Abstract
DNA methylation is closely involved in the regulation of cellular differentiation, including chondrogenic differentiation of mesenchymal stem cells. Recent studies showed that Ten–eleven translocation (TET) family proteins converted 5-methylcytosine (5mC) to 5-hydroxymethylcytosine, 5-formylcytosine and 5carboxylcytosine by oxidation. These reactions constitute potential mechanisms for active demethylation of methylated DNA. However, the relationship between the DNA methylation patterns and the effects of TET family proteins in chondrocyte differentiation is still unclear. In this study, we showed that DNA hydroxylation of 5mC was increased during chondrocytic differentiation of C3H10T1/2 cells and that the expression of Tet1 was particularly enhanced. Moreover, knockdown experiments revealed that the downregulation of Tet1 expression caused decreases in chondrogenesis markers such as type 2 and type 10 collagens. Furthermore, we found that TET proteins had a site preference for hydroxylation of 5mC on the Insulin-like growth factor 1 (Igf1) promoter in chondrocytes. Taken together, we showed that the expression of Tet1 was specifically facilitated in chondrocyte differentiation and Tet1 can regulate chondrocyte marker gene expression presumably through its hydroxylation activity for DNA.
Abbreviations: 5mC, 5-methylcytosine; 5hmC, 5-hydroxymethylcytosine; Tet, ten–eleven translocation; Col2, collagen 2; Col10, collagen 10; Igf1, insulin-like growth factor
Keywords: Chondrocyte differentiation, Hydroxymethylcytosine, TET1, Col2, Col10, Igf1
Highlights
-
•
The level of 5hmc increases concomitant with chondrocyte differentiation.
-
•
Among the TET family proteins, the expression of TET1 is specifically facilitated in chondrocyte differentiation.
-
•
Knock-down of TET1 leads to decrease in the expression of chondrocyte differentiation marker.
-
•
TET proteins have a site preference for hydroxylation of 5mC on the Igf1 promoter.
1. Introduction
DNA methylation is a well-known epigenomic mark involved in gene silencing [1], [2]. Methylation of DNA affects the transcription of genes related to various cellular processes such as differentiation. Moreover, the variation of DNA methylation between tissues has been termed “tissue-dependent differentially methylated regions” (T-DMRs). This methylation diversity reportedly contributes to tissue-specific gene expression [3].
Recently, Ten–eleven translocation (TET) family proteins (TET1/2/3) were identified as the family of enzymes that sequentially converted 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) [4], [5]. This series of reactions is considered a potential mechanism for active demethylation of 5mC [6]. The physiological function of each TET protein has distinguishing features. TET1 has been reported to be involved in the genetic regulation of memory formation [7], [8] and cancer development [9]. TET2 plays a critical role in hematopoietic differentiation and its mutations have been frequently observed in myeloid malignancies [10]. TET3 is expressed particularly in oocytes and is indispensable for paternal DNA demethylation after fertilization [11]. As described herein, although TET proteins have similar enzymatic activities, their biological significance includes various cellular processes in different organs.
DNA methylation is a key factor for cell fate decisions, such as differentiation from mesenchymal stem cells into adipocytes, myocytes and chondrocytes [12]. Many reports have demonstrated that there are alterations of DNA methylation states during adipocyte and myocyte differentiation [13], [14], and specific methylation of CpG dinucleotides is required for proper adipocyte differentiation [15]. Furthermore, the state of DNA methylation reportedly changes during chondrocyte differentiation [16].
Chondrocytes are established through subsequent differentiation steps, consisting of resting, proliferative and hypertrophic chondrocytes [17]. During differentiation, various transcription factors, for instance, SRY-box containing gene 9 (Sox9) play important roles and are essential for normal early stage chondrocyte differentiation [18]. Furthermore, a recent study revealed that epigenetic regulators, such as ERG-associated protein with a SET domain (ESET) and autoimmune regulator (Aire) also controlled chondrocyte differentiation through histone modification [19], [20]. However, it is still unclear how TET family proteins are involved in this process.
In this paper, we evaluated the relationship between chondrocyte differentiation and the hydroxylation of methylated DNA by TET proteins using the mesenchymal stem cell line C3H10T1/2. We found that the amount of hydroxyl-methylated DNA increased during chondrocyte differentiation. From knockdown experiments, we found that TET1 had a role in the expression of chondrocyte marker genes.
2. Materials and methods
2.1. Cell culture, cell differentiation, and Alcian blue staining
Mouse C2C12 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Wako, Osaka, Japan) supplemented with 20% fetal bovine serum (FBS, Gibco, USA) and antibiotics. Mouse 3T3-L1 and C3H10T1/2 cells were maintained in DMEM containing 10% FBS and antibiotics. The latter two strains were grown at 37 °C in 5% CO2. For myogenic differentiation, the culture medium of C2C12 cells was exchanged for DMEM containing 0.5% FBS. In accord with a previous report [21], adipocyte differentiation of 3T3-L1 cells was induced using AdipoInducer Reagent (Takara Bio Inc., Otsu, Japan). For chondrocyte differentiation, C3H10T1/2 cells were placed in high-density micromass cultures with 100 ng/mL of recombinant human bone morphogenetic protein 2 (BMP2, OSTEOPHARMA, Osaka, Japan) as previously reported [19].
2.2. Genomic DNA preparation, dot blotting and densitometric analysis
Genomic DNA purification and dot blotting were carried out as previously described [22]. Briefly, the cells were lysed with genome extraction buffer (50 mM Tris–HCl, 20 mM EDTA, 100 mM NaCl, and 1% SDS, pH 7.9) with 0.15 mg/mL Proteinase K. After phenol/chloroform extraction and isopropanol precipitation, 2 μg DNA of each samples were subjected to dot blot assay using 5hmC antibody (Active Motif, USA). As a standard for comparison, 1 μg DNA derived from mouse embryonic stem cells (mESc) was blotted in the same membranes. Densitometric analysis was carried out by ImageJ.
2.3. RNA isolation, complementary DNA (cDNA) synthesis, polymerase chain reaction (PCR), and statistics
RNA isolation, cDNA synthesis and PCR were conducted as previously described [22]. The sequences of the primer sets are shown in Supplemental Table 1. All values are means ± standard deviations from three independent experiments. We used a two-tailed Student’s t test to analyze the differences between two groups.
2.4. DNA methylation analysis using restriction enzymes and glucosyltransferase
For the measurement of DNA methylation state, the genomic DNA was digested with either MspI (that recognized and cleaved CCGG sequences independent of its methylation) or HapII (that recognized the same sequences as MspI but could not cleave methylated DNA). These digested DNAs ware subjected to quantitative PCR (qPCR) analysis using primers designed for the Insulin-like growth factor 1 (Igf1) promoter. To determine the level of 5hmC, we sequentially treated the genomic DNA with MspI and T4-β-glucosyltransferase (BGT, New England Biolabs, USA) following the manufacturer’s instructions. The data were normalized to the qPCR intensities of the uncut samples.
2.5. Preparation of cell extracts, Western blotting and antibodies
As previously described [22], cell lysates were prepared in TNE buffer (20 mM Tris–HCl, 137 mM NaCl, two mM EDTA, 1% NP-40, and protease inhibitor, pH 7.9) with sonication. Western blotting analysis was performed using α-Tet1 (Millipore, USA), α-Tet2 (Sigma, USA), α-Collagen2 (Col2, Santa Cruz, USA), and α-actin (Santa Cruz).
2.6. Plasmids for shRNA expression, retroviral production and infection
For short hairpin RNA (shRNA) expression plasmids, oligonucleotides were inserted into a pSUPER.retro.hygro vector that was generated from pSUPER.retro.puro (Oligoengine) and had the resistance gene to hygromycin instead of puromycin. The following target sequences were used in reference to previous studies. For mouse Tet1 shRNA (shTet1-1), 5′-GCAGATGGCCGTGACACAAAT-3′ [23]; for mouse Tet1 shRNA (shTet1-2), 5′-GAATTACAGTTGTTACGGA-3′ [24] ; for Escherichia coli LacZ shRNA (shLacZ), 5′-GCCCATCTACACCAACGTAAC-3′ [25]. The shRNA-expressing retroviruses for Tet1 or LacZ (control) were generated as previously described [26]. C3H10T1/2 cells were suspended in medium containing the virus with 10 μg/mL hexadimethrine bromide and then centrifuged at 1000g for 1 h at room temperature. The next day, the medium for these cells was exchanged for DMEM with 500 μg/mL hygromycin. Two weeks after viral infection, the surviving cells were subjected to differentiation.
3. Results
3.1. Hydroxylation of methylated cytosine was accompanied by the progression of chondrocyte differentiation of 10T1/2 cells
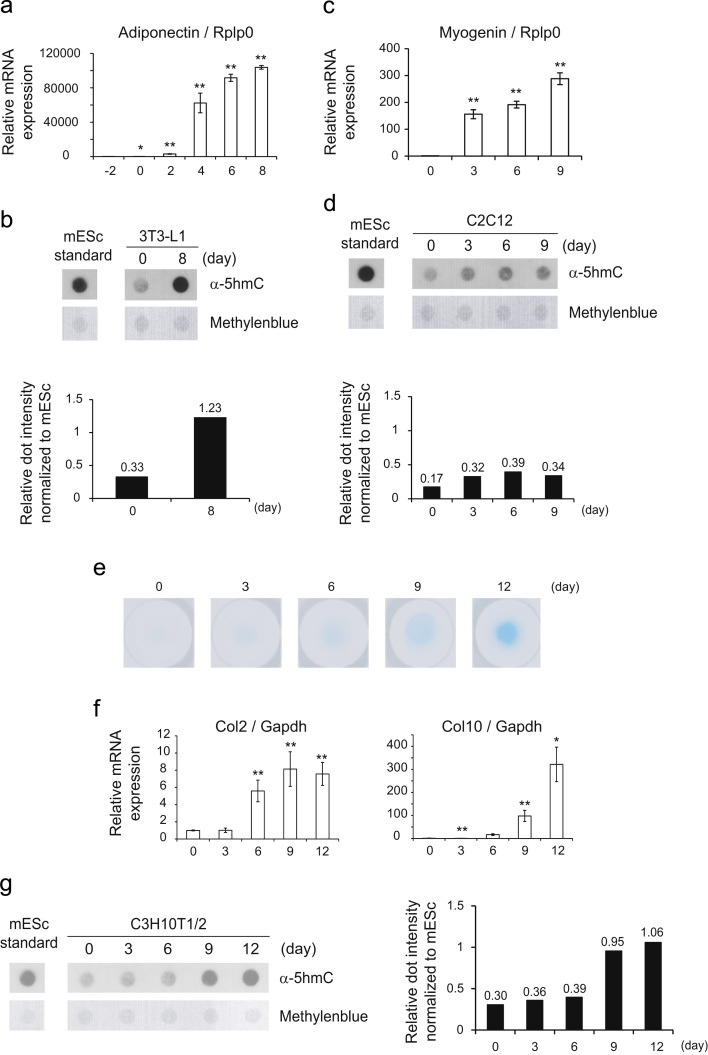
The level of DNA methylation fluctuates during cell differentiation processes. We reasoned that 5hmC might also show differentiation-dependent dynamics. Thus, we investigated the state of hydroxylation of methylated DNA using cultured mesenchymal stem cells. First, 3T3-L1 cells were differentiated to adipocytes, and we observed increased expression of an adipocyte differentiation marker, the gene for adiponectin (Fig. 1a). DNA hydroxymethylation showed about 4-fold increased dot intensity during differentiation (Fig. 1b), consistent with a previous study by Fujiki’s group [27]. Next, we analyzed the change of DNA hydroxymethylation during differentiation from myoblasts to myotubes using the C2C12 mouse mesenchymal stem cell line. Although myogenin gene (Myog) mRNA expression (a differentiation marker of myotubes) was increased from 3 days (Fig. 1c), the remarkable change of hydroxylation of methylated DNA was not detected (Fig. 1d). Finally, we examined the alteration of methylated DNA hydroxylation during chondrocyte differentiation using C3H10T1/2 cells. C3H10T1/2 cells were differentiated to chondrocytes by incubation in micromass cultures with BMP2. These micromass-cultured C3H10T1/2 cells showed a positive reaction to Alcian blue staining, indicating successful chondrocyte differentiation (Fig. 1e). Then, a reverse transcription qPCR (RT-qPCR) assessment showed that the expression of the Col2 gene, a marker gene for proliferative chondrocytes, peaked at 9 days. Collagen 10 (Col10) expression, a marker gene for hypertrophic chondrocytes, peaked at 12 days (Fig. 1f). These results indicated that micromass-cultured C3H10T1/2 differentiated to proliferative chondrocytes by 9 days and to hypertrophic chondrocytes at around 12 days. A dot blot analysis of differentiating chondrocytes indicated that the level of 5hmC about 3-fold increased with chondrogenesis (Fig. 1g) as well as with adipocyte differentiation (Fig. 1b). In the remainder of this study, we focused on chondrocyte differentiation of C3H10T1/2 cells and carried out the following experiments.
Fig. 1.
Altered amounts of 5hmC in cell differentiation. (a) Two days after reaching confluence, 3T3-L1 cells were stimulated by insulin, dexamethasone, 3-isobutyl-1-methylxanthine and troglitazone for 3 days. RT-qPCR was conducted using the primers for adiponectin as a differentiation marker, and for Rplp0 as a control. RT-qPCR intensities were normalized to Rplp0 expression. (b) The genomic DNA was subjected to dot blotting using anti-5hmC antibody. Methylene blue staining was carried out for loading control of DNA. Relative dot intensity normalized to a dot derived from mESc in the same membrane was indicated. (c) C2C12 cells were cultured in medium with low-concentration FBS (0.5%). RT-qPCR was conducted as in (a). Myogenin expression was validated as a differentiation marker, and Gapdh was used as a control. (d) Dot blotting was carried out as in (b). (e) C3H10T1/2 cells underwent micromass cultivation with recombinant human BMP2. Alcian blue staining was used to confirm their differentiation into chondrocytes. (f) The mRNA expression of collagen 2 (Col2), and collagen 10 (Col10) were measured by RT-qPCR as differentiation markers. (g) The isolated DNA was subjected to dot blotting. Data are represented as means ± standard deviations (n=3), ratio to non-differentiation group. *p<0.05; **p<0.01.
3.2. The expression of Tet1 was increased in chondrocytes, and Tet1’s role in chondrocyte differentiation
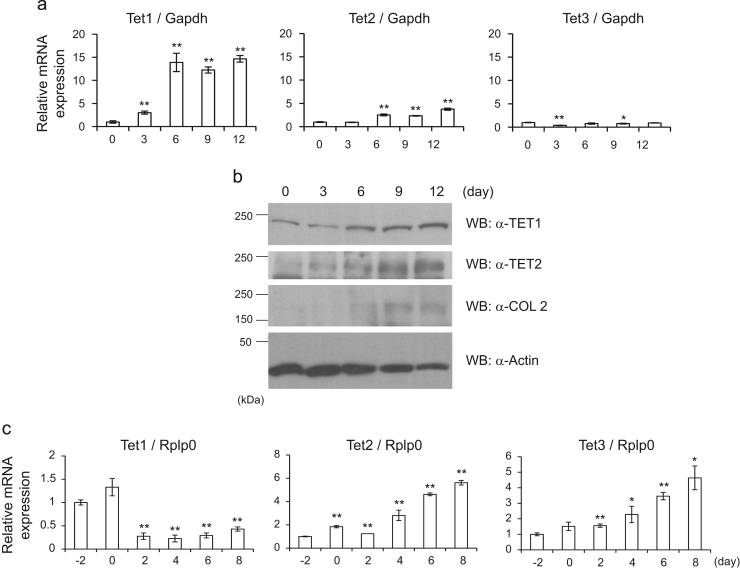
Because hydroxylation of methylated DNA was governed by Tet proteins, we measured the expression levels of Tet gene mRNAs in C3H10T1/2 cells. In the early stage of chondrocyte differentiation, the expression of Tet1 and Tet2 mRNAs was elevated. In particular, Tet1’s expression was robustly upregulated fifteen-fold, although the expression of Tet3 was not dynamically altered (Fig. 2a). Moreover, Western blotting showed that the amounts of Tet1 and Tet2 proteins increased in differentiated chondrocytes (Fig. 2b). In contrast to chondrocyte differentiation, the dramatic increase of Tet1 mRNA was not observed in adipocyte differentiation (Fig. 2c), although the expression Tet2 and Tet3 was slightly increased. These results suggested that Tet1 expression was enhanced specifically in chondrocyte differentiation and not in adipocyte differentiation.
Fig. 2.
Expression changes of the Tet family of genes in differentiation of chondrocytes and adipocytes. (a) RT-qPCR was carried out using primers for TET1, TET2 and TET3 as in Fig. 1(f). (b) Cell lysates were extracted at each time point, and they were subjected to Western blotting with the indicated antibodies. COL2 and actin were detected as a differentiation marker and a loading control, respectively. (c) 3T3-L1 cells were differentiated into adipocytes as in Fig. 1(a), and RT-qPCR was conducted. Data present means ± standard derivations (n=3). * p<0.05; ** p<0.01.
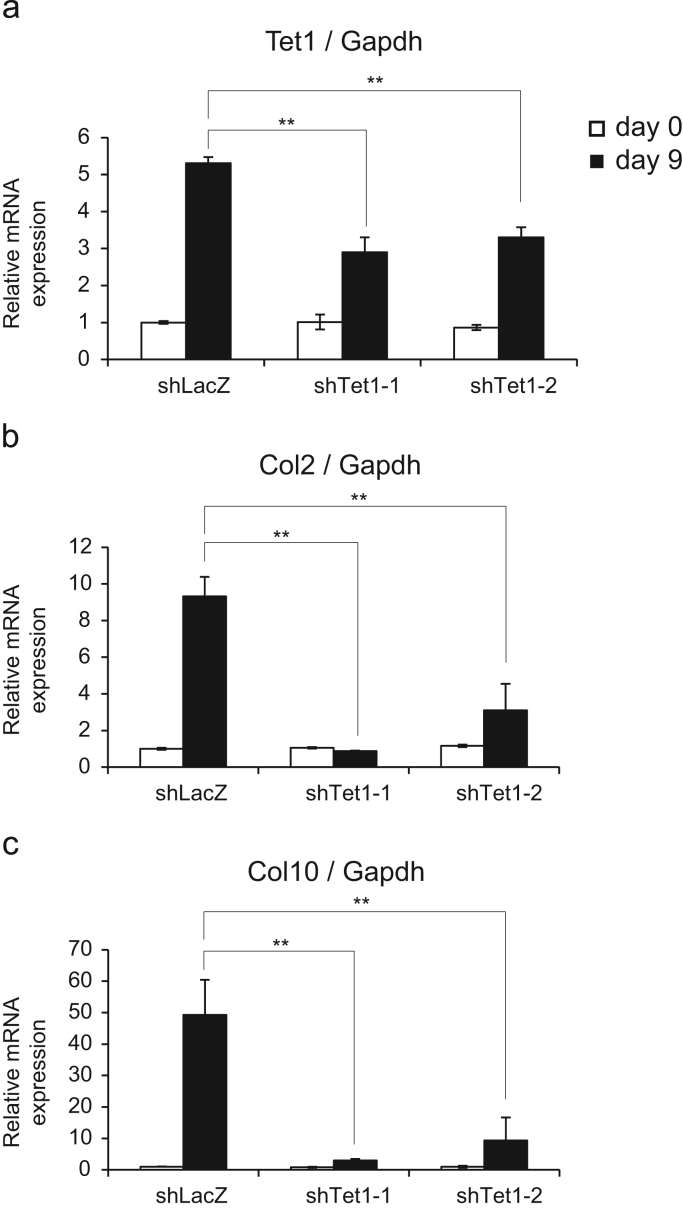
From these results, we hypothesized that Tet1 might have a function in transcriptional regulation of chondrocyte differentiation-related genes via the hydroxylation of its promoter DNA. To address this hypothesis, we investigated the effect of Tet1-knockdown on Col2 and Col10 expression in chondrocyte differentiation. Retroviral shRNA against Tet1 reduced the expression to approximately 50% at 9 days (Fig. 3a). Under this condition, the expression of Col2 (Fig. 3b) and Col10 (Fig. 3c) was significantly decreased by shTet1. This result suggested that Tet1 might play a role in the progression of chondrocyte differentiation.
Fig. 3.
Tet1 knockdown in C3H10T1/2 cells downregulated the expression of Col2 and Col10 during chondrocyte differentiation. (a) (b) (c) Tet1 knockdown cells were generated using retroviral shRNA. For a control, a shRNA against LacZ was introduced into the cells. After these cells were differentiated to chondrocytes, RT-qPCR using primers for Tet1 (a), Col2 (b), and Col10 (c) were conducted. Data are presented as means ± standard deriviations (n=3), ratio to shLacZ at day zero samples. **p< 0.01.
3.3. Locus-specific hydroxylation of methylated DNA during chondrocyte differentiation
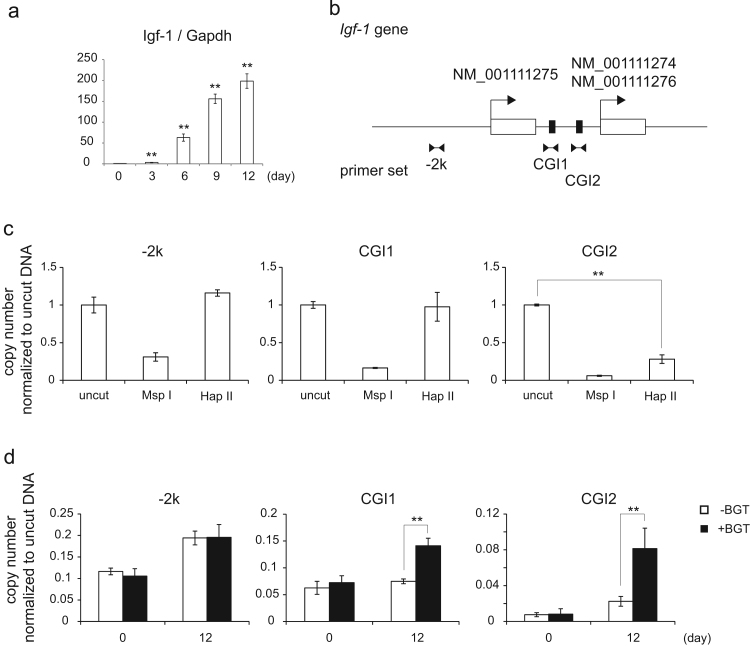
In this study, we showed that the hydroxylation of methylated DNA occurred during chondrocyte differentiation (Fig. 1g). However, precisely where this hydroxylation occurred was still unclear. Therefore, we analyzed the change in the hydroxylation level of the Igf1 gene promoter as a model for upregulated genes and the promotion of chondrocyte differentiation. Igf1 expression is governed by growth hormone (GH), which is secreted from the liver, muscle, adipose tissue, osteoblasts and chondrocytes [28]. Our experiment showed that the expression of the Igf1 gene was accompanied by chondrocyte differentiation of C3H10T1/2 cells (Fig. 4a). Next, we investigated the alteration of DNA methylation status of the Igf1 gene in chondrocyte differentiation. According to the NCBI database, the promoter of the Igf1 gene has alternative transcription start sites for Igf1 mRNA (NM_001111275 or NM_001111274/NM_00111276), and there are two CpG islands in this region (Fig. 4b). We designed two primers for quantitative PCR in these CpG islands (named CGI1, and CGI2 as indicated in Fig. 4b), and a primer 2000 base-pairs upstream from the NM_001111275 transcription start site (−2k) as a control. Every primer amplified the fragments containing the CCGG nucleotides. Restriction enzymes MspI and HapII recognized the CCGG sequences and digested the CpG dinucleotides. MspI could digest CCGG independent of the methylation status of CpG. On the other hand, HapII could digest CpG only when it was not methylated. The qPCR analysis combined with the digestion of genomic DNA revealed that the −2k and CGI1 regions were heavily methylated, whereas the CGI2 region was hardly methylated in undifferentiated C3H10T1/2 cells (Fig. 4c). We then examined the hydroxylation level of methylated cytosine in the three regions using the BGT reaction and MspI digestion. BGT could transfer the glycosyl group to only 5hmC, and glycosylated 5hmC was not digested by MspI. This experiment showed that CpG dinucleotides in CGI1 and CGI2 regions were hydroxylated in differentiated cells although the −2k region was not (Fig. 4d). Furthermore, we found that the extent of hydroxylation in CGI2 is much higher than that in CGI1. These results showed that the hydroxylation of methylated DNA by TET proteins had a site preference in the Igf1 promoter.
Fig. 4.
The hydroxylation of methylated DNA of the Igf1 promoter in chondrocyte differentiation. (a) RT-qPCR was carried out using a primer for Igf1 as in Fig. 1(f). Data present means ± standard deriviations (n=3). ** p<0.01. (b) Schematic representation of the Igf1 promoter. There are two transcription start sites (arrows) for NM_001111275, and NM_001111274/6. The Igf1 promoter has two CpG islands (solid boxes) between the first exons (empty boxes). For the following experiments, primers were designed for 2,000 base pairs upstream from the transcription start site (−2k) and at two CpG islands (CGI1 and CGI2). (c) Genomic DNA derived from undifferentiated C3H10T1/2 cells was digested by MspI or HapII or not treated. qPCR was conducted using −2k, CGI1 and CGI2 primers. qPCR intensity was normalized to uncut DNA samples. Data present means ± standard deriviations (n=3). **p<0.01 (d) DNA was extracted from undifferentiated cells (day 0) or differentiated cells (day 12) and was reacted with/without BGT. The glycosylated DNA was digested by MspI and qPCR was conducted as in (c). Data present means ± standard deriviations (n=3). **p<0.01.
4. Discussion
The relationship between epigenomic status and cell identity has been studied extensively. With regard to DNA methylation, when C3H10T1/2 cells were treated with 5-azacytidine, which inhibits DNA methyltransferase (DNMT) activity, there was a decrease in DNA methylation, and cells differentiated into chondrocytes, adipocytes and myocytes [12], [29]. Moreover, the hydroxylation of methylated DNA by Tet family proteins led to the removal of the methyl groups from cytosine, according to recent work [4], [5]. Based on these past reports, we postulated that Tet proteins play a role in epigenetic regulation during cell differentiation.
The expression pattern of the Tet family of genes varies with specific organs, and it was found that Tet1’s expression was abundant in embryonic stem cells [23]. In this report, we showed that the expression of Tet1 mRNA and protein were specifically enhanced during the differentiation of chondrocytes (Fig. 2a, b). Recently, some reports suggested that Tet1 is highly expressed in chondrocytes [30], [31]. For example, de Andres’s group has shown that the expression of Tet1 in human fetal bone cells was higher than in adult chondrocytes [30], and Haseeb’s group has revealed that Interleukin 1β (IL-1β) and Tumor necrosis factor α (TNF-α), known as pro-inflammatory cytokines, reduced the expression of Tet1 and the amount of 5hmC in human primary chondrocytes [31]. Our observations regarding the increased level of Tet1 expression in chondrocytes agreed with these reports. Together, these findings emphasize the significance of Tet1 to bone physiology.
The Igf1 gene is expressed in the liver, chondrocytes and elsewhere [28]. So far, the mechanisms regulating Igf1 transcription have primarily been studied in the liver. It has been well documented that GH induces Igf1 expression via the receptor signaling pathway [28]. The mechanisms regulating Igf1 transcription in chondrocytes have also been reported [32]. However, the epigenetic alterations of the Igf1 promoter in chondrocytes have remained unclear. In this study, we revealed that the expression of Igf1 mRNA was elevated in C3H10T1/2 cells, a line used as a chondrocyte differentiation model (Fig. 4a). Moreover, a specific CpG island (CGI2) showed a low methylation level, whereas other regions (−2k and CGI1) were more highly methylated in C3H10T1/2 cells (Fig. 4c). Furthermore, the hydroxylation of 5mC was detected in both CGI1 and CGI2. Of note, the amount of 5hmC in CGI2 was increased four-fold (Fig. 4d). These results suggested that the epigenetic states of the Igf1 promoter might change during chondrocyte differentiation.
Our results demonstrated that during chondrocyte differentiation, hydroxylation by Tet proteins took place in specific regions (Fig. 4d). Tet1 and Tet3 have a CXXC domain that recognizes the non-methylated CpG sequences [33]. Though Tet2 does not have it, it was reported that Tet2 could interact with Inhibition of the Dvl and Axin complex (IDAX) containing the CXXC domain [34]. In this study, we showed that CGI2 region was not frequently methylated before differentiation, and that the 5mC of CGI2 was hydroxylated after chondrocyte differentiation. These results support the hypothesis that Tet proteins recognized non-methylated CpG via its CXXC domain and probably hydroxylated the remaining portion of the methylated CpG.
In this study, we revealed DNA hydroxymethylation of Igf1 promoter in differentiated chondrocyte (Fig. 4d). This result suggested that Igf1 was one of the Tet1’s target genes. On the other hand, we showed the necessity of Tet1 for chondrocyte differentiation (Fig. 3). However, we did not clarify the reason why the down-regulation of Tet1 led to reduce the differentiation efficiency to chondrocyte. To answer this question we need to explore the target genes of TET1 by the genome-wide analysis in future efforts. This experiment would identify the target gene of Tet1 regulating the chondrocyte differentiation, and reveal more detailed mechanism of chondrocyte differentiation regulated by Tet1.
Acknowledgment
We thank Dr. Shigeaki Kato (Soma Central Hospital) for helpful discussion. We also thank Dr. Katsunori Fujiki (Tokyo University) for providing the antibodies against TET1 and TET2, Dr. Yuan Si (Johns Hopkins University) for technical assistance and OSTEOPHARMA for providing us rhBMP2. This work was supported by Grant-in-Aid for JSPS Fellows (No. 13J07784) and Platform for Drug Discovery, Informatics, and Structural Life Science (PDIS).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.11.009.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yagi S., Hirabayashi K., Sato S., Li W., Takahashi Y., Hirakawa T., Wu G., Hattori N., Ohgane J., Tanaka S., Liu X.S., Shiota K. DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 2008;18:1969–1978. doi: 10.1101/gr.074070.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C.X., Zhang K., He C., Xu G.L. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2012;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastor W.A., Aravind L., Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell. Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaas G.A., Zhong C., Eason D.E., Ross D.L., Vachhani R.V., Ming G.L., King J.R., Song H., Sweatt J.D. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rudenko A., Dawlaty M.M., Seo J., Cheng A.W., Meng J., Le T., Faull K.F., Jaenisch R., Tsai L.H. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takai H., Masuda K., Sato T., Sakaguchi Y., Suzuki T., Koyama-Nasu R., Nasu-Nishimura Y., Katou Y., Ogawa H., Morishita Y., Kozuka-Hata H., Oyama M., Todo T., Ino Y., Mukasa A., Saito N., Toyoshima C., Shirahige K., Akiyama T. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014;9:48–60. doi: 10.1016/j.celrep.2014.08.071. [DOI] [PubMed] [Google Scholar]
- 10.Ko M., Huang Y., Jankowska A.M., Pape U.J., Tahiliani M., Bandukwala H.S., An J., Lamperti E.D., Koh K.P., Ganetzky R., Liu X.S., Aravind L., Agarwal S., Maciejewski J.P., Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu T.P., Guo F., Yang H., Wu H.P., Xu G.F., Liu W., Xie Z.G., Shi L., He X., Jin S.G., Iqbal K., Shi Y.G., Deng Z., Szabo P.E., Pfeifer G.P., Li J., Xu G.L. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 12.Taylor S.M., Jones P.A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- 13.Yokomori N., Tawata M., Onaya T. DNA demethylation during the differentiation of 3T3-L1 cells affects the expression of the mouse GLUT4 gene. Diabetes. 1999;48:685–690. doi: 10.2337/diabetes.48.4.685. [DOI] [PubMed] [Google Scholar]
- 14.Lucarelli M., Fuso A., Strom R., Scarpa S. The dynamics of myogenin site-specific demethylation is strongly correlated with its expression and with muscle differentiation. J. Biol. Chem. 2001;276:7500–7506. doi: 10.1074/jbc.M008234200. [DOI] [PubMed] [Google Scholar]
- 15.Londono Gentile T., Lu C., Lodato P.M., Tse S., Olejniczak S.H., Witze E.S., Thompson C.B., Wellen K.E. Vol. 33. 2013. DNMT1 is regulated by ATP-citrate lyase and maintains methylation patterns during adipocyte differentiation; pp. 3864–3878. (Mol. Cell Biol.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmermann P., Boeuf S., Dickhut A., Boehmer S., Olek S., Richter W. Correlation of COL10A1 induction during chondrogenesis of mesenchymal stem cells with demethylation of two CpG sites in the COL10A1 promoter. Arthritis Rheum. 2008;58:2743–2753. doi: 10.1002/art.23736. [DOI] [PubMed] [Google Scholar]
- 17.Kronenberg H.M. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 18.Akiyama H., Chaboissier M.C., Martin J.F., Schedl A., de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Si Y., Inoue K., Igarashi K., Kanno J., Imai Y. Autoimmune regulator, Aire, is a novel regulator of chondrocyte differentiation. Biochem. Biophys. Res. Commun. 2013;437:579–584. doi: 10.1016/j.bbrc.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Lawson K.A., Teteak C.J., Zou J., Hacquebord J., Ghatan A., Zielinska-Kwiatkowska A., Fernandes R.J., Chansky H.A., Yang L. Mesenchyme-specific knockout of ESET histone methyltransferase causes ectopic hypertrophy and terminal differentiation of articular chondrocytes. J. Biol. Chem. 2013;288:32119–32125. doi: 10.1074/jbc.M113.473827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okuno Y., Ohtake F., Igarashi K., Kanno J., Matsumoto T., Takada I., Kato S., Imai Y. Epigenetic regulation of adipogenesis by PHF2 histone demethylase. Diabetes. 2013;62:1426–1434. doi: 10.2337/db12-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito R., Katsura S., Shimada H., Tsuchiya H., Hada M., Okumura T., Sugawara A., Yokoyama A. TET3-OGT interaction increases the stability and the presence of OGT in chromatin. Genes Cells. 2014;19:52–65. doi: 10.1111/gtc.12107. [DOI] [PubMed] [Google Scholar]
- 23.Ito S., D'Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koh K.P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C.A., Mostoslavsky G., Lahesmaa R., Orkin S.H., Rodig S.J., Daley G.Q., Rao A. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell. Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokoyama A., Takezawa S., Schule R., Kitagawa H., Kato S. Transrepressive function of TLX requires the histone demethylase LSD1. Mol. Cell. Biol. 2008;28:3995–4003. doi: 10.1128/MCB.02030-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokoyama A., Igarashi K., Sato T., Takagi K., Otsuka I.M., Shishido Y., Baba T., Ito R., Kanno J., Ohkawa Y., Morohashi K., Sugawara A. Identification of myelin transcription factor 1 (MyT1) as a subunit of the neural cell type-specific lysine-specific demethylase 1 (LSD1) complex. J. Biol. Chem. 2014;289:18152–18162. doi: 10.1074/jbc.M114.566448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiki K., Shinoda A., Kano F., Sato R., Shirahige K., Murata M. PPARgamma-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nat. Commun. 2013;4:2262. doi: 10.1038/ncomms3262. [DOI] [PubMed] [Google Scholar]
- 28.Chia D.J. Minireview: mechanisms of growth hormone-mediated gene regulation. Mol. Endocrinol. 2014;28:1012–1025. doi: 10.1210/me.2014-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones P.A., Taylor S.M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 30.de Andres M.C., Kingham E., Imagawa K., Gonzalez A., Roach H.I., Wilson D.I., Oreffo R.O. Epigenetic regulation during fetal femur development: DNA methylation matters. Plos ONE. 2013;8:e54957. doi: 10.1371/journal.pone.0054957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haseeb A., Makki M.S., Haqqi T.M. Modulation of ten-eleven translocation 1 (TET1), Isocitrate Dehydrogenase (IDH) expression, alpha-Ketoglutarate (alpha-KG), and DNA hydroxymethylation levels by interleukin-1beta in primary human chondrocytes. J. Biol. Chem. 2014;289:6877–6885. doi: 10.1074/jbc.M113.512269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu S., Morrison A., Sun H., De Luca F. Nuclear factor-kappaB (NF-kappaB) p65 interacts with Stat5b in growth plate chondrocytes and mediates the effects of growth hormone on chondrogenesis and on the expression of insulin-like growth factor-1 and bone morphogenetic protein-2. J. Biol. Chem. 2011;286:24726–24734. doi: 10.1074/jbc.M110.175364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H., Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko M., An J., Bandukwala H.S., Chavez L., Aijo T., Pastor W.A., Segal M.F., Li H., Koh K.P., Lahdesmaki H., Hogan P.G., Aravind L., Rao A. Modulation of TET2 expression and 5-methylcytosine oxidation by the CXXC domain protein IDAX. Nature. 2013;497:122–126. doi: 10.1038/nature12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material