Abstract
The importance of H2S in biology and medicine has been widely recognized in recent years, and protein S-sulfhydration is proposed to mediate the direct actions of H2S bioactivity in the body. Thioredoxin 1 (Trx1) is an important reducing enzyme that cleaves disulfides in proteins and acts as an S-denitrosylase. The regulation of Trx1 on protein S-sulfhydration is unclear. Here we showed that Trx1 facilitates protein S-desulfhydration. Overexpression of Trx1 attenuated the basal level and H2S-induced protein S-sulfhydration by direct interaction with S-sulfhydrated proteins, i.e., glyceraldehyde 3-phosphate dehydrogenase and pyruvate carboxylase. In contrast, knockdown of Trx1 mRNA expression by short interfering RNA or blockage of Trx1 redox activity with PX12 or 2,4-dinitrochlorobenzene enhanced protein S-sulfhydration. Mutation of cysteine-32 but not cysteine-35 in the Trp–Cys32–Gly–Pro–Cys35 motif eliminated the binding of Trx1 with S-sulfhydrated proteins and abolished the S-desulfhydrating effect of Trx1. All these data suggest that Trx1 acts as an S-desulfhydrase.
Abbreviations: DNCB, 2,4-dinitrochlorobenzene; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GSNO, S-nitrosoglutathione; PC, pyruvate carboxylase; siRNA, short interfering RNA; Trx1, Thioredoxin 1; TrxR, thioredoxin reductase; Txnip, Trx-interacting protein
Keywords: Hydrogen sulfide, S-sulfhydration, S-desulfhydration, Thioredoxin 1
Highlights
-
•
Thioredoxin 1 facilitates protein S-desulfhydration.
-
•
Thioredoxin 1 directly interacts with S-sulfhydrated proteins.
-
•
Cysteine-32 in Thioredoxin 1 is required for protein S-desulfhydration.
1. Introduction
Hydrogen sulfide (H2S) can S-sulfhydrate proteins by yielding a hydropersulfide moiety (–SSH) in the active cysteine residues [1], [2], [3]. It is predicted that S-sulfhydration can alter protein conformation and the final function and activity of target proteins [2], [4]. This particular redox modification of cysteine by S-sulfhydration has been achieved using proteomics techniques by coupling a specific enrichment strategy (biotin-switch assay and/or tag-switch assay) with high-throughput mass spectrometry analysis [1], [5], [6]. So far, there have been a dozen proteins observed to be modified by H2S through S-sulfhydration. KATP channel is a direct target for H2S in regulating vasodilation and cardioprotection [7]. In fact, H2S induces S-sulfhydration of Kir6.1 subunit of KATP channels, and dithiothreitol reverses this S-sulfhydration [4]. H2S-induced S-sulfhydration of phosphatase PTP1B alters endoplasmic reticulum stress response [8]. H2S S-sulfhydrates p65 and mediates the anti-apoptotic effect of NF-κB [6]. H2S modulates cellular redox signaling via direct S-sulfhydration of Keap1/Nrf2 system, p66, and electrophiles, contributing to the beneficial effects of H2S on cellular aging and heart failure [9], [10], [11]. S-sulfhydration of MEK1 leads to PARP1 activation and induces DNA damage repair in endothelia cells [12]. More recently, we observed that S-sulfhydration of IRF-1 and pyruvate carboxylase induce mitochondrial DNA replication and gluconeogenesis [13], [14].
Like protein phosphorylation and S-nitrosylation, S-sulfhydration is proposed to mediate or modulate transduction of myriad cellular signals [1], [2], [3]. While decades of research have established the generality and broad physiological importance of protein phosphorylation and S-nitrosylation, our current understanding of the fundamental biology and chemistry of S-sulfhydration as a protein signaling modality is still in its infancy [15], [16]. Is H2S S-sulfhydration of cysteine enzyme-regulated or a spontaneous process? How is the biochemical stability of S-sulfhydrated proteins? Which critical factors are involved and what conditions are required for the formation of S-sulfhydration? Is there any possibility for desulfhydration and/or transsulfhydration and how are they regulated? Little is known about the nature of or even necessity for enzymatic mechanisms that may directly add or remove SH groups from cysteine thiols.
In the present study, we demonstrated that thioredoxin 1 (Trx1), an oxidoreductase found in both prokaryotes and eukaryotes, is essential for protein S-desulfhydration. Overexpression of Trx1 attenuated the basal level and H2S-induced protein S-sulfhydration by direct interaction with S-sulfhydrated proteins, i.e., glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and pyruvate carboxylase (PC). In contrast, blockage of Trx1 activity strengthened protein S-sulfhydration. Furthermore, mutation of cyseine-32 in Trx1 eliminated the binding of Trx1 with S-sulfhydated proteins and abolished the S-desulfhydrating effect of Trx1 on both GAPDH and PC.
2. Materials and methods
2.1. Cell culture and transfection
HepG2 (a human hepatocellular liver carcinoma cell line) and HEK293 cells were purchased from American Type Culture Collection (Manassas, VA) and maintained in Dulbecco's Modified Eagle's medium (Sigma, Oakville, ON) supplemented with 10% fetal bovine serum (Clontech, Mountain View, CA) and 1% penicillin–streptomycin solution (Sigma). For cell transfection, wild-type Trx1 or mutant Trx1 was transfected into HEK293 or HepG2 cells using Lipofectamine™ 2000 reagent as described by the manufacturer's protocol (Invitrogen, Burlington, ON). Pre-designed Trx1-targeted short interfering RNA (Trx1-siRNA) was purchased from Ambion (Austin, TX). Negative siRNA (Neg-siRNA), a 21-nucleotide RNA duplex with no known sequence homology with all the genes, was also from Ambion. Transfection of siRNA into HEK293 cells was achieved using the siPORTTM lipid transfection agent from Ambion as we previously described [10].
2.2. Construction of Trx1 mutants
The plasmid Myc-TRX was purchased from Addgene (MA, USA) [17]. Single mutation of cyteine-32 (Trx1-C32) and cysteine-35 (Trx1-C35) to serine were conducted using the Quick Change Site-Directed Mutagenesis kit (Stratagene, CA, USA) [12]. The oligonucleotides using for mutagenesis were 5′- CTTCTCAGCCACGTGGAGTGGGCCTTGCAAAATG-3′ (forward) and 5′-CATTTTGCAAGG-CCCACTCCACGTGGCTGAGAAG-3′ (reverse) for cysteine-32, and 5′-CGTGGTGTGGGCC-TAGCAAAATGATCAAGC-3′ (forward) and 5′-GCTTGATCATTTTGCTAGGCCCACACCACG-3′ for cysteine-35. The correct mutant was confirmed by DNA sequencing at the Paleo-DNA laboratory in Lakehead University, ON, Canada.
2.3. Western immunoblotting
The cells were harvested and lysed in a cell lysis buffer including protease inhibitor cocktail (Sigma). Extracts were separated by centrifugation at 14,000g for 15 min at 4 °C. SDS-PAGE and Western blot analysis were performed. Briefly, equal amount of proteins were boiled in 1× SDS sample buffer and run in a 10% SDS-PAGE gel, and transferred onto pure nitrocellulose blotting membranes (Pall Corporation). The membrane was blocked with 3% nonfat dry milk solution in PBS at room temperature for 2 h and rinsed three times with PBS before incubating with primary antibodies. After that, the immunoblots were probed with anti-biotin (1:500 dilution; Santa Cruz Biotechnology), anti-Trx1 (1:500 dilution; Santa Cruz Biotechnology), anti-PC (1:500 dilution; Santa Cruz Biotechnology) and horseradish peroxidase (HRP)-conjugated rabbit anti-mouse secondary antibody (1:10,000 dilution; Sigma). Anti-GAPDH primary antibody (1:10,000 dilution; Sigma) and HRP-conjugated rabbit anti-mouse secondary antibody (1:5,000 dilution; Sigma) were employed to normalize protein expression after the membranes were stripped by incubating in a buffer containing 100 mM β-mercaptoethanol, 2% SDS, and 62.5 mM Tris–HCl (pH 6.8). The blots were developed using chemiluminescence (ECL Western Blotting Detection Reagents, Amersham Biosciences).
2.4. Biotin switch assay of S-sulfhydration
Biotin switch assay was carried-out as described previously with some modifications [1], [10]. Briefly, the cells were homogenized in HEN buffer (250 mM HEPES (pH 7.7), 1 mM EDTA, and 0.1 mM Neocuproine) supplemented with 100 μM deferoxamine and centrifuged at 13,000×g for 30 min at 4 °C. The lysates were added to blocking buffer (HEN buffer adjust to 2.5% SDS and 20 mM methyl methanethiosulfonate (MMTS)) at 50 °C for 20 min with frequent vortexing. The MMTS was then removed by acetone and the proteins were precipitated at −20 °C for 20 min. Proteins were resuspended in HENS buffer (HEN buffer containing 1% SDS) and 4 mM biotin-N-[6-(biotinamido) hexyl]-3′-(2′-pyridyldithio) propinamide (HPDP) in DMSO without ascorbic acid. After incubation for 2 h at 25 °C, biotinylated proteins were purified by streptavidin–agarose beads, which were then washed with HENS buffer. The biotinylated proteins were eluted by SDS-PAGE sample buffer and subjected to Western blotting analysis with antibody against biotin, GAPDH or PC. For quantitation of protein S-sulfhydration, samples were run on blots alongside the same amount of lysates (total protein) without processing biotin switch assay and then subjected to western blotting by using the same antibody. The ratio of S-sulfhydrated protein signal to total protein signal was then densitometrically analyzed using the software Image J.
2.5. Co-Immunoprecipitation (Co-IP)
The cells were lysed in IP lysis/wash buffer (25 mM Tris, 0.15 M NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, pH 7.4) including protease inhibitor cocktail (Sigma) [10]. The proteins (500 µg) were then incubated with 5 μg anti-PC antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or anti-GAPDH (Santa Cruz Biotechnology) for overnight at 4 °C. Protein A/G agarose was incubated with lysates for 1 h with gentle end-over-end mixing. Agarose resin was washed three times with IP lysis/wash buffer. The resin was then washed one time with saline solution (0.15 M NaCl). Immune complex was eluted with 2× SDS PAGE sample buffer (62.5 mM Tris–Cl, pH 6.8, 2% SDS, 10% glycerol, 50 mM DTT, and 0.01% bromophenol blue) at 100 °C for 5 min. The samples were subjected to Western immunoblotting analysis.
2.6. Statistical analysis
Data were presented as means±SEM, representing at least three independent experiments. Statistical comparisons were made using Excel 2007 (Microsoft, Redmond, WA) with Student's t-test to evaluate the difference between two groups, and the difference between multiple groups were analyzed by using SigmaPlot 12.0 (Systat Software Inc. San Jose, CA) with ANOVA and post-hoc Tukey test. Significance level was set at p<0.05.
3. Results
3.1. Trx1 suppresses protein S-sulfhydration
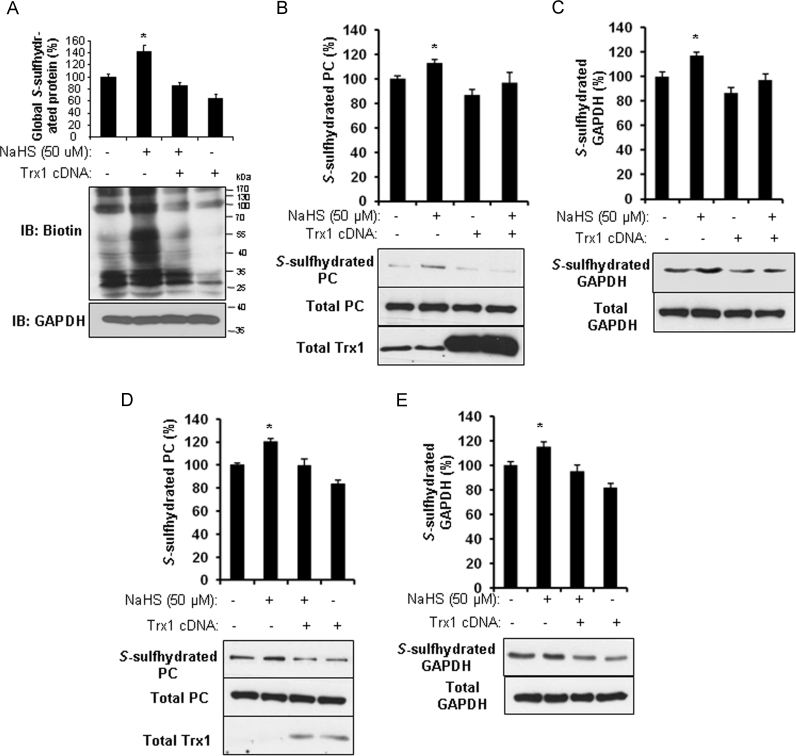
Protein S-sulfhydration can be detected by the modified biotin switch assay [1]. Following the procedures descried in the Methods, the S-sulfhydrated thiols were labeled with biotin–HPDP and then detected by western blotting with the antibody against either biotin or specific protein. We first detected the basal labeling of global proteins using an antibody against biotin, and the signaling became even stronger after treatment with 50 µM NaHS (a well-known H2S donor) for 2 hours in HEK293 cells. We further found that Trx1 overexpression in HEK293 cells markedly decreases H2S-induced protein S-sulfhydration (Fig. 1A). Probing with antibodies to specific proteins, such as PC (Fig. 1B) and GAPDH (Fig. 1C), it clearly showed that both GAPDH and PC proteins were basically S-sulfhydrated and further strengthened by exogenously applied NaHS, while transfection of HEK293 cells with Trx1 cDNA significantly reversed the basal and H2S-initiated S-sulfhydration of PC and GAPDH, suggesting Trx1 acts as an S-desulfhydrase. Identical results were further validated in another cell line, HepG2 cells (Fig. 1D and E). Trx1 cDNA alone slightly inhibited the basal level of protein S-sulfhydration.
Fig. 1.
Trx1 acts as an S-desulfhydrase. A, B, and C, Trx1 overexpression inhibited the S-sulfhydration level of global proteins, PC and GAPDH in HEK293 cells. HEK293 cells were transfected with Trx1 cDNA for 24 h, and then treated with 50 µM NaHS for 2 h. After that, the cells were collected for biotin switch assay using antibody against biotin (A), PC (B), and GAPDH (C). *p<0.05 versus all other groups. n=4. D and E, Trx1 overexpression inhibited PC and GAPDH S-sulfhydration in HepG2 cells. HepG2 cells were transfected with Trx1 cDNA for 24 h, and then treated with 50 µM NaHS for 2 h. After that, the cells were collected for biotin switch assay using antibody against PC (D) and GAPDH (E). *p<0.05 versus all other groups. n=4.
3.2. Inhibition of Trx1 activity stimulates protein S-sulfhydration
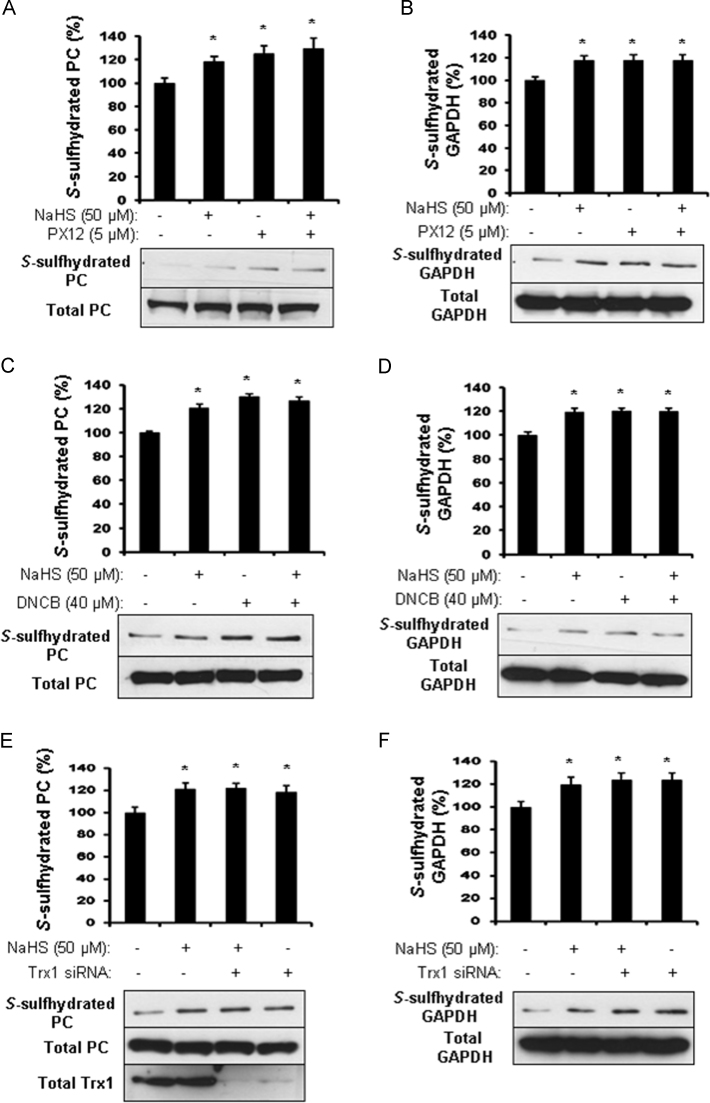
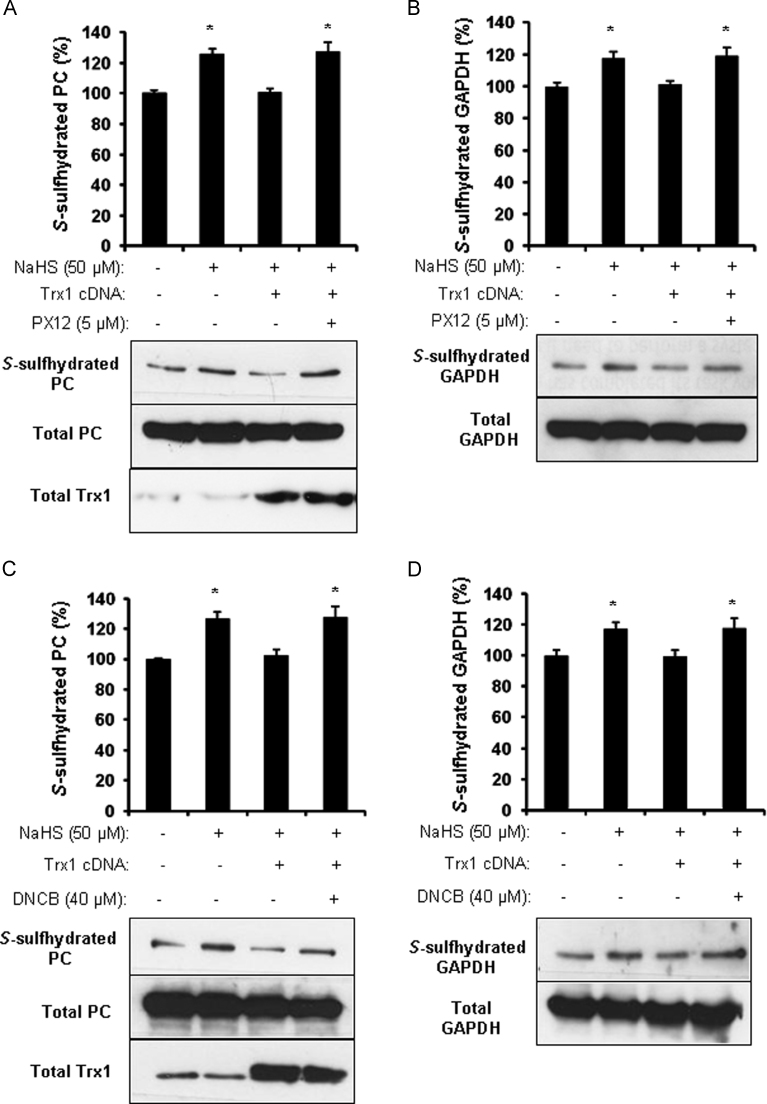
PX12, an irreversible and competitive inhibitor of Trx1, significantly enhanced the S-sulfhydration of PC and GAPDH when compared with the control HepG2 cells (Fig. 2A and B). Similarly, 2,4-dinitrochlorobenzene (DNCB), an irreversible inhibitor of thioredoxin reductase (TrxR), also increased S-sulfhydrated PC and GAPDH (Fig. 2C and D), suggesting that thioredoxin reductase cooperates with Trx1 contributing to protein S-desulfhydration. Either PX12 or DNCB did not further increase H2S-stimulated PC and GAPDH S-sulfhydration (Fig. 2A–D). And next we tried to knockdown Trx1 in HEK293 cells by siRNA transfection. Compared with the negative siRNA-transfected cells, the basal level of PC and GAPDH S-sulhydration were significantly increased (Fig. 2E and F). Furthermore, we observed that PX12 or DNCB significantly reverses the inhibitory role of Trx1 overexpression on S-desulfhydration of PC and GAPDH (Fig. 3A–D).
Fig. 2.
Inhibition of Trx system enhances protein S-sulfhydration. A and B, PX12 induces PC and GAPDH S-sulfhydration. HepG2 cells were firstly treated with 5 µM PX12 for 4 h, and then 50 µM NaHS was added for additional 2 h. After that, the cells were collected for biotin switch assay using antibody against PC (A) and GAPDH (B). *p<0.05 versus control. n=4. C and D, DNCB induces PC and GAPDH S-sulfhydration. HepG2 cells were treated with 40 µM DNCB or 30 min and/or 50 µM NaHS for 2 h. After that, the cells were collected for biotin switch assay using antibody against PC (C) and GAPDH (D). *p<0.05 versus control. n=4. E and F, knockdown of Trx1 stimulates PC and GAPDH S-sulfhydration. HEK293 cells were transfected with Trx1-specific siRNA or negative siRNA for 48 h following incubation with 50 µM NaHS for 2 h. After that, the cells were collected for biotin switch assay using antibody against PC (E) and GAPDH (F). *p<0.05 versus control. n=3.
Fig. 3.
PX12 and DNCB reverse the inhibitory effect of Trx1 on protein S-desulfhydration. A and B, PX12 abolished the inhibitory effect of Trx1 on PC and GAPDH S-desulfhydration. HEK293 cells were firstly transfected with Trx1 cDNA for 24 h following incubation with 5 µM PX12 for 4 h and/or 50 µM NaHS for 2 h. After that, the cells were collected for biotin switch assay using antibody against biotin PC (A) and GAPDH (B). *p<0.05 versus control. n=4. C and D, DNCB reversed the inhibitory effect of Trx1 on PC and GAPDH S-desulfhydration. HEK293 cells were firstly transfected with Trx1 cDNA for 24 h following incubation with 40 µM DNCB or 30 min and/or 50 µM NaHS for 2 h. After that, the cells were collected for biotin switch assay using antibody against biotin PC (C) and GAPDH (D). *p<0.05 versus control. n=4.
3.3. Cysteine-32 is requisite for Trx1 desulfhydration of proteins
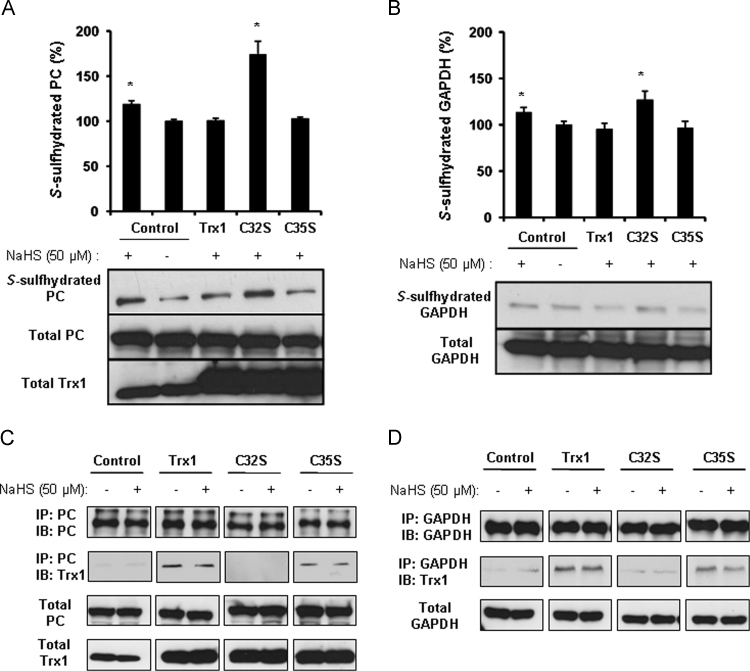
Trx1 has a highly conserved active site, Trp–Cys32–Gly–Pro–Cys35, which is required for its redox activity [18], [19]. Here we found that mutation of cysteine-32 but not cysteine-35 in the Trp-Cys32–Gly–Pro–Cys35 motif eliminates the S-desulfhydration activity of Trx1 on both PC and GAPDH (Fig. 4A and B), suggesting cysteine-32 is requisite for Trx1 S-desulfhydration of proteins. Trx1 directly interacted with PC and GAPDH, which was not altered in the presence of NaHS. Interestedly, mutation of cysteine-32 but not cysteine-35 abolished the binding of Trx1 with PC (Fig. 4C) or GAPDH (Fig. 4B).
Fig. 4.
Cysteine-32 in Trx1 is responsible for Trx1 S-desulfhydrating proteins. A and B, cysteine-32 mutation abolished the inhibitory role of Trx1 on protein S-desulfhydration. HEK293 cells were transfected with Trx1 cDNA and mutated Trx1 cDNA (C32S and C35S) for 24 h following incubation with 50 µM NaHS for 2 h. After that, the cells were collected for biotin switch assay using antibody against PC (A) and GAPDH (B). *<0.05 versus the control without NaHS treatment. n=4. C and D, cysteine-32 mutation disassociated Trx1 binding with S-sulfhydrated PC and GAPDH. HEK293 cells were transfected with Trx1 cDNA and mutated Trx1 cDNA (C32S and C35S) for 24 h following incubation with 50 µM NaHS for 2 h. After that, the cells were collected for co-immunoprecipitation assay as indicated in the figure. n=3.
4. Discussion
H2S regulates cellular processes largely through S-sulfhydration of active cysteine residues within proteins [2], [3]. S-sulfhydration of proteins has been demonstrated to be involved in a broad range of signaling and metabolic pathways, including cell death and differentiation, enzymatic activity, protein localization, protein–protein interactions, and protein stability, etc [1], [4], [6], [8], [10], [11], [12], [20], [21]. Given the importance of protein S-sulfhdyration in both health and diseases, much less is known about the systems governing protein S-desulfhydration. In our present report, we demonstrated that Trx1 is essential for breakage of hydropersulfide group from the cysteine thiol in S-sulfhdyrated proteins by direct interaction with Trx1 at cysteine-32.
Trx1, a ubiquitous 12 kDa proteins with general anti-oxidative properties, is involved in a large number of cellular functions such as cell proliferation and differentiation, redox and metabolic pathways, gene transcription, embryogenesis, etc [22], [23], [24], [25], [26]. In recent years, Trx1 has been suggested to mediate cysteine S-denitrosylation by reducing inter- and intramolecular disulfide bonds in proteins [26], [27]. By analogy to S-nitrosylation, the extent of S-sulfhydration of a given protein must be governed by the equilibrium between S-sulfhydration and S-desulfhydration reactions [26]. Protein S-sulfhydration is mainly dependent on the activity of H2S-producing enzymes [6], [28]. Here we showed that Trx1 is clearly involved in protein S-desulfhydration, because overexpression of Trx1 suppressed but blockage of Trx1 activity enhanced protein S-sulfhydration. Trx1 catalyzes thiol-disulfide oxidoreductions by using redox-active cysteine residues present in a Trp–Cys32–Gly–Pro–Cys35 sequence motif [25], [27], [29]. The active cysteine residues form a disulfide in oxidized Trx1, while the disulfide is broken into reduced Trx1 with the aid of TrxR using electrons from NADPH [24], [30]. In our study, inhibition of TrxR activity by DNCB must block the transition of reduced Trx1 to oxidized Trx1, which diminishes the inhibitory role of Trx1 on protein S-desulfhydration. These findings indicate that protein S-desulfhydration is indeed mediated by the Trx1/TrxR system.
We further provided evidence that Trx1 contributes to protein S-desulfhydration via protein-to-protein association between its substrate and Trx1 at cysteine-32. Co-immunoprecipitation results demonstrated that Trx1 directly interacts with S-sulfhydrated proteins, and single mutation of cysteine-32 to serine abolished the inhibitory role of Trx1 on protein S-desulfhydration. Mutation of cysteine-32 also eliminated the binding of Trx1 with S-sulfhydrated proteins. In contrary, mutation of cysteine-35 in the same Trp–Cys32–Gly–Pro–Cys35 sequence motif failed to change the S-desulfhydration activity of Trx1. There may be several reasons for this difference. Firstly, formation of the cysteine-32/cystine-35 disulfide bridge leads to a rotation of the side-chain of cysteine-32 away from cysteine-35 in the reduced form [19], [24]. Mutation of cysteine-32 will induce formation of new disulfide between cysteine-35 and other cysteine residues within Trx1, which would change the position of cysteine 32 and block the interaction of cysteine-32 with S-sulfhydrated proteins. Secondly, the thiol of cysteine-32 has a low pKa value in comparison with cysteine-35, and cysteine-32 is more easily attacked by S-sulfhydrated proteins for disulfide reduction [25], [29], [30]. Thirdly, Trx-interacting protein (Txnip), a constitutively expressed protein, forms a mixed disulfide with the active site cysteine-32 of Trx1 [18], [26], [31]. Mutation of cysteine-32 would interrupt the interaction of Txnip with Trx1, thereby affecting the oxidoreductase activity of Trx1. More studies are required to elucidate the precise interaction of Trx1 cysteine-32 and S-sulfhydrated proteins. In addition, we cannot exclude the possibility that other cysteine residues within Trx1 are involved in this process.
Two major pathways for enzymatic denitrosylation have recently been recognized [24]. Besides with Trx system, S-nitrosoglutathione (GSNO) reductase also can lower the levels of protein S-nitrosothiols [19], [26], [32]. Unlike Trx1, which directly target specific S-nitrosylated proteins for cysteine reduction, GSNO reductase indirectly decreases cell protein S-nitrosylation through GSNO metabolism. In addition, glutathione, the most abundant intracellular thiols, plays a major role in protein S-denitrosylation by GSNO reductase. We had data showed that the supplemental of glutathione has no effect on protein S-desulfhydration (data not shown). Therefore, GSNO reductase would not act as an S-desulfhydrase. The dynamic and regulation of protein S-sulfhydration/desulfhydration must be very complicated; many other factors may be also involved through non enzymatic processes, such as extreme oxidative stress, energy depletion, and competition with NO for the same cysteine residue(s), all of which need to be further tested.
Based on the results above, we conclude that the S-desulfhydration of proteins is associated with the Trx1/TrxR system, and cysteine-32 within Trx1 is responsible for the direct interaction of Trx1 and S-sulfhydrated proteins following the breakage of hydropersulfide group. Further investigation on the role of Trx system in relation to protein S-desulfhydation and their biological relevance needs to be explored.
Acknowledgment
This study was supported by a grant-in-aid (#000205) from the Heart and Stroke Foundation of Canada and Laurentian University start-up funding (#1092582).
Footnotes
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.bbrep.2015.11.012.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Mustafa A.K., Gadalla M.M., Sen N., Kim S., Mu W., Gazi S.K., Barrow R.K., Yang G., Wang R., Snyder S.H. H2S signals through protein S-sulfhydration. Sci. Signal. 2009;2 doi: 10.1126/scisignal.2000464. ra72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mustafa A.K., Gadalla M.M., Snyder S.H. Signaling by gasotransmitters. Sci. Signal. 2009;2:re2. doi: 10.1126/scisignal.268re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang G. Protein S-sulfhydration as a major source of H2S bioactivity. Recept. Clin. Investig. 2014;1:e337. [Google Scholar]
- 4.Mustafa A.K., Sikka G., Gazi S.K., Steppan J., Jung S.M., Bhunia A.K., Barodka V.M., Gazi F.K., Barrow R.K., Wang R., Amzel L.M., Berkowitz D.E., Snyder S.H. Hydrogen sulfide as endothelium-derived hyperpolarizing factor sulfhydrates potassium channels. Circ. Res. 2011;109:1259–1268. doi: 10.1161/CIRCRESAHA.111.240242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang D., Macinkovic I., Devarie-Baez N.O., Pan J., Park C.M., Carroll K.S., Filipovic M.R., Xian M. Detection of protein S-sulfhydration by a tag-switch technique. Angew. Chem. Int. Ed. Engl. 2014;53:575–581. doi: 10.1002/anie.201305876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen N., Paul B.D., Gadalla M.M., Mustafa A.K., Sen T., Xu R., Kim S., Snyder S.H. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol. Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao W., Zhang J., Lu Y., Wang R. The vasorelaxant effect of H2S as a novel endogenous gaseous KATP channel opener. EMBO J. 2001;20:6008–6016. doi: 10.1093/emboj/20.21.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan N., Fu C., Pappin D.J., Tonks N.K. H2S-induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Sci. Signal. 2011;4:ra86. doi: 10.1126/scisignal.2002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Z.Z., Shi M.M., Xie L., Wu Z.Y., Li G., Hua F., Bian J.S. Sulfhydration of p66Shc at cysteine59 mediates the anti-oxidant effect of Hydrogen Sulfide. Antioxid. Redox Signal. 2014;21:2531–2542. doi: 10.1089/ars.2013.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang G., Zhao K., Ju Y., Mani S., Cao Q., Puukila S., Khaper N., Wu L., Wang R. Hydrogen sulfide protects against cellular senescence via S-sulfhydration of Keap1 and activation of Nrf2. Antioxid. Redox Signal. 2013;18:1906–1919. doi: 10.1089/ars.2012.4645. [DOI] [PubMed] [Google Scholar]
- 11.Nishida M., Sawa T., Kitajima N., Ono K., Inoue H., Ihara H., Motohashi H., Yamamoto M., Suematsu M., Kurose H., van der Vliet A., Freeman B.A., Shibata T., Uchida K., Kumagai Y., Akaike T. Hydrogen sulfide anion regulates redox signaling via electrophile sulfhydration. Nat. Chem. Biol. 2012;8:714–724. doi: 10.1038/nchembio.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao K., Ju Y., Li S., Altaany Z., Wang R., Yang G. S-sulfhydration of MEK1 leads to PARP-1 activation and DNA damage repair. EMBO Rep. 2014;15:792–800. doi: 10.1002/embr.201338213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S., Yang G. Hydrogen sulfide maintains mitochondrial DNA replication via demethylation of TFAM. Antioxid. Redox Signal. 2015;23:630–642. doi: 10.1089/ars.2014.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ju Y., Untereiner A., Wu L., Yang G. H2S-induced of pyruvate carboxylase contributes to gluconeogenesis in liver cells. Biochem. Biophys. Acta Gen. Subj. 2015;1850:2293–2303. doi: 10.1016/j.bbagen.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Benhar M., Forrester M.T., Hess D.T., Stamler J.S. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roskoski R., Jr. Src kinase regulation by phosphorylation and dephosphorylation. Biochem. Biophys. Res. Commun. 2005;331:1–14. doi: 10.1016/j.bbrc.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Liu H., Nishitoh H., Ichijo H., Kyriakis J.M. Activation of apoptosis signal-regulating kinase 1 (ASK1) by tumor necrosis factor receptor-associated factor 2 requires prior dissociation of the ASK1 inhibitor thioredoxin. Mol. Cell. Biol. 2000;20:2198–2208. doi: 10.1128/mcb.20.6.2198-2208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forrester M.T., Seth D., Hausladen A., Eyler C.E., Foster M.W., Matsumoto A., Benhar M., Marshall H.E., Stamler. J.S. Thioredoxin-interacting protein (Txnip) is a feedback regulator of S-nitrosylation. J. Biol. Chem. 2009;284:36160–36166. doi: 10.1074/jbc.M109.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahl M.C., Irmler A., Hecker B., Schirmer R.H., Becker K. Comparative structural analysis of oxidized and reduced thioredoxin from Drosophila melanogaster. J. Mol. Biol. 2005;345:1119–1130. doi: 10.1016/j.jmb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y., Yang R., Liu X., Zhou Y., Qu C., Kikuiri T., Wang S., Zandi E., Du J., Ambudkar I.S., Shi S. Hydrogen Sulfide Maintains Mesenchymal Stem Cell Function and Bone Homeostasis via Regulation of Ca(2+) Channel Sulfhydration. Cell Stem Cell. 2014;15:66–78. doi: 10.1016/j.stem.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altaany Z., Ju Y., Yang G., Wang R. The coordination of S-sulfhydration, S-nitrosylation, and phosphorylation of endothelial nitric oxide synthase by hydrogen sulfide. Sci. Signal. 2014;7:ra87. doi: 10.1126/scisignal.2005478. [DOI] [PubMed] [Google Scholar]
- 22.Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y., Kawabata M., Miyazono K., Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell D.A., Morton S.U., Fernhoff N.B., Marletta M.A. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11609–11614. doi: 10.1073/pnas.0704898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sengupta R., Holmgren A. The role of thioredoxin in the regulation of cellular processes by S-nitrosylation. Biochim. Biophys. Acta. 2012;1820:689–700. doi: 10.1016/j.bbagen.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Kelleher Z.T., Sha Y., Foster M.W., Foster W.M., Forrester M.T., Marshall H.E. Thioredoxin-mediated denitrosylation regulates cytokine-induced nuclear factor κB (NF-κB) activation. J. Biol. Chem. 2014;289:3066–3072. doi: 10.1074/jbc.M113.503938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu C., Kavalier A., Lukyanov E., Gross S.S. S-sulfhydration/desulfhydration and S-nitrosylation/denitrosylation: a common paradigm for gasotransmitter signaling by H2S and NO. Methods. 2013;62:177–181. doi: 10.1016/j.ymeth.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benhar M., Forrester M.T., Stamler J.S. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell. Biol. 2009;10:721–732. doi: 10.1038/nrm2764. [DOI] [PubMed] [Google Scholar]
- 28.Vandiver M.S., Paul B.D., Xu R., Karuppagounder S., Rao F., Snowman A.M., Ko H.S., Lee Y.I., Dawson V.L., Dawson T.M., Sen N., Snyder S.H. Sulfhydration mediates neuroprotective actions of parkin. Nat. Commun. 2013;4:1626. doi: 10.1038/ncomms2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qu Z.W., Miao W.Y., Hu S.Q., Li C., Zhuo X.L., Zong Y.Y., Wu W.P., Zhang G.Y. N-methyl-d-aspartate receptor-dependent denitrosylation of neuronal nitric oxide synthase increase the enzyme activity. PLoS One. 2012;7:e52788. doi: 10.1371/journal.pone.0052788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaffagnini M., Morisse S., Bedhomme M., Marchand C.H., Festa M., Rouhier N., Lemaire S.D., Trost P. Mechanisms of nitrosylation and denitrosylation of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. J. Biol. Chem. 2013;288:22777–22789. doi: 10.1074/jbc.M113.475467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C C., Parrott A.M., Liu T., Jain M.R., Yang Y., Sadoshima J., Li H. Distinction of thioredoxin transnitrosylation and denitrosylation target proteins by the ICAT quantitative approach. J. Proteom. 2011;74:2498–24509. doi: 10.1016/j.jprot.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anand P., Stamler J.S. Enzymatic mechanisms regulating protein S-nitrosylation: implications in health and disease. J. Mol. Med. (Berl.) 2012;90:233–244. doi: 10.1007/s00109-012-0878-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material