The authors regret to say that they found a minor mistake in their article.
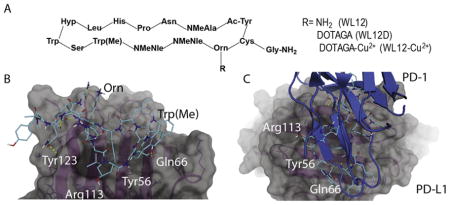
[To assess the binding mode of WL12 to PD-L1, we used the co-crystal structure of human PD-L1 bound to PD-1 (PDB ID: 4ZQK) to dock WL12 in the place of PD-1. Given the structural complexity of the macrocycle, WL12, we first performed a conformational search and the conformers of WL12 were docked into the PD-1 binding site on PD-L1 using Glide. WL12 forms a beta sheet like structure with two hydrogen bonds made between the backbone of the two macrocycle strands (Fig. 1B). This conformation is supported by circular dichroism experiments (Fig. S2). An overlay of the structure of PD-1 with the bound WL12 reveals the similarities in binding mode between the two. The two beta strands of PD-1 that form the binding interface with PD-L1 overlap with the pseudo strands of WL12 (Fig. 1C). The L-leucine of WL12 inserts into the same, small hydrophobic pocket as Ile134 of PD-1, and one of the two norleucine residues aligns with Ile126 of PD-1. In addition to these hydrophobic interactions, a number of hydrogen bonds are present between WL12 and PD-L1. The carboxamide of the asparagine on WL12 forms a hydrogen bond with Tyr123, the glycine amide forms hydrogen bonds with the backbone of Gly120 and the serine hydroxyl interacts with Gln66. The ornithine residue is exposed and does not participate in binding with PD-L1. This suggests that conjugation of a suitable label via amine-coupling methods would not disrupt WL12 binding to PD-L1.]
The authors would like to apologise for any inconvenience caused.


