial for controlling gene expression through recruitment of various chromatin-modifying enzymes that alter chromatin dynamics during myogenesis. The chromatin modifying enzymes are also recruited at regions of muscle gene regulation, coordinating transcription regulators to influence gene expression. In particular, the reversible methylation status of histone N-terminal tails provides the important regulatory mechanisms in either activation or repression of muscle genes. In this report, we review the recent literatures to deduce mechanisms underlying the epigenetic regulation of gene expression with a focus on histone methylation modification during embryo myogenesis and adult muscle regeneration. Recent results from different histone methylation/demethylation modifications have increased our understanding about the highly intricate layers of epigenetic regulations involved in myogenesis and cross-talk of histone enzymes with the muscle-specific transcriptional machinery.
Abbreviations: SCs, satellite cells; MRFs, myogenic regulatory factors; bHLH, basic helix-loop-helix; MEF2, myocyte enhancer factor 2; Shh, sonic hedgehog; BMP4, bone morphogenic protein 4; p38 MAPK, p38 mitogen-activated protein kinase; H3K4, methylation of histone H3 lysine 4; H3K9, methylation of histone H3 lysine 9; H3K27, methylation of histone H3 lysine 27; PRC2, polycomb repressive complex 2; LSD1, lysine specific demethyltransferase 1; KDMs, lysine demethyltransferases; UTX, ubiquitously transcribed tetratricopeptide repeat, X chromosome; ChIP, chromatin immunoprecipitation; TSS, transcription start sites
Keywords: Myogenesis, Muscle progenitor cells, Muscle differentiation, Muscle regeneration, Epigenetic, Histone methylation/demethylation modification
Highlights
-
•
Myogenesis is influenced by regulation of transcription factors, signal pathways and post-transcriptional modifications.
-
•
Histone methylation modifications as “on/off” switches regulated myogenic lineage commitment and differentiation.
-
•
The myogenic regulatory factors and histone methylation modifications established dynamic regulatory mechanism.
1. Introduction
Embryo myogenesis or adult muscle regeneration is the programming of a population of muscle progenitors, embryonic or fetal myoblasts and satellite cells (SCs) into committing to the myogenic lineages and of myoblasts into differentiating into mature myofibers. The formation of skeletal muscles involves both genetic and epigenetic changes that culminate in alterations in gene expression [1]. Chromatin-modifying enzymes and remodeling complexes orchestrate the pattern of gene expression and reprogram the myogenic lineage toward terminal differentiation [1], [2]. The transcriptional regulation of muscle specification has been well characterized, and the role of histone acetylation modification in control of muscle-specific gene expression has been studied extensively, however, less is known about the role of histone methylation modification in this process [3]. Here, we review the potential roles of histone modifications during myogenesis with a focus on H3 lysine 27 tri-methylation (H3K27me3), H3 lysine 4 tri-methylation (H3K4me3) and H3 lysine 9 di/tri-methylation (H3K9me2/3) markers at myogenic gene regulatory regions in myoblasts and satellite cells.
2. Myogenesis: gene regulatory networks and transcriptional mechanisms
2.1. Development of embryo myogenesis
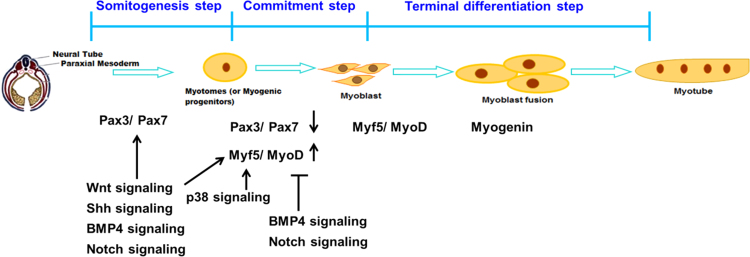
Skeletal muscle is initiated in the somite (epithelial spheres along the anterior–posterior axis of the embryo) which derives from paraxial mesoderm adjacent to the neural tube and notochord [2], [4], [5]. The newly formed dorsal somites rapidly differentiate into dermomyotomes, which are the source of muscle precursor cells. Cells from the dorsomedial part of the somites adjacent to the neural tubes migrate under the dermomyotomes to form the myotomes [6]. The myotomes are committed and differentiate into myoblasts, and final maturation of myotubes fuse into myofibers (Fig. 1). The epaxial (dorso-medial) part of the dermomyotomes and myotomes generate the back muscles while the hypaxial (ventro-lateral) somites generate the rest of the trunk and limb muscles [7], [8], [9].
Fig. 1.
A schematic representation of developmental myogenesis. It shows that the somites give rise to muscle progenitor cells, progenitor cells determinate and proliferate as myoblasts, and myoblasts differentiate into myotubes. During this process, the action of both positive and negative signals control the spatio-temporal expression of muscle genes. And gene regulatory networks are performed by myogenic transcriptional factors.
2.2. Genetic regulatory networks in myogenesis
2.2.1. Regulation of transcription regulators in myogenesis
The paired-homeobox family of transcription factors Pax3 and Pax7 are important upstream regulators of the myogenic process in the embryo myogenesis. In Splotch mice, because of mutation of Pax3, cells fail to develop the hypaxial domain of the somite and thus lack limb musculature and other muscle masses in the body. However, epaxial-derived muscles are less affected [6], [10], [11], [12]. In the chick embryo, Pax3:Pax7-positive cells are maintained as proliferating cells and do not express myogenic regulatory factors or muscle proteins. However, they can give rise to skeletal muscle cells leading to subsequent skeletal muscle differentiation and producing muscle satellite cells [13]. Pax3:Pax7 double mutants die at early fetal stages. In the absence of both Pax3 and Pax7 proteins, muscle progenitor cells do not activate the myogenic determination genes to enter the myogenic program [14].
The myogenic regulatory factors (MRFs) of MyoD, Myf5, myogenin and Mrf4 genes have highly conserved basic helix-loop-helix domain (bHLH) structure and are expressed in the skeletal muscle lineage [15], [16]. When myogenesis continues, Pax3 expression decreases gradually while MRF family gene expression increases significantly [17]. The high expression of Myf5 gene forms the primitive muscle structure containing committed muscle cells [15], [18]. Subsequently, MyoD gene is activated after the onset of Myf5 expression in the dermomyotome [4], [19]. Two of these factors are muscle determination genes. They are the major factors that activate the myogenic program in muscle and non-muscle cells [20]. In contrast to MyoD and Myf5, myogenin is a key gene for activating the muscle differentiation program. The number of skeletal muscle fibers are reduced in mice carrying mutations in myogenin [21], [22]. The role of Mrf4 is more complicated in myogenesis. Mrf4 closely linked with Myf5 gene in mice is activated in myotomes following the Myf5 expression, and its expression is reinitiated in differentiating muscle cells stage. Overexpression of Mrf4 can compensate for the muscle fibers in myogenin double mutations embryo [23], [24].
The myocyte enhancer factor 2 family alone has no myogenic activity but they assist the MRFs through transcriptional cooperation to mediate expression of muscle-specific genes [25], [26]. MyoD family of MRFs have MEF2 binding sites, so MEF2 could provide a positive feedback loop and cooperative interactions between many MRF proteins [27]. The other families of transcription factors for myogenesis are the Six family of homeobox proteins Six1 (the sine oculis-related homeobox 1) and Six4 and their transcriptional cofactors eyes-absent homologs Eya1 and Eya2. Six proteins associate with the Eya1 and Eya2, generating a regulatory cascade that directs dermomyotomal progenitor cells toward the myogenic lineage. Hence, they are critical in the migration of hypaxial myogenic precursor cells from the somite to the limb and serve as upstream regulators of Pax3, MyoD and myogenin during somitogenesis [28], [29], [30].
2.2.2. Regulation of extracellular signals in myogenesis
During embryogenesis, the adjacent structures in embryo release both positive (Wnt, Sonic hedgehog (Shh) and noggin) and negative (BMP4 and Notch) signals that control the spatial and temporal expression pattern (Fig. 1). All of these signals regulate the amount of myogenic regulatory factors for balancing between proliferation and differentiation status during muscle development [2], [15].
Wnt signaling is involved in forming the dermomyotome and myotome [15], [31]. Wnt1 and Wnt3 are produced by the dorsal neural tube, and Wnt4, Wnt6 and Wnt7a are produced by the surface ectoderm [32]. The expression of the Pax3 and Myf5 is increased by Wnt1 and Wnt3 [33], while the expression of MyoD is reduced by Wnt6 and Wnt7a [34]. Moreover, Wnt signaling activates the program of myogenesis through the TCF/β-catenin pathway [8]. β-catenin acts as a Wnts receptor and is present in the myotome depending on Wnt1 or Wnt3a. β-catenin interacts with Shh and is up-regulated in the dorsal somite prior to MyoD activation.
Shh signaling regulates myogenesis mechanism through the down-regulation of Pax3/7 and up-regulation of Myf5/MyoD expression, leading to myogenic cells to withdraw from proliferation and start differentiation [38], [39], [40]. Whereas Shh signaling is controlled by Wnt signaling through the regulation of Gli2 and Gli3 [35].
BMP4 (bone morphogenic protein 4) can up-regulate Pax3 expression in the embryonic trunk [36]. Overexpression of BMP4 in the somite can inhibit MyoD expression, resulting in the inhibition of myotome formation [37]. Overexpression of BMP2b increases expression of Pax3 and the number of Pax7-positive myogenic precursor cells and delays muscle differentiation via decreasing MyoD expression in the zebrafish embryo [38]. As BMP4's antagonist, noggin lies downstream of the Shh and Wnt signaling pathways and physically interacts with it and counteracts the effects of BMP4 on myotome formation [39], [40].
The activity of Notch signaling influences cell differentiation, proliferation, and apoptotic programs [41]. Stimulation of Notch signaling in Pax3-positive myogenic progenitor cells inhibits muscle differentiation and causes a reduction in myogenic cells that migrate into the limbs [42]. MyoD cooperated with the DNA binding protein RBP-J and the transcriptional repressor Hes1 (hairy and Enhancer of split) can suppress Notch signaling activity [43]. Hes1 also effectively inhibits MyoD, inducing myogenic conversion of C3H10T1/2 cells into muscle cells [44]. Two-hybrid assays indicate that constitutively active Notch is able to block MEF2C DNA binding ability [45].
The p38 MAPK (p38 mitogen-activated protein kinase) signaling is an intracellular signaling pathway that activates myogenic cell lines to affect the muscle transcription program. In mice, (1) p38 is rapidly activated during myocyte differentiation. Conversely, inhibition of p38 activity prevents the differentiation in myogenic cell lines and human primary myocytes, (2) overexpression of p38 stimulates the transcriptional activity of MyoD and in embryonic fibroblasts of p38 double mutant mice, the efficiency of MyoD-dependent myogenic conversion is reduced, and (3) p38 signaling activates MEF2C transcription factor through selective phosphorylation of MEF2C on Thr293 residue in differentiating myocytes [46], [47], [48].
2.3. Adult muscle regeneration: common mechanisms and mediators
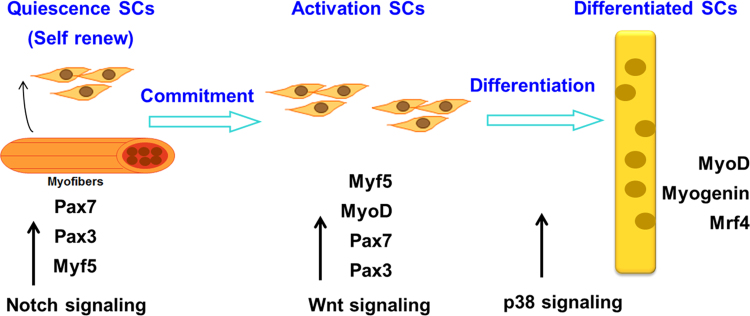
Adult muscle regeneration or de novo myogenesis in vertebrates depends on the injured tissue recruiting an undifferentiated myogenic progenitor cells or muscle stem cells to the site of injury. In adult skeletal muscle, this function is provided by the satellite cells [4], [15], [49], [50]. Responding to injury, quiescent satellite cells are activated and then proliferate as myoblast cells and differentiate to form new muscle fibers or returned to quiescence to maintain the stem cell pool (self-renewal).
2.3.1. Regulation of transcription regulators in adult muscle regeneration
Similarly, in embryo myogenesis, Pax7 as a satellite cell marker is expressed in quiescent satellite cells in muscles from throughout the body [4], [14], [19], [49], [51], [52], [53] and Pax7 is co-expressed with MyoD in their proliferating myoblast progeny [54]. Pax3 is expressed in satellite cell in muscles from approximately 50% of forelimb muscles and most ventral trunk muscles [49]. Constitutive expression of Pax3 or Pax7 in satellite cells results in an increased proliferative rate and prevents differentiation [54]. Interestingly, in Pax7 mutant cells, the cell number progressively decreases in both Pax3-expressing and Pax3-non-expressing muscles. It has been shown that Pax7 cannot be compensated by Pax3 during the postnatal development of skeletal muscles [49]. In addition, recent studies analyzing the role of Pax7 in adult satellite cells have demonstrated that temporary deletion of Pax7 in adult satellite cells results in normal muscle regeneration after injury, indicating that Pax7 is not required for de novo myogenesis in the adult [55], but long-term Pax7 abrogation leads to loss of adult satellite cells, resulting in impaired muscle regeneration after injury [56], [57].
Myf5 gene is already expressed in quiescent satellite cells. MyoD gene is expressed in the activated satellite cells and, along with down-regulation of Pax7 expression, subsequently differentiates with the expression of myogenin gene [49], [50], [58], [59], [60]. However, overexpressing Pax7 in myoblasts represses MyoD expression and inhibits myogenesis. Ectopic expression of Pax7 can efficiently block the MyoD-dependent conversion of mesenchymal stem cells (10T1/2 cells) to the muscle lineage [61]. These studies suggest that satellite cells are heterogeneous population cells. They have maintained stem cells self-renewal ability and given rise to committed myogenic progenitors which later undergoes differentiation. The molecular mechanisms may involve in a reciprocal inhibition between Pax7 and the muscle regulatory factors (Fig. 2).
Fig. 2.
A schematic representation of adult muscle regeneration. Upon muscle injury, quiescent satellite cells are activated and divided asymmetrically, generating a self-renewing cell and a committed progenitor which begins to express Myf5. Next, activated satellite cells express MyoD and down-regulate Pax7. Then activated satellite cells will differentiate and fuse to form new myofibers during adult muscle regeneration.
2.3.2. Regulation of extracellular signaling in adult muscle regeneration
Studies with adult muscle regeneration indicate that Wnt signaling induces CD45+stem cells purified from regenerating muscle to form determined myoblasts but fails to induce CD45+stem cells purified from uninjured muscle to form myogenic cells. Regenerating muscles with rejection of Wnt antagonists sFRP2/3 inhibits CD45+stem cells proliferation and myogenic specification. β-catenin, as a key downstream transcriptional co-activator of Wnt signaling, is strongly up-regulated in satellite cells and during muscle regeneration. Other studies indicate that activation of the Wnt/β-catenin signaling pathway causes changes in the structure of chromatin at the MyoD and Myf5 promoters, leading to increases in the expression of Myf5 and MyoD in satellite cells and in the number of proliferative Pax7+/Myf5+ and Pax7+/MyoD+ cells in skeletal muscle. All of these findings suggest that Wnt signaling plays a critical role in the regulation of satellite cells in adult muscle regeneration [62], [63], [64], [65].
Notch-1 is expressed in satellite cells, and its activity is essential for maintaining the expression of Pax7. Attenuation of Notch signaling by increasing expression of inhibitor Numb leads to the commitment of satellite cells to the myoblasts fate [66], [67].
3. The epigenetic modifications in myogenesis and adult muscle regeneration
3.1. Epigenetic code
The concept of “Epigenetics” was first introduced by Conrad Waddington in 1942. After decades of research, epigenetics has been found to involve DNA methylation, histone modification, chromatin remodeling, and non-coding RNAs (ncRNAs) in mammalian animals [68]. For nucleosomes, the basic unit of chromatin, the amino residues on histone tails (histone amino termini or histone N-termini) are subject to diverse arrays of covalent post translational modifications, including acetylation, methylation, phosphorylation, and ubiquitination. Distinct histone modifications generate synergistic or antagonistic interaction for chromatin-associated proteins, which in turn determine dynamic transitions between transcriptionally active or silent states of chromatin [69], [70]. Histone modifications influence chromatin packaging and are read by adaptor molecules, chromatin modifying enzymes, transcription factors and transcriptional repressors, and thus contribute to the regulation of transcription. Epigenetic histone modifications have been best characterized for histones H3 and H4 [71]. Histone methylation occurs on arginine and lysine amino acids residues. Mono-, di- or tri-methylation has been discovered on histone H3 and H4 [68]. H3K4me3 has been associated with euchromatin and facilitates active transcription by recruiting the RNA polymerase II complex at gene promoter/enhancer regions [72]. In contrast, H3K27me3 is mutually exclusive with H3K4me3, which is generally considered a repression marker for transcription [73]. H3K9me2 or H3K9me3 is linked to silencing of gene expression at euchromatin and heterochromatin [74].
3.2. Epigenetic modifications in embryo development
3.2.1. Epigenetic modifications in embryonic stem (ES) cells
Embryo development proceeds from a cascade of gene activation and repression. Differentially expressed embryonic genes are regulated by various patterns of histone modifications on unmethylated DNA, which create a developmentally permissive chromatin state and confer a specific chromatin configuration on gene regulatory and coding regions [71], [75]. The analysis of histone modifications in ES cells has generated genome-wide maps of H3K27me3 and H3K4me3 [76]. ChIP assays for zebrafish embryos show that all embryonic gene promoters have severe deacetylation on H3K9 and H4 relative to H3K4me3. Deacetylation is accompanied by enrichment in H3K9me3 and H3K27me3, suggesting that embryonic genes are associated with repressing histone modifications in fibroblasts [75]. ChIP-chip experiments form developmental transition from late fetus to lamb during late ovine skeletal muscle development reveal that H3K27me3 modification is associated with genes, transcription start sites (TSS) and CpG islands and with transcriptional silencing. Most modified genes have no changes during the muscle transition, indicating that H3K27me3 does not have a large role in late muscle maturation [77]. These findings suggest that H3K27me3 represses developmental genes at initial embryonic stages.
Recent data from studies in ES cells show that the function of H3K4me3 in ES cells is poising the expression of embryonic genes that allows for the proper activation of lineage regulators upon differentiation, but its mechanism remains unclear. How H3K4me3 and H3K27me3 markers are established in ES cells is still unclear. The existing evidence shows that some developmental regulatory genes are already marked by H3K4me3 and H3K27me3 in sperm [76], [78].
A prevalent model in stem cell biology suggests that the loss of pluripotency entails global increase in heterochromatin and coincident shutdown of lineage-unrelated genes [79]. H3K9me2 appears to be the most abundant heterochromatic modification and occupies large parts (>50%) of the genome in ES cells. Analysis of the H3K9me2 in ES cells shows that active regions are mutually exclusive with H3K9me2, and the majority of H3K4me2 regions are mutually exclusive with H3K9me2 [79], [80]. Moreover, G9a has been implicated in embryonic development, based on the embryonic lethality of G9a knockout mice [81].
3.2.2. Bivalency of H3K4 and H3K27 methylation
Bivalent domains were first identified at the TSS of key regulators of lineage determination in cultured ES cells [65]. ChIP-chip studies in human and mouse ES cells show that developmental genes are maintained in a bivalent status. These bivalent promoters are maintained in a repressive chromatin state through H3K27me3, however, the presence of an active chromatin modification H3K4me3 suggests that these genes are poised for rapid induction [82], [83].
In ES cells, Polycomb group (PcG) proteins predominantly suppress H3K4-methylated CpG island promoters. Polycomb repressive complex 2 (PRC2) and polycomb repressive complex 1 (PRC1) seem to cooperate to counteract H3K4 methylation. PRC2 contributes to silencing by recruiting H3K4 histone demethyltransferases, thereby regulating the homeostasis of the bivalent state. Furthermore, PRC2 facilitates PRC1 targeting by providing the H3K27me3 binding site [83]. Similarly, the studies of Orford and colleagues show that H3K4me2+/me3- or H3K4me2+/me3+ CpG island promoters are repressed at the progenitor stage by the presence of H3K27me3, but non-CpG island tissue-specific genes are amenable to marking by H3K4me2 “only” [83], [84]. During myoblasts differentiation, modifications of H3-K4 by Set7 and H3-K9 by Suv39h1 are mutually exclusive at the promoters/enhancers of myogenic differentiation genes [85].
Pax3 is the only myogenic transcription factor with bivalent domains in quiescent satellite cells, which correlates well with the low expression levels of Pax3 in satellite cells of adult mice. In contrast, Pax7 is only marked by H3K4me3 at its TSS. On the other hand, myogenin is not marked by either H3K4me3 or H3K27me3 in quiescent satellite cells, but shows significant enrichment of the H3K4me3 mark at its TSS upon activation [86].
3.3. Histone methylation modifications in myogenesis
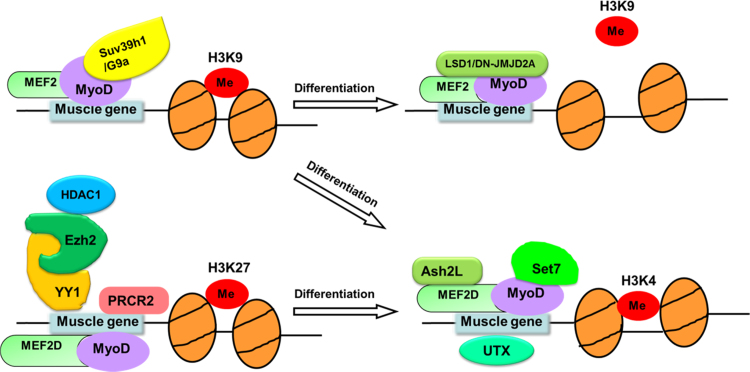
The transition from proliferation into differentiation of muscle cells is accompanied by a number of chromatin-associated complexes with histone modifications through dynamic changes in chromatin to balance activation or repression at regulation of muscle genes transcription. Histone methylation modifications as “on/off” switches play an important role in lineage commitment and differentiation (Fig. 3). Generally, transient and long-term gene silencing are performed by tri-methyl histone H3 on lysine 9 and 27, and gene activation is regulated by tri-methylation of H3K4.
Fig. 3.
Regulation of histone methyltransferases in muscle differentiation. In proliferation of myoblasts and quiescent satellite cells, the cooperation of myogenic genes and histone methytransferases represses expression of muscle differentiation genes and maintains H3K27 or H3K9 methylation on muscle-specific gene promoters. Once myoblasts and quiescent satellite cells are activated, the cooperation of myogenic genes and histone methytransferases are released by histone demethyltransferases or pro-myogenic triggering signaling cascades (e.g. p38 signaling). The H3K4 tri-methylation status exists at the chromatin region of muscle-specific gene promoters in differentiated muscle cells.
3.3.1. H3K27 methylation modification
Tri-methyl lysine modifications on histones are the most stable epigenetic marks and control chromatin-mediated regulation of gene expression [87], [88]. H3K27me3 is known to regulate muscle differentiation through silencing muscle-specific genes and is widely distributed throughout the genome in both myoblasts and myotubes, including promoters, gene bodies, and intergenic regions [89]. While PcG proteins have histone lysine methyltransferase activity, PRC2 catalyzes tri-methylation of histone H3 lysine 27 that is associated with transcriptional silencing [90], [91], [92]. The global mapping analysis reveals that the PcG proteins bind a large number of tissue-specific differentiation genes such as myogenin and CKM, which are specifically induced in muscle differentiation [92]. The Ezh2 protein is one component of PRC2 [93]. Ezh2 is expressed in the myotome compartment of mouse somite and in proliferating satellite cells and is down-regulated in terminally differentiated muscle cells. This coincides with activation of muscle gene expression in differentiation of myoblasts. In undifferentiated myoblasts, endogenous Ezh2 is associated with the transcriptional regulator YY1 and histone deacetylase HDAC1. And the complex will occupy genomic regions of silent muscle-specific genes [91].
3.3.2. H3K4 methylation modification
Trithorax (TrxG) group proteins are known for antagonizing polycomb group proteins repressing. These two group of proteins establish the transcription patterns that maintain the repressed or active transcription states of developmentally important genes [94]. TrxG proteins with a SET domain catalyze tri-methylation of H3 lysine 4 [94]. H3K4me3, as an active marker, is almost exclusively found in promoter regions of actively transcribed genes [87], [88]. Chromatin immunoprecipitation experiments find that the promoter region of myogenin gene becomes enriched in H3K4me3 during C2C12 cell differentiation. As Ash2L protein is a component of H3K4 methyltransferase complexes and is expressed in C2C12 cells, protein-protein interaction analysis indicates that MEF2 (MEF2D and MEF2C) is associated with Ash2L and MLL complexes (including MLL2, Set1 and MLL3) which is recruited to the myogenin promoter in differentiated C2C12 cells and the nucleosomes containing H3K4me3 during C2C12 cell differentiation. And this process needs phosphorylation of MEF2D by p38-α via p38 signaling pathway [95], [96], [97].
Mixed lineage leukemia 5 (MLL5) is an H3K4 methyltransferase encoding a SET and PHD domain protein homologous to Drosophila Trithorax and yeast SET3, but appears to lack intrinsic HMT activity. In quiescent myoblasts, MLL5 regulates both the cell cycle and differentiation via a hierarchy of chromatin and transcriptional regulators. Knocking down MLL5 delays entry of quiescent myoblasts into S phase, but hastens S-phase completion by inducing the expression of cyclin E and cyclin A (CycA). Meanwhile, CycA mRNA is induced throughout G0/G1, with activation of the cell cycle regulated element (CCRE) in the CycA promoter. Comparing with normal cells, defective differentiation in MLL5 knockdown myoblasts correlates with reduced expression of Pax7 throughout the cell cycle, suppressed expression of Myf5 in G0, with delayed G1 induction, altered function of MyoD, and inefficient activation of myogenin [98].
Set7, the H3K4 specific methyltransferase, catalyzes H3K4 methylation in vitro and in vivo and potentiates transcription activation. Set7 contains a SET domain, but lacks the pre- and post-SET domains [99]. Studies show that the expression of Set7 is dynamically increased during normal skeletal muscle differentiation. Knockdown of Set7 by siRNA represses the expression of MyoD, myogenin and MyHC and reduces the number of myotubes. Moreover, Set7 physically interacts with MyoD and Set7 association and the H3K4 monomethylation status on the regulatory regions of myogenic differentiation genes substantially increased during C2C12 cells differentiation. All of these findings suggest that Set7 has a role in skeletal muscle development and is required for the terminal differentiation of skeletal muscle myoblasts [85].
3.3.3. H3K9 methylation modification
Suv39h1/KMT1A is a H3K9 specific methyltransferase catalyzing di- or tri-methylation of histone H3 lysine 9 [100], [101], that is associated with Heterochromatin protein 1 (HP1) in gene silencing [102], [103]. Studies reveal that Suv39h1 has multi-roles in regulating muscle genes transcription during muscle cell differentiation. Mal et al. demonstrate that Suv39h1 interacts with MyoD and represses MyoD-dependent muscle gene expression. This repression requires Suv39h1 association with MyoD and sustains methylation of H3K9 on myogenin promoter [104]. Ait-Si-Ali et al. demonstrate that Suv39h1 depleted myoblasts have a poor capacity to fuse into myotubes and are unable to express early and late muscle differentiation markers, such as myogenin, MCK and MyHC [105].
G9a is a member of SET domain containing Suv39 family [106] and it mainly catalyzes mono- or di-methylation of histone H3 lysine 9 in euchromatin [81], [107]. G9a is also involved in catalyzing histone H3 lysine 9 tri-methylation [108] and histone H3 lysine 27 methylation [106]. Recently, G9a is reported to have a dynamical expression mode during skeletal muscle cells differentiation. G9a can inhibit skeletal muscle cells differentiation in a methyltransferase activity-dependent manner by mediating H3K9me2 around MyoD target gene promoters. In addition, G9a catalyzes MyoD methylation at K104 site in a methyltransferase activity-dependent manner to constrain its transcriptional activity [107].
Sharp-1, a DNA-binding transcription and basic helix-loop-helix transcription factor, is a potent repressor of skeletal muscle differentiation via negatively affecting MyoD function by competition for binding to E-box sites, formation of inactive heterodimers, and inhibition of its transcriptional activity. Sharp-1 is expressed widely in a number of cell types, including developing skeletal muscles. Current evidence demonstrates that G9a associates with and enhances Sharp-1-mediated transcriptional repression and meanwhile overexpression of Sharp-1 correlates with increased H3K9me2 on the myogenin promoter and enhanced MyoD-G9a association and MyoD methylation in C2C12 cells [109].
3.4. Histone demethylation modifications in myogenesis
3.4.1. H3K9 demethylation modification
Histone demethyltransferases regulates gene expression by removing histone methyl group marks [110]. LSD1 (Lysine Specific Demethylase 1), a nuclear homolog of amine oxidases, is a lysine specific demethyltransferase which is responsible for removing methyl group from mono-, di-methylation of H3K4 and H3K9 [111], [112], [113]. Therefore, LSD1 can repress H3K4 transcription activation and activate H3K9 transcription repression [114]. Current studies have proven that expression of LSD1 is increasing during myogenic cells differentiation. LSD1 is directly associated with MyoD and MEF2 on their target promoters to activate myogenic genes in vivo. Abrogation of LSD1 can cause myoblasts without differentiation ability. In short, it suggests that catalytic activity of LSD1 is vital for skeletal muscle cells differentiation [115].
Another histone lysine demethyltransferase (KDM) JHDM2A, a jumonji (or JmjC) domain containing protein that specifically removes methyl group from H3K9 [116], [117]. JHDM2A mediates the demethylation of mono- and di-methyl H3K9 in vitro and in vivo [117]. A new isoform of JMJD2A, named DN-JMJD2A, lacks the N-terminal demethyltransferase domain whose expression is up-regulated during muscle cells differentiation [3]. DNA microarray analysis indicates that the expression of muscle-specific genes as myogenin and CKM are strongly decreased by knockdown DN-JMJD2A. ChIP analysis indicates that DN-JMJD2A is recruited to myogenin promoter and DN-JMJD2A can promote the demethylation of H3K9me2 at myogenin promoter [3]. Moreover, DN-JMJD2A promotes MyoD induced-myogenic conversion of NIH3T3 cells into muscle cells [3].
Another JmjC domain containing demethyltransferases (Jmjd), Jmjd2C, one of Jmjd2 family members, demethylates histone H3K9me2 and H3K9me3. It has been reported that Jmjd2C increases MyoD transcriptional activity to facilitate skeletal muscle differentiation by increasing MyoD stability through inhibiting G9a-dependent MyoD degradation [118].
3.4.2. H3K27demethylation modification
The UTX (ubiquitously transcribed tetratricopeptide repeat, X chromosome) protein, belongs to a subfamily of JmjC domain-containing proteins. As mammalian H3K27 specific demethyltransferases, the UTX removes di- and tri-methyl group fromH3K27me2/3 [119], [120], [121]. UTX is enriched around the transcription starting sites of many HOX genes in primary human fibroblasts, and UTX is present in the MLL2/3/4 containing histone H3K4 methyltransferase complexes altering histone methylation dynamics. These findings suggest that UTX is essential for H3K27 methylation regulation and is also important for the stringent regulation of transcription during cellular differentiation [119], [120], [121]. In differentiating myoblasts, UTX is recruited to the transcriptional regulatory region of both the myogenin and CKM genes with removal of the repressive H3K27me3 mark at both genes. Knockdown of UTX in C2C12 cells prevents the formation of multinucleated, MyHC-positive myotubes [122]. The recent findings demonstrate that UTX with an important role in mediating demethylation of H3K27me3 at muscle-specific genes during myogenesis. In addition, histone chaperoneSpt6 is reported to promote UTX chromatin enrichment and reduce H3K27me3 level at myogenin regulation regions [123].
3.5. Histone methylation modifications in satellite cell regeneration
In proliferating satellite cells, histones associated with the promoters of genes specifically involved in muscle differentiation are hypo-acetylated and contain H3K9me2, H3K9me3 and H3K27me3 residues [65]. Pax7 is a key mark in satellite cells and has predominant function during the postnatal development of skeletal muscles. Studies on protein–protein interactions provide a post-translational modification mechanism for regulating Pax7 and Pax7 target genes Myf5 and MyoD function in satellite cells.
3.5.1. H3K27 methylation modification
H3K27 methyltransferases play important roles in the epigenetic regulation of stem cell quiescence. Ezh2 is expressed in quiescent satellite cells and required for homeostasis of the adult satellite cell pool [124]. When proliferating myoblasts receive differentiation signals, phosphorylation of the enhancer of Ezh2 recruits the Trithorax group/Ash2L complex through p38-phosphorylated MEF2D and induces transition from a transcriptionally permissive H3K4me3 mark to a repressive H3K27me3 mark on the Pax7 promoter to down-regulate its expression. Myogenic genes that are highly expressed in proliferating or differentiated cells, such as MyoD and myogenin, are also marked by the repressive H3K27me3 mark [97], [125], [126]. While in activated satellite cells, the level of H3K27me3 increases significantly across the genome, whereas theH3K4me3 mark is retained [65], [127].
Mice with satellite cell-specific inactivation of Ezh2 have reduced muscle mass, a decreased number of satellite cells and exhibit defective regeneration after injury. These differences have been attributed to a failure of the proliferative progenitor population to expand (Ezh2 maintains a key phase of muscle satellite cell expansion but does not regulate terminal differentiation.) and to impair maintenance and/or return to quiescence (self-renewal) after injury [124].
3.5.2. H3K4 methylation modification
Yeast two-hybrid screening, tandem affinity purification (TAP) and mass spectrometry experiments indicate that Pax7 is associated with the Wdr5–Ash2L–MLL2 histone methyltransferases complex which directs methylation of H3K4 at the Myf5 promoter, resulting in H3K4 tri-methylation of surrounding chromatin [96], [128], [129]. The comparative microarray analysis, siRNA, fluorescence-activated cell sorting (FACS) and ChIP confirm that Myf5 is directly regulated by Pax7 in C2C12 cells which are derived from satellite cells [128]. Currently protein–protein interaction experiments indicate that Pax7 is a substrate of CARM1, and CARM1 could specifically methylate multiple arginine in the N-terminus of Pax7. Methylated Pax7 directly binds to C-terminal cleavage forms of MLL1/2 and recruits Ash2L:MLL1/2:Wdr5:Rbbp5 histone H3K4 methyltransferase complex to regulatory enhancers and the proximal promoter of Myf5 [95], [130]. Together, these experiments provide important epigenetic mechanisms that Pax7 stimulates transcriptional activation of MyoD family of target genes to regulate entry into the myogenic developmental program by inducing chromatin methylation modification.
3.5.3. H3K9 methylation modification
The p38 signaling is a trigger in satellite cells during muscle regeneration. p38-γ is highly expressed in skeletal muscles. During adult myogenesis, p38-γ phosphorylates MyoD on Ser199 and Ser200, which results in enhanced occupancy of MyoD on the promoter of myogenin together with markedly decreased transcriptional activity. This repression is associated with extensive methylation of histone H3K9 together with recruitment of the Suv39h1/KMT1A methyltransferase to the myogenin promoter. Notably, a MyoD S199A/S200A mutant exhibits markedly reduced binding to Suv39h1/KMT1A. Therefore, p38-γ signaling directly induces the assembly of a repressive MyoD transcriptional complex [131].
3.6. Other methylation modifications and histone chaperones in myogenesis
CARM1, referred to as PRMT4 (protein arginine N methyltransferase 4), is defined as a type of the S-adenosyl-L-methionine-dependent methyltransferases, which catalyzes the methylation of arginine residues on specific proteins [132]. The expression of CARM1 is detected in somites during embryogenesis and in the nuclei of muscle cells. Protein-protein interaction assay systems demonstrate that CARM1 directly interacts with the C-terminal region of MEF2C in vivo and CARM1 cooperates with the steroid receptor co-activator (SRC) cofactor GRIP-1 to stimulate MEF2C. Inhibition of CARM1 dramatically reduces the myogenin and MEF2 protein level and morphological differentiation of proliferating myoblasts into multinucleated myotubes. CARM1/PRMT4 is necessary for skeletal muscle differentiation [133].
Spt6 (Suppressor of Ty 6), as a transcription elongation factor and histone chaperone, interacts with RNA polymerase II (Pol II) to reassemble nucleosomes during Pol II elongation [134] and interacts with histones and primarily with histone H3 in controlling chromatin structure, respectively [135], [136], [137]. Spt6 is considered critical for somite formation in zebrafish embryogenesis. Genetic and biochemical experiments demonstrate that Spt6 is essential for the transcriptional response to activation of the Notch pathway: her1 and her7, two of Notch pathway key target genes for somite formation, are suppressed by disruption of Spt6 function in panSBU2 (Zebrafish Spt6 mutants) embryos [138]. Spt6 associated with Pol II is recruited at chromatin regions of muscle-specific genes and is required for appropriate muscle-specific genes expression in myoblasts (e.g., MyoD and des) and differentiated myotubes (e.g., myogenin, MyH3, MyoD, des and Mylpf) [123].
Histone chaperone HIRA (HIR histone cell cycle regulation defective homolog A) is a human homolog of the S. cerevisiae HIR1 and HIR2 (histone regulatory) [139], [140], [141]. HIR1 and HIR2 proteins act as co-repressors to assist a repressor that repress histone gene transcription in S. cerevisiae [142]. HIRA is classified as DNA replication-independent chaperone and specifically chaperones at H3.3 in mammalian variants [139]. It has been confirmed that HIRA interacts with MEF2 and stimulates MEF2 target genes transcription during muscle differentiation [143].
3.7. Methylation modification at myogenic regulators
As a myogenic regulator, MyoD can regulate transcription at gene promoters. Moreover, MyoD can also bind enhancers and mediates the recruitment of several chromatin-modifying enzymes, such as Set7, which catalyzes histone H3K4me1 to active enhancers [65], [144], [145], or Suv39h1, which catalyzes histone H3K9me2/3 to inhibit MyoD transcriptional activity [104]. In addition, MyoD can also be as a substrate that catalyzed by chromatin-modifying enzyme. Unlike Suv39h1, G9a not only mediates H3K9me2 but also directly methylates MyoD at K104 to inhibit its transcriptional activity [107]. Similarly, MEF2 activity is also regulated by posttranslational modifications, such as methylation. Recently studies find that MEF2D is methylated and demethylated by G9a and LSD1 at its K267, respectively, which effects the dynamic regulation of MEF2D transcriptional activity and the expression of its target genes during skeletal muscle differentiation [146].
4. Conclusions
Chromatin modifiers mediate dynamic modifications of histone tails that are vital to reprogramming cells toward terminal differentiation. Genetic and epigenetic mechanisms ensure that complex developmental programs are correctly executed [83].
In this report, we review the recent literature to deduce mechanisms underlying a complex interplay between myogenic regulatory factors and histone methylation modifications and reveal the reciprocal regulation between histone methyltransferases and demethyltransferases in the control of gene expression during skeletal muscle myogenesis and regeneration (Fig. 3). We also elucidate that histone chaperones play critical roles in controlling of muscle gene regulatory programs in this review. In addition, we find that histone methyltransferases not only mediate dynamic modifications of histone tails for influencing myogenic gene expression during skeletal myogenesis but also catalyze myogenic regulatory factors methylation at lysine site in controlling of gene transcription activity. To date, little is known about the role of histone methylation modifications in adult muscle regeneration. However, a similar regulatory mechanism from myogenesis might regulate the availability and composition of chromatin modifying-complexes that promote muscle gene transcription in adult muscle regeneration. Likewise, we predict that histone methylation-modifying enzymes mediate dynamic modifications of histone tails that are vital to programming myogenic precursors (satellite cells) toward terminal differentiation in adult muscle regeneration.
The networks established by myogenic regulatory factors and chromatin-associated enzymatic complexes will provide an important paradigm to understand the epigenetic regulation of skeletal muscle development and regeneration, and help to decipher the molecular pathways that control the transition from skeletal muscle progenitors to fibers—a current challenge in regenerative medicine [147].
Moreover, the current approach of ChIP assays is suitable for chromatin from small cell numbers, but limited by the number cell samples. Thus, establishing chromatin state maps across genome (genome-wide changes in the epigenetic landscape) is an open avenue for investigation. While most of the findings outlined here are based on cell culture studies, in vivo models or animal experiments need to be explored. Moreover, chromatin plasticity and epigenetic changes during muscle differentiation have yet to be revealed.
Acknowledgements
We are grateful to members of our laboratories for critical reading of the manuscript and constructive discussion. This work was supported by National Science R&T Program (2013BAD20B00, 2011BAD28B01), the Hubei Province Technology Support Program (2014ABA025).
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.04.009.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Bharathy N., Ling B.M., Taneja R. Epigenetic regulation of skeletal muscle development and differentiation. Subcell. Biochem. 2013;61:139–150. doi: 10.1007/978-94-007-4525-4_7. [DOI] [PubMed] [Google Scholar]
- 2.Palacios D., Puri P.L. The epigenetic network regulating muscle development and regeneration. J. Cell. Physiol. 2006;207:1–11. doi: 10.1002/jcp.20489. [DOI] [PubMed] [Google Scholar]
- 3.Verrier L., Escaffit F., Chailleux C., Trouche D., Vandromme M. A new isoform of the histone demethylase JMJD2A/KDM4A is required for skeletal muscle differentiation. PLoS Genet. 2011;7:e1001390. doi: 10.1371/journal.pgen.1001390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckingham M., Rigby P.W. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell. 2014;28:225–238. doi: 10.1016/j.devcel.2013.12.020. [DOI] [PubMed] [Google Scholar]
- 5.Christ B., Ordahl C.P. Early stages of chick somite development. Anat. Embryol. 1995;191:381–396. doi: 10.1007/BF00304424. [DOI] [PubMed] [Google Scholar]
- 6.Cossu G., Tajbakhsh S., Buckingham M. How is myogenesis initiated in the embryo? Trends Genet. 1996;12:218–223. doi: 10.1016/0168-9525(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 7.Bismuth K., Relaix F. Genetic regulation of skeletal muscle development. Exp. Cell Res. 2010;316:3081–3086. doi: 10.1016/j.yexcr.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 8.Buckingham M. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 2001;11:440–448. doi: 10.1016/s0959-437x(00)00215-x. [DOI] [PubMed] [Google Scholar]
- 9.Buckingham M., Bajard L., Chang T., Daubas P., Hadchouel J., Meilhac S., Montarras D., Rocancourt D., Relaix F. The formation of skeletal muscle: from somite to limb. J. Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bober E., Franz T., Arnold H.H., Gruss P., Tremblay P. Pax-3 is required for the development of limb muscles: a possible role for the migration of dermomyotomal muscle progenitor cells. Development. 1994;120:603–612. doi: 10.1242/dev.120.3.603. [DOI] [PubMed] [Google Scholar]
- 11.Daston G., Lamar E., Olivier M., Goulding M. Pax-3 is necessary for migration but not differentiation of limb muscle precursors in the mouse. Development. 1996;122:1017–1027. doi: 10.1242/dev.122.3.1017. [DOI] [PubMed] [Google Scholar]
- 12.Tremblay P., Dietrich S., Mericskay M., Schubert F.R., Li Z., Paulin D. A crucial role for Pax3 in the development of the hypaxial musculature and the long-range migration of muscle precursors. Dev. Biol. 1998;203:49–61. doi: 10.1006/dbio.1998.9041. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Yair R., Kalcheim C. Lineage analysis of the avian dermomyotome sheet reveals the existence of single cells with both dermal and muscle progenitor fates. Development. 2005;132:689–701. doi: 10.1242/dev.01617. [DOI] [PubMed] [Google Scholar]
- 14.Relaix F., Rocancourt D., Mansouri A., Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 15.Bentzinger C.F., Wang Y.X., Rudnicki M.A. Building muscle: molecular regulation of myogenesis. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Megeney L.A., Rudnicki M.A. Determination versus differentiation and the MyoD family of transcription factors. Biochem. Cell Biol. 1995;73:723–732. doi: 10.1139/o95-080. [DOI] [PubMed] [Google Scholar]
- 17.Yusuf F., Brand-Saberi B. The eventful somite: patterning, fate determination and cell division in the somite. Anat. Embryol. 2006;211(Suppl. 1):S21–S30. doi: 10.1007/s00429-006-0119-8. [DOI] [PubMed] [Google Scholar]
- 18.Kiefer J.C., Hauschka S.D. Myf-5 is transiently expressed in nonmuscle mesoderm and exhibits dynamic regional changes within the presegmented mesoderm and somites I–IV. Dev. Biol. 2001;232:77–90. doi: 10.1006/dbio.2000.0114. [DOI] [PubMed] [Google Scholar]
- 19.Yokoyama S., Asahara H. The myogenic transcriptional network. Cell. Mol. Life Sci. 2011;68:1843–1849. doi: 10.1007/s00018-011-0629-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berkes C.A., Tapscott S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Nabeshima Y., Hanaoka K., Hayasaka M., Esumi E., Li S., Nonaka I. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- 22.Hasty P., Bradley A., Morris J.H., Edmondson D.G., Venuti J.M., Olson E.N., Klein W.H. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- 23.Zhu Z., Miller J.B. MRF4 can substitute for myogenin during early stages of myogenesis. Dev. Dyn. 1997;209:233–241. doi: 10.1002/(SICI)1097-0177(199706)209:2<233::AID-AJA9>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 24.Sumariwalla V.M., Klein W.H. Similar myogenic functions for myogenin and MRF4 but not MyoD in differentiated murine embryonic stem cells. Genesis. 2001;30:239–249. doi: 10.1002/gene.1070. [DOI] [PubMed] [Google Scholar]
- 25.Molkentin J.D., Black B.L., Martin J.F., Olson E.N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 26.Black B.L., Olson E.N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 27.Potthoff M.J., Olson E.N. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 28.Kawakami K., Sato S., Ozaki H., Ikeda K. Six family genes--structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 29.Grifone R., Demignon J., Giordani J., Niro C., Souil E., Bertin F., Laclef C., Xu P.X., Maire P. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev. Biol. 2007;302:602–616. doi: 10.1016/j.ydbio.2006.08.059. [DOI] [PubMed] [Google Scholar]
- 30.Grifone R., Demignon J., Houbron C., Souil E., Niro C., Seller M.J., Hamard G., Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132:2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- 31.Geetha-Loganathan P., Nimmagadda S., Scaal M., Huang R., Christ B. Wnt signaling in somite development. Ann. Anat. 2008;190:208–222. doi: 10.1016/j.aanat.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Parr B.A., Shea M.J., Vassileva G., McMahon A.P. Mouse Wnt genes exhibit discrete domains of expression in the early embryonic CNS and limb buds. Development. 1993;119:247–261. doi: 10.1242/dev.119.1.247. [DOI] [PubMed] [Google Scholar]
- 33.Ikeya M., Takada S. Wnt signaling from the dorsal neural tube is required for the formation of the medial dermomyotome. Development. 1998;125:4969–4976. doi: 10.1242/dev.125.24.4969. [DOI] [PubMed] [Google Scholar]
- 34.Tajbakhsh S., Borello U., Vivarelli E., Kelly R., Papkoff J., Duprez D., Buckingham M., Cossu G. Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development. 1998;125:4155–4162. doi: 10.1242/dev.125.21.4155. [DOI] [PubMed] [Google Scholar]
- 35.Borycki A., Brown A.M., Emerson C.P., Jr. Shh and Wnt signaling pathways converge to control Gli gene activation in avian somites. Development. 2000;127:2075–2087. doi: 10.1242/dev.127.10.2075. [DOI] [PubMed] [Google Scholar]
- 36.Amthor H., Christ B., Patel K. A molecular mechanism enabling continuous embryonic muscle growth – a balance between proliferation and differentiation. Development. 1999;126:1041–1053. doi: 10.1242/dev.126.5.1041. [DOI] [PubMed] [Google Scholar]
- 37.Pourquie O., Fan C.M., Coltey M., Hirsinger E., Watanabe Y., Breant C., Francis-West P., Brickell P., Tessier-Lavigne M., Le Douarin N.M. Lateral and axial signals involved in avian somite patterning: a role for BMP4. Cell. 1996;84:461–471. doi: 10.1016/s0092-8674(00)81291-x. [DOI] [PubMed] [Google Scholar]
- 38.Patterson S.E., Bird N.C., Devoto S.H. BMP regulation of myogenesis in zebrafish. Dev. Dyn. 2010;239:806–817. doi: 10.1002/dvdy.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmerman L.B., De Jesus-Escobar J.M., Harland R.M. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 40.Hirsinger E., Duprez D., Jouve C., Malapert P., Cooke J., Pourquie O. Noggin acts downstream of Wnt and Sonic Hedgehog to antagonize BMP4 in avian somite patterning. Development. 1997;124:4605–4614. doi: 10.1242/dev.124.22.4605. [DOI] [PubMed] [Google Scholar]
- 41.Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 42.Mayeuf-Louchart A., Lagha M., Danckaert A., Rocancourt D., Relaix F., Vincent S.D., Buckingham M. Notch regulation of myogenic versus endothelial fates of cells that migrate from the somite to the limb. Proc. Natl. Acad. Sci. USA. 2014;111:8844–8849. doi: 10.1073/pnas.1407606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuroda K., Tani S., Tamura K., Minoguchi S., Kurooka H., Honjo T. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J. Biol. Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 44.Sasai Y., Kageyama R., Tagawa Y., Shigemoto R., Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 45.Wilson-Rawls J., Molkentin J.D., Black B.L., Olson E.N. Activated notch inhibits myogenic activity of the MADS-Box transcription factor myocyte enhancer factor 2C. Mol. Cell Biol. 1999;19:2853–2862. doi: 10.1128/mcb.19.4.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keren A., Tamir Y., Bengal E. The p38 MAPK signaling pathway: a major regulator of skeletal muscle development. Mol. Cell. Endocrinol. 2006;252:224–230. doi: 10.1016/j.mce.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 47.Zetser A., Gredinger E., Bengal E. p38 Mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J. Biol. Chem. 1999;274:5193–5200. doi: 10.1074/jbc.274.8.5193. [DOI] [PubMed] [Google Scholar]
- 48.Wu Z., Woodring P.J., Bhakta K.S., Tamura K., Wen F., Feramisco J.R., Karin M., Wang J.Y., Puri P.L. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol. Cell. Biol. 2000;20:3951–3964. doi: 10.1128/mcb.20.11.3951-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Relaix F., Montarras D., Zaffran S., Gayraud-Morel B., Rocancourt D., Tajbakhsh S., Mansouri A., Cumano A., Buckingham M. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biressi S., Bjornson C.R., Carlig P.M., Nishijo K., Keller C., Rando T.A. Myf5 expression during fetal myogenesis defines the developmental progenitors of adult satellite cells. Dev. Biol. 2013;379:195–207. doi: 10.1016/j.ydbio.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuang S., Kuroda K., Le Grand F., Rudnicki M.A. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sousa-Victor P., Munoz-Canoves P., Perdiguero E. Regulation of skeletal muscle stem cells through epigenetic mechanisms. Toxicol. Mech. Methods. 2011;21:334–342. doi: 10.3109/15376516.2011.557873. [DOI] [PubMed] [Google Scholar]
- 53.Oustanina S., Hause G., Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collins C.A., Gnocchi V.F., White R.B., Boldrin L., Perez-Ruiz A., Relaix F., Morgan J.E., Zammit P.S. Integrated functions of Pax3 and Pax7 in the regulation of proliferation, cell size and myogenic differentiation. PLoS ONE. 2009;4:e4475. doi: 10.1371/journal.pone.0004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lepper C., Conway S.J., Fan C.M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gunther S., Kim J., Kostin S., Lepper C., Fan C.M., Braun T. Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell. 2013;13:590–601. doi: 10.1016/j.stem.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Maltzahn J., Jones A.E., Parks R.J., Rudnicki M.A. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl. Acad. Sci. USA. 2013;110:16474–16479. doi: 10.1073/pnas.1307680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gayraud-Morel B., Chretien F., Flamant P., Gomes D., Zammit P.S., Tajbakhsh S. A role for the myogenic determination gene Myf5 in adult regenerative myogenesis. Dev. Biol. 2007;312:13–28. doi: 10.1016/j.ydbio.2007.08.059. [DOI] [PubMed] [Google Scholar]
- 59.Beauchamp J.R., Heslop L., Yu D.S., Tajbakhsh S., Kelly R.G., Wernig A., Buckingham M.E., Partridge T.A., Zammit P.S. Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J. Cell Biol. 2000;151:1221–1234. doi: 10.1083/jcb.151.6.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yablonka-Reuveni Z., Rivera A.J. Temporal expression of regulatory and structural muscle proteins during myogenesis of satellite cells on isolated adult rat fibers. Dev. Biol. 1994;164:588–603. doi: 10.1006/dbio.1994.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olguin H.C., Olwin B.B. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Dev. Biol. 2004;275:375–388. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Polesskaya A., Seale P., Rudnicki M.A. Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell. 2003;113:841–852. doi: 10.1016/s0092-8674(03)00437-9. [DOI] [PubMed] [Google Scholar]
- 63.Otto A., Schmidt C., Luke G., Allen S., Valasek P., Muntoni F., Lawrence-Watt D., Patel K. Canonical Wnt signalling induces satellite-cell proliferation during adult skeletal muscle regeneration. J. Cell Sci. 2008;121:2939–2950. doi: 10.1242/jcs.026534. [DOI] [PubMed] [Google Scholar]
- 64.Fujimaki S., Hidaka R., Asashima M., Takemasa T., Kuwabara T. Wnt protein-mediated satellite cell conversion in adult and aged mice following voluntary wheel running. J. Biol. Chem. 2014;289:7399–7412. doi: 10.1074/jbc.M113.539247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Segales J., Perdiguero E., Munoz-Canoves P. Epigenetic control of adult skeletal muscle stem cell functions. FEBS J. 2015;282:1571–1588. doi: 10.1111/febs.13065. [DOI] [PubMed] [Google Scholar]
- 66.Conboy I.M., Rando T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 2002;3:397–409. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 67.Sun D., Li H., Zolkiewska A. The role of Delta-like 1 shedding in muscle cell self-renewal and differentiation. J. Cell Sci. 2008;121:3815–3823. doi: 10.1242/jcs.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang G., Pradhan S. Mammalian epigenetic mechanisms. IUBMB Life. 2014;66:240–256. doi: 10.1002/iub.1264. [DOI] [PubMed] [Google Scholar]
- 69.Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 70.Won J., Kim T.K. Histone modifications and transcription factor binding on chromatin ChIP-PCR assays. Methods Mol. Biol. 2006;325:273–283. doi: 10.1385/1-59745-005-7:273. [DOI] [PubMed] [Google Scholar]
- 71.Collas P. Epigenetic states in stem cells. Biochim. Biophys. Acta. 2009;1790:900–905. doi: 10.1016/j.bbagen.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 72.Pekowska A., Benoukraf T., Zacarias-Cabeza J., Belhocine M., Koch F., Holota H., Imbert J., Andrau J.C., Ferrier P., Spicuglia S. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim D.H., Tang Z., Shimada M., Fierz B., Houck-Loomis B., Bar-Dagen M., Lee S., Lee S.K., Muir T.W., Roeder R.G., Lee J.W. Histone H3K27 trimethylation inhibits H3 binding and function of SET1-like H3K4 methyltransferase complexes. Mol. Cell. Biol. 2013;33:4936–4946. doi: 10.1128/MCB.00601-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 75.Lindeman L.C., Winata C.L., Aanes H., Mathavan S., Alestrom P., Collas P. Chromatin states of developmentally-regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int. J. Dev. Biol. 2010;54:803–813. doi: 10.1387/ijdb.103081ll. [DOI] [PubMed] [Google Scholar]
- 76.Vastenhouw N.L., Schier A.F. Bivalent histone modifications in early embryogenesis. Curr. Opin. Cell Biol. 2012;24:374–386. doi: 10.1016/j.ceb.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Byrne K., McWilliam S., Vuocolo T., Gondro C., Cockett N.E., Tellam R.L. Genomic architecture of histone 3 lysine 27 trimethylation during late ovine skeletal muscle development. Anim. Genet. 2014;45:427–438. doi: 10.1111/age.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hammoud S.S., Nix D.A., Zhang H., Purwar J., Carrell D.T., Cairns B.R. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lienert F., Mohn F., Tiwari V.K., Baubec T., Roloff T.C., Gaidatzis D., Stadler M.B., Schubeler D. Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genet. 2011;7:e1002090. doi: 10.1371/journal.pgen.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stancheva I. Revisiting heterochromatin in embryonic stem cells. PLoS Genet. 2011;7:e1002093. doi: 10.1371/journal.pgen.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shpargel K.B., Starmer J., Yee D., Pohlers M., Magnuson T. KDM6 demethylase independent loss of histone H3 lysine 27 trimethylation during early embryonic development. PLoS Genet. 2014;10:e1004507. doi: 10.1371/journal.pgen.1004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hublitz P., Albert M., Peters A.H. Mechanisms of transcriptional repression by histone lysine methylation. Int. J. Dev. Biol. 2009;53:335–354. doi: 10.1387/ijdb.082717ph. [DOI] [PubMed] [Google Scholar]
- 84.Orford K., Kharchenko P., Lai W., Dao M.C., Worhunsky D.J., Ferro A., Janzen V., Park P.J., Scadden D.T. Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Dev. Cell. 2008;14:798–809. doi: 10.1016/j.devcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tao Y., Neppl R.L., Huang Z.P., Chen J., Tang R.H., Cao R., Zhang Y., Jin S.W., Wang D.Z. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J. Cell Biol. 2011;194:551–565. doi: 10.1083/jcb.201010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu L., Cheung T.H., Charville G.W., Hurgo B.M., Leavitt T., Shih J., Brunet A., Rando T.A. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yun M., Wu J., Workman J.L., Li B. Readers of histone modifications. Cell Res. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vermeulen M., Eberl H.C., Matarese F., Marks H., Denissov S., Butter F., Lee K.K., Olsen J.V., Hyman A.A., Stunnenberg H.G., Mann M. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 89.Asp P., Blum R., Vethantham V., Parisi F., Micsinai M., Cheng J., Bowman C., Kluger Y., Dynlacht B.D. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc. Natl. Acad. Sci. USA. 2011;108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Margueron R., Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Caretti G., Di Padova M., Micales B., Lyons G.E., Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bracken A.P., Dietrich N., Pasini D., Hansen K.H., Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Levine S.S., King I.F., Kingston R.E. Division of labor in polycomb group repression. Trends Biochem. Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 94.Ringrose L., Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 95.Dou Y., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 96.Steward M.M., Lee J.S., O’Donovan A., Wyatt M., Bernstein B.E., Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat. Struct. Mol. Biol. 2006;13:852–854. doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- 97.Rampalli S., Li L., Mak E., Ge K., Brand M., Tapscott S.J., Dilworth F.J. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat. Struct. Mol. Biol. 2007;14:1150–1156. doi: 10.1038/nsmb1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sebastian S., Sreenivas P., Sambasivan R., Cheedipudi S., Kandalla P., Pavlath G.K., Dhawan J. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc. Natl. Acad. Sci. USA. 2009;106:4719–4724. doi: 10.1073/pnas.0807136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang H., Cao R., Xia L., Erdjument-Bromage H., Borchers C., Tempst P., Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 100.Fritsch L., Robin P., Mathieu J.R., Souidi M., Hinaux H., Rougeulle C., Harel-Bellan A., Ameyar-Zazoua M., Ait-Si-Ali S. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol. Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 101.Esteve P.O., Patnaik D., Chin H.G., Benner J., Teitell M.A., Pradhan S. Functional analysis of the N- and C-terminus of mammalian G9a histone H3 methyltransferase. Nucleic Acids Res. 2005;33:3211–3223. doi: 10.1093/nar/gki635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lachner M., O’Sullivan R.J., Jenuwein T. An epigenetic road map for histone lysine methylation. J. Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- 103.Fuks F., Hurd P.J., Deplus R., Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mal A.K. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 2006;25:3323–3334. doi: 10.1038/sj.emboj.7601229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ait-Si-Ali S., Guasconi V., Fritsch L., Yahi H., Sekhri R., Naguibneva I., Robin P., Cabon F., Polesskaya A., Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tachibana M., Sugimoto K., Fukushima T., Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 107.Ling B.M., Bharathy N., Chung T.K., Kok W.K., Li S., Tan Y.H., Rao V.K., Gopinadhan S., Sartorelli V., Walsh M.J., Taneja R. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc. Natl. Acad. Sci. USA. 2012;109:841–846. doi: 10.1073/pnas.1111628109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yokochi T., Poduch K., Ryba T., Lu J., Hiratani I., Tachibana M., Shinkai Y., Gilbert D.M. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc. Natl. Acad. Sci. USA. 2009;106:19363–19368. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ling B.M., Gopinadhan S., Kok W.K., Shankar S.R., Gopal P., Bharathy N., Wang Y., Taneja R. G9a mediates Sharp-1-dependent inhibition of skeletal muscle differentiation. Mol. Biol. Cell. 2012;23:4778–4785. doi: 10.1091/mbc.E12-04-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kooistra S.M., Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 111.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 112.Metzger E., Wissmann M., Yin N., Muller J.M., Schneider R., Peters A.H., Gunther T., Buettner R., Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 113.El Mansouri F.E., Nebbaki S.S., Kapoor M., Afif H., Martel-Pelletier J., Pelletier J.P., Benderdour M., Fahmi H. Lysine-specific demethylase 1-mediated demethylation of histone H3 lysine 9 contributes to interleukin 1beta-induced microsomal prostaglandin E synthase 1 expression in human osteoarthritic chondrocytes. Arthritis Res. Ther. 2014;16:R113. doi: 10.1186/ar4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Laurent B., Ruitu L., Murn J., Hempel K., Ferrao R., Xiang Y., Liu S., Garcia B.A., Wu H., Wu F., Steen H., Shi Y. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol. Cell. 2015;57:957–970. doi: 10.1016/j.molcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Choi J., Jang H., Kim H., Kim S.T., Cho E.J., Youn H.D. Histone demethylase LSD1 is required to induce skeletal muscle differentiation by regulating myogenic factors. Biochem. Biophys. Res. Commun. 2010;401:327–332. doi: 10.1016/j.bbrc.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 116.Okada Y., Scott G., Ray M.K., Mishina Y., Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 117.Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 118.Jung E.S., Sim Y.J., Jeong H.S., Kim S.J., Yun Y.J., Song J.H., Jeon S.H., Choe C., Park K.T., Kim C.H., Kim K.S. Jmjd2C increases MyoD transcriptional activity through inhibiting G9a-dependent MyoD degradation. Biochim. Biophys. Acta. 2015;1849:1081–1094. doi: 10.1016/j.bbagrm.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 119.Hong S., Cho Y.W., Yu L.R., Yu H., Veenstra T.D., Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 121.Lan F., Bayliss P.E., Rinn J.L., Whetstine J.R., Wang J.K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E., Roberts T.M., Chang H.Y., Shi Y. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 122.Seenundun S., Rampalli S., Liu Q.C., Aziz A., Palii C., Hong S., Blais A., Brand M., Ge K., Dilworth F.J. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J. 2010;29:1401–1411. doi: 10.1038/emboj.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang A.H., Zare H., Mousavi K., Wang C., Moravec C.E., Sirotkin H.I., Ge K., Gutierrez-Cruz G., Sartorelli V. The histone chaperone Spt6 coordinates histone H3K27 demethylation and myogenesis. EMBO J. 2013;32:1075–1086. doi: 10.1038/emboj.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Juan A.H., Derfoul A., Feng X., Ryall J.G., Dell’Orso S., Pasut A., Zare H., Simone J.M., Rudnicki M.A., Sartorelli V. Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells. Genes Dev. 2011;25:789–794. doi: 10.1101/gad.2027911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dilworth F.J., Blais A. Epigenetic regulation of satellite cell activation during muscle regeneration. Stem Cell Res. Ther. 2011;2:18. doi: 10.1186/scrt59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Palacios D., Mozzetta C., Consalvi S., Caretti G., Saccone V., Proserpio V., Marquez V.E., Valente S., Mai A., Forcales S.V., Sartorelli V., Puri P.L. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell. 2010;7:455–469. doi: 10.1016/j.stem.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McKinnell I.W., Ishibashi J., Le Grand F., Punch V.G., Addicks G.C., Greenblatt J.F., Dilworth F.J., Rudnicki M.A. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell Biol. 2008;10:77–84. doi: 10.1038/ncb1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Diao Y., Guo X., Li Y., Sun K., Lu L., Jiang L., Fu X., Zhu H., Sun H., Wang H., Wu Z. Pax3/7BP is a Pax7-and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. Cell Stem Cell. 2012;11:231–241. doi: 10.1016/j.stem.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 130.Kawabe Y., Wang Y.X., McKinnell I.W., Bedford M.T., Rudnicki M.A. Carm1 regulates Pax7 transcriptional activity through MLL1/2 recruitment during asymmetric satellite stem cell divisions. Cell Stem Cell. 2012;11:333–345. doi: 10.1016/j.stem.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gillespie M.A., Le Grand F., Scime A., Kuang S., von Maltzahn J., Seale V., Cuenda A., Ranish J.A., Rudnicki M.A. p38-{gamma}-dependent gene silencing restricts entry into the myogenic differentiation program. J. Cell Biol. 2009;187:991–1005. doi: 10.1083/jcb.200907037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bedford M.T. Arginine methylation at a glance. J. Cell Sci. 2007;120:4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 133.Chen S.L., Loffler K.A., Chen D., Stallcup M.R., Muscat G.E. The coactivator-associated arginine methyltransferase is necessary for muscle differentiation: CARM1 coactivates myocyte enhancer factor-2. J. Biol. Chem. 2002;277:4324–4333. doi: 10.1074/jbc.M109835200. [DOI] [PubMed] [Google Scholar]
- 134.Duina A.A. Histone chaperones Spt6 and FACT: similarities and differences in modes of action at transcribed genes. Genet. Res. Int. 2011;2011:625210. doi: 10.4061/2011/625210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hartzog G.A., Wada T., Handa H., Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kato H., Okazaki K., Iida T., Nakayama J., Murakami Y., Urano T. Spt6 prevents transcription-coupled loss of posttranslationally modified histone H3. Sci. Rep. 2013;3:2186. doi: 10.1038/srep02186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bortvin A., Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 138.Kok F.O., Oster E., Mentzer L., Hsieh J.C., Henry C.A., Sirotkin H.I. The role of the SPT6 chromatin remodeling factor in zebrafish embryogenesis. Dev. Biol. 2007;307:214–226. doi: 10.1016/j.ydbio.2007.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Elsasser S.J., D’Arcy S. Towards a mechanism for histone chaperones. Biochim. Biophys. Acta. 1819;2013:211–221. doi: 10.1016/j.bbagrm.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lamour V., Lecluse Y., Desmaze C., Spector M., Bodescot M., Aurias A., Osley M.A., Lipinski M. A human homolog of the S. cerevisiae HIR1 and HIR2 transcriptional repressors cloned from the DiGeorge syndrome critical region. Hum. Mol. Genet. 1995;4:791–799. doi: 10.1093/hmg/4.5.791. [DOI] [PubMed] [Google Scholar]
- 141.Lorain S., Demczuk S., Lamour V., Toth S., Aurias A., Roe B.A., Lipinski M. Structural Organization of the WD repeat protein-encoding gene HIRA in the DiGeorge syndrome critical region of human chromosome 22. Genome Res. 1996;6:43–50. doi: 10.1101/gr.6.1.43. [DOI] [PubMed] [Google Scholar]
- 142.Sherwood P.W., Tsang S.V., Osley M.A. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:28–38. doi: 10.1128/mcb.13.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang J.H., Choi J.H., Jang H., Park J.Y., Han J.W., Youn H.D., Cho E.J. Histone chaperones cooperate to mediate Mef2-targeted transcriptional regulation during skeletal myogenesis. Biochem. Biophys. Res. Commun. 2011;407:541–547. doi: 10.1016/j.bbrc.2011.03.055. [DOI] [PubMed] [Google Scholar]
- 144.Blum R., Vethantham V., Bowman C., Rudnicki M., Dynlacht B.D. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev. 2012;26:2763–2779. doi: 10.1101/gad.200113.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Blum R., Dynlacht B.D. The role of MyoD1 and histone modifications in the activation of muscle enhancers. Epigenetics. 2013;8:778–784. doi: 10.4161/epi.25441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Choi J., Jang H., Kim H., Lee J.H., Kim S.T., Cho E.J., Youn H.D. Modulation of lysine methylation in myocyte enhancer factor 2 during skeletal muscle cell differentiation. Nucleic Acids Res. 2014;42:224–234. doi: 10.1093/nar/gkt873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Saccone V., Puri P.L. Epigenetic regulation of skeletal myogenesis. Organogenesis. 2010;6:48–53. doi: 10.4161/org.6.1.11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material