Abstract
Background
Estrone (E1), the major circulating estrogen in postmenopausal women, promotes estrogen-receptor positive (ER+) breast tumor growth and proliferation. Two major reactions contribute to E1 plasma concentrations, aromatase (CYP19A1) catalyzed E1 synthesis from androstenedione and steroid sulfatase (STS) catalyzed hydrolysis of estrone conjugates (E1Cs). E1Cs have been associated with breast cancer risk and may contribute to tumor progression since STS is expressed in breast cancer where its activity exceeds that of aromatase.
Methods
We performed genome-wide association studies (GWAS) to identify SNPs associated with variation in plasma concentrations of E1Cs, E1, and androstenedione in 774 postmenopausal women with resected early-stage ER+ breast cancer. Hormone concentrations were measured prior to aromatase inhibitor therapy.
Results
Multiple SNPs in SLCO1B1, a gene encoding a hepatic influx transporter, displayed genome-wide significant associations with E1C plasma concentrations and with the E1C/E1 ratio. The top SNP for E1C concentrations, rs4149056 (p = 3.74E–11), was a missense variant that results in reduced transporter activity. Patients homozygous for the variant allele had significantly higher average E1C plasma concentrations than did other patients. Furthermore, three other SLCO1B1 SNPs, not in LD with rs4149056, were associated with both E1C concentrations and the E1C/E1 ratio and were cis-eQTLs for SLCO1B3. GWAS signals of suggestive significance were also observed for E1, androstenedione, and the E1/androstene-dione ratio.
Conclusion
These results suggest a mechanism for genetic variation in E1C plasma concentrations as well as possible SNP biomarkers to identify ER+ breast cancer patients for whom STS inhibitors might be of clinical value.
Keywords: Estrone conjugates, SLCO1B1, SLCO1B3, Genome-wide association studies, Breast cancer, Steroid sulfatase inhibitors
Introduction
Estrogens are important drivers of the growth and proliferation of normal mammary tissue [1, 2] and of estrogen-receptor positive (ER+) breast cancer [3]. Estrone (E1) and estradiol (E2) are the two major active estrogens. E1 is more abundant than E2 in postmenopausal women, the group with the highest incidence of breast cancer in the United States [4]. E1 activates ER, and potentiates estrogenic effects such as the growth and survival of hormone-responsive breast cancer. However, since E1 can be converted to E2 by 17-beta-hydroxysteroid dehydrogenase, the estrogenic effects observed for E1 may be driven by E2 as it has a much higher affinity for ER [5]. There are two major precursors for E1, androstenedione and estrone-3-sulfate (E1S). In post-menopausal women, androstenedione is converted to E1 by aromatase (CYP19A1) in adipose tissue, the adrenal glands, liver, and in breast tumors. E1 can then be conjugated to form E1S [6]. E1S is biologically inactive, but is a “reservoir” for active estrogen since it can be hydrolyzed to E1 by steroid sulfatase (STS) [7]. Circulating E1S concentrations are an order of magnitude higher than those of E1 in post-menopausal women [8–10]. The major site of sulfation and desulfation of estrone is the liver, and this cycling of estrone is believed to be an intracellular mechanism for regulating estrogen activity [11–13].
Epidemiological studies have demonstrated a positive association between circulating E1 or E1S concentrations and breast cancer risk [14–18]. Furthermore, E1 concentrations in breast tumors are higher than in the plasma of postmenopausal women [10, 19, 20]. This may be due to the in situ biosynthesis of estrogens from both androgens and conjugated estrogen precursors. Aromatase inhibitors (AI), the major endocrine therapy for ER+ tumors in the adjuvant setting [21], target only one source of estrogens, i.e., their synthesis from androgens. That leaves another source uninhibited, i.e., synthesis from E1S catalyzed by STS. Therefore, drugs that target this source of estrogens, i.e., STS inhibitors [22–25], have been developed as an additional approach to the treatment of ER+ breast cancer.
In this era of “Precision Medicine,” understanding factors that govern individual variation in plasma concentrations of estrogens might help us understand their role in individual variation in risk for breast cancer occurrence and/or recurrence. Together with factors such as body mass index (BMI), age, and diet [14], genetics also influences circulating estrogen concentrations. Polymorphisms in genes such as CYP19A1, ESR1, and SHBG have all been shown to affect circulating estrogen concentrations [26–28]. However, application of an agnostic approach such as GWAS might help us to identify additional genetic factors. For example, Liu et al. [29] in a GWAS for E2 plasma concentrations in post-menopausal women with ER+ breast cancer, identified a novel genome-wide significant SNP signal in TSPYL5, and demonstrated that TSPYL5 encoded a transcription factor that regulated the expression of CYP19A1 in a SNP-dependent fashion. Most published GWAS for estrogens in post-menopausal women have focused on E2 [29, 30], rather than the more abundant E1, and its precursor E1S. No GWAS results have been published for plasma E1, E1S, or androstenedione in postmenopausal women. We hypothesized that genetic factors might contribute to variation in the concentrations of plasma hormones in the estrone biosynthesis and metabolism pathways, so we performed a comprehensive series of GWAS for plasma concentrations of these hormones in postmenopausal women with ER+ breast cancer prior to adjuvant AI therapy. We also performed GWAS of ratios of hormones and their precursors as phenotypes to “isolate” specific reactions in this pathway. The studies described subsequently identified genome-wide significant SNP signals associated with variation in plasma E1C and the E1C/E1 ratio across the SLCO1B1 gene, a gene encoding OATP1B1, an E1S transporter. These observations might ultimately help us identify individualized approaches for the therapy and/or prevention of ER+ breast cancer.
Materials and methods
The Mayo/MDA/MSK AI pharmacogenomics study
Details of the Mayo/MDA/MSK study were described previously [29, 31, 32]. Briefly, our study population consisted of 774 postmenopausal women with resected early-stage ER+ breast cancer. These patients were recruited from three Mayo Clinic locations (i.e., Rochester, MN, Jacksonville FL, and Scottsdale, AZ) as well as from MD Anderson Cancer Center and Memorial Sloan Kettering Cancer Center. Demographic information about the study population is listed in Table 1. Prior to the initiation of anastrozole therapy, plasma samples were collected and assayed for E2, E1, and total E1 conjugates (E1C), which included both sulfate and glucuronide conjugates (with E1 sulfate as the predominant conjugate), testosterone, and androstenedione using gas chromatography–tandem mass spectrometry. DNA from the subjects was genotyped for over 600,000 SNPs using the Ilumina Human610-Quad BeadChip. SNPs with minor allele frequencies (MAFs) <0.01 and/or Hardy–Weinberg equilibrium p values <1.0E–06 were excluded from the analyses. Approximately 8 million SNPs were imputed using reference populations from the 2010 release of the 1000 Genomes Project. Reference SNPs with MAF values <0.005 were removed prior to imputation performed using BEAGLE v3.3.1 [33]. Imputed SNPs with dosage r2 values <0.3 and/or MAF values <0.01 were excluded from analyses. See supplemental information for details.
Table 1.
Patient Characteristics for 774 postmenopausal women with ER+ breast cancer
| Total | N = 774 |
|---|---|
| Mean age, years (range) | 67.9 (39.7–96.0) |
| Mean BMI, kg/m2 (range) | 27.97 (16.0–57.2) |
| Current smoker | |
| No | 720 (93%) |
| Yes | 43 (5.6%) |
| Unknown | 11 (1.4%) |
| ER/PgR status | |
| Positive/negative | 152 (20%) |
| Positive/positive | 614 (79%) |
| Negative/positive | 7 (1.0%) |
| Positive/unknown | 1 (0.1%) |
| HER2 status | |
| Negative | 669 (86.4%) |
| Positive | 91 (11.8%) |
| Unknown | 14 (1.8%) |
| Prior tamoxifen | |
| No | 692 (89%) |
| Yes | 82 (11%) |
| Prior hormone replacement therapy | |
| No | 295 (38.1%) |
| Yes | 364 (47.0%) |
| Unknown | 115 (14.9%) |
| Prior chemotheraphy | |
| No | 463 (60%) |
| Yes | 311 (40%) |
| Race | |
| European ancestry | 707 (91%) |
| African ancestry | 52 (7%) |
| Asian ancestry | 15 (2%) |
| Recruitment site | |
| MCA | 78 (10%) |
| MCF | 32 (4%) |
| MCR | 182 (24%) |
| MDA | 349 (45%) |
| MSK | 133 (17%) |
Recruitment sites MCA Mayo Clinic Arizona, MCF Mayo Clinic Florida, MCR Mayo Clinic Rochester, MDA MD Anderson Cancer Center, MSK Memorial Sloan Kettering Cancer Center
Statistical analyses
The phenotypes analyzed were plasma concentrations of E1, E1C, androstenedione, and the ratios of E1/androstenedione and E1C/E1. These values were analyzed as continuous quantitative variables. Van Der Waerden transformations [34] were applied to phenotypes with skewed distributions to satisfy the assumption of a Gaussian distribution. To identify potential confounders, univariate linear regression analyses were performed with the hormone phenotypes and clinical variables such as BMI, age at diagnosis, recruitment site, smoking history, and eigenvectors from a principal component analysis (PCA) of population substructure. Covariates with the association of p values of 0.01 or lower were included in a multivariate linear regression model for the GWAS. For each phenotype, two analyses were performed, one using an unadjusted model without covariates, and another in which we adjusted for relevant covariates including eigenvectors from a PCA substructure analyses. The best model was determined by examining Quantile–Quantile (Q–Q) plots of the observed association of p values for adherence to the null distribution. This strategy was used for the genotyped SNP analyses. The best model emerging from the application of this approach was then applied to the analyses of the imputed SNPs. We used a p value of 5.0E–08 as the threshold for genome-wide significance. All analyses were performed using R statistical computing software (v3.0.2) and Plink (v1.07) [35].
Results
Associations with clinical variables
Associations among E1, E1C, androstenedione, E1C/E1, and E1/androstenedione and relevant clinical variables are listed in Table 2. E1 and E1C displayed significant positive associations with BMI as did the E1/androstenedione ratio. E1C/E1 was not associated with BMI. Age at the time of diagnosis was positively associated with E1 and the E1/androstenedione ratio, and was negatively associated with E1C/E1. Age did not appear to be associated with the plasma concentrations of androstenedione or E1C. Smoking status was only associated with E1C concentrations, with smokers having a lower mean E1C concentration than did non-smokers (p = 0.01). Women with prior exposure to hormone replacement therapy had lower mean E1C concentrations than did those who had not been treated with those agents (p = 0.01). With regard to race, the only significant association observed in our data involved the E1/androstenedione ratio. African-American women had a higher mean E1/androstenedione than did patients of European ancestry (p = 1.0E–03) (Table 2).
Table 2.
Association between plasma hormone concentrations or ratios of concentrations and clinical and demographic variables among 774 postmenopausal women with ER+ breast cancer
| Variable | Androstenedione | Estrone | Estrone Conjugates | E1C/E1 | E1/Androstenedione | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| (E1) | (E1C) | |||||||||
|
|
|
|
|
|
||||||
| β | P | β | P | β | P | β | P | β | P | |
| BMI | 0.01 | 0.03 | 0.06 | 6.60E–24 | 0.04 | 3.30E–13 | 0.001 | 0.84 | 0.05 | 8.50E–19 |
| Age at treatment | −0.01 | 0.2 | 0.01 | 3.00E–03 | 0 | 0.91 | −0.01 | 0.01 | 0.02 | 1.60E–05 |
| Her2 status | ||||||||||
| Positive versus negative | 0.09 | 0.42 | 0.02 | 0.87 | −0.11 | 0.33 | −0.15 | 0.2 | −0.13 | 0.26 |
| Prior chemotherapy | ||||||||||
| Yes versus no | −0.09 | 0.2 | −0.11 | 0.14 | −0.02 | 0.8 | 0.07 | 0.35 | −0.05 | 0.52 |
| Prior tamoxifen use | ||||||||||
| Yes versus no | 0.05 | 0.64 | −0.08 | 0.51 | 0.14 | 0.23 | 0.3 | 0.01 | −0.18 | 0.12 |
| Prior HRT use | ||||||||||
| Yes versus no | −0.15 | 0.05 | −0.15 | 0.05 | −0.21 | 0.01 | −0.12 | 0.15 | 0.01 | 0.95 |
| Current smoker | ||||||||||
| Yes versus no | 0.12 | 0.43 | −0.25 | 0.11 | −0.43 | 0.01 | −0.33 | 0.04 | −0.41 | 0.01 |
| STRUCTURE-defined race | ||||||||||
| European ancestry | reference | |||||||||
| African ancestry | −0.26 | 0.07 | 0.19 | 0.18 | 0.01 | 0.94 | −0.15 | 0.29 | 0.48 | 1.00E–03 |
| Asian ancestry | −0.23 | 0.39 | −0.13 | 0.62 | −0.06 | 0.81 | −0.05 | 0.85 | 0.14 | 0.6 |
β values represent the coefficient from univariate linear regression analyses of the Van der Waerden transformed hormone quantitative phenotypes. Positive β values represent the positive associations between phenotype and covariate. Negative β values represent negative associations between phenotype and covariate. Bolded P values are statistically significant
E1C GWAS and the SLCO1B1 SNP signal
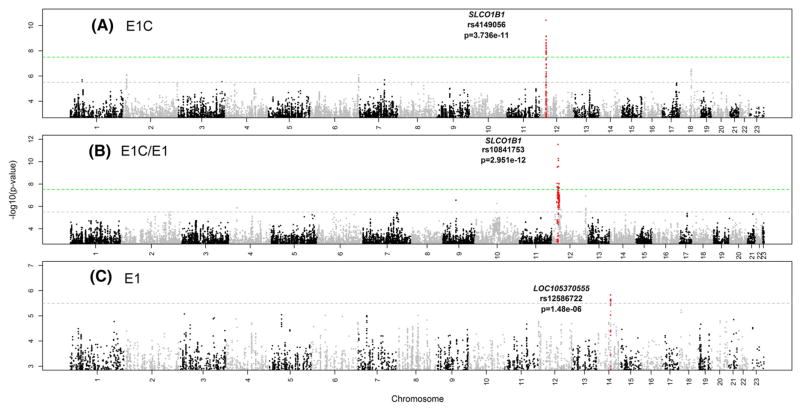
In the GWAS for circulating E1C concentrations (Q–Q plots in Supplementary Fig. 1), we observed a genome-wide significant SNP signal across SLCO1B1, a gene that encodes a solute carrier (SLC) transporter that is highly expressed in liver (Figs. 1A, 2A, Supplementary Table 1). The minor allele (C) of the top SNP, rs4149056 (MAF = 0.14), was associated with increased E1C concentrations with a p value of 3.74E–11 (Table 3). This non-synonymous (ns) SNP (521T>C) resulted in an amino acid change from valine to alanine (Val174Ala) at residue 174 of the OATP1B1 protein encoded by SLCO1B1, resulting in a transporter with reduced activity [36]. OATP1B1 is involved in hepatic uptake from the systemic circulation of both endogenous molecules such as E1C and xenobiotics such as statins [37]. Given the reduced activity conferred by this SNP, patients with the C allele had higher concentrations of E1C in the circulation compared to those with the T allele (CC: mean = 873.6 pg/mL, TT: mean = 285.6 pg/mL) (Fig. 3). Higher circulating E1C results in greater systemic exposure to E1C by tissues such as the breast.
Fig. 1.
Manhattan plots showing GWAS results for plasma concentrations of (A). Estrone Conjugates (E1C), (B). the ratio of E1C/E1, (C). Estrone (E1) in 774 postmenopausal women with ER+ breast cancer prior to the initiation of endocrine therapy. Dashed lines represent thresholds for genome-wide significance (p ≤ 5.0E–08, green) and suggestive significance (p ≤ 5.0E–06, gray)
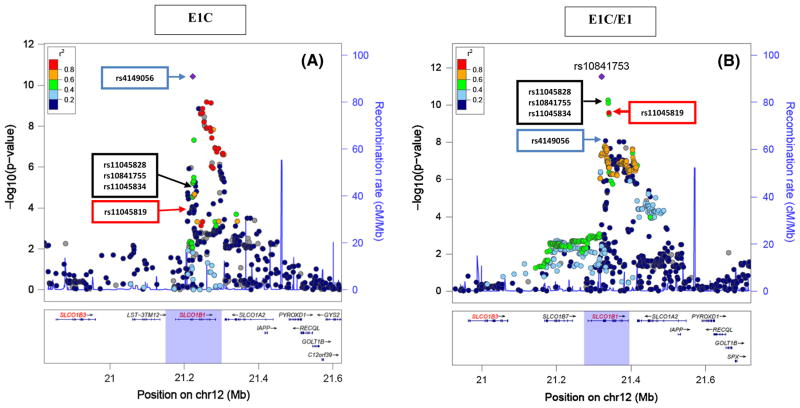
Fig. 2.
Regional plots of the genome-wide significant SNP signal across SLCO1B1 from (A). GWAS of E1C which shows the top SNP, rs4149056 (blue arrow), a missense variant that results in a plasma membrane transporter with reduced activity, (B) GWAS of E1C/E1 which shows the top SNP, rs10841753 (purple point), which is an intronic variant that is in high LD (R2 = 1.00, CEU population, distance = 8443 bp), with another missense variant, rs11045819 (red arrow) in populations of European descent, but it is a signal independent from rs4149056 (R2 = 0.013, CEU population, distance = 101179 bp). The black arrow highlights 3 SNPs (rs11045828, rs10841755, and rs11045834) that are cis-eQTLs for SLCO1B3 and are associated with breast cancer risk
Table 3.
List of top and selected SNP signals from the GWAS analyses
| Phenotype | SNP | CHR | BP | Minor allele |
MAF | Effect size |
Direction of effecta |
SE | P | Gene | Gene location |
SNP function |
Cis-eQTL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E1C | rs4149056 | 12 | 21222816 | C | 0.136 | 0.50 | ↑ | 0.075 | 3.74E–11 | SLCO1B1 | Exon | Missense | – |
| E1C | rs2900478 | 12 | 21368797 | A | 0.14 | 0.48 | ↑ | 0.077 | 6.87E–10 | SLCO1B1 | Intron | – | – |
| E1C | rs4149083 | 12 | 21380630 | T | 0.14 | 0.48 | ↑ | 0.077 | 7.29E–10 | SLCO1B1 | Intron | – | – |
| E1C | rs12367888 | 12 | 21347021 | T | 0.15 | 0.45 | ↑ | 0.073 | 1.40E–09 | SLCO1B1 | Intron | – | – |
| E1C | rs58310495 | 12 | 21357711 | T | 0.15 | 0.45 | ↑ | 0.073 | 1.41E–09 | SLCO1B1 | Intron | – | – |
| E1C/E1 | rs10841753 | 12 | 21321370 | C | 0.208 | −0.43 | ↓ | 0.061 | 2.95E–12 | SLCO1B1 | Intron | – | – |
| E1C/E1 | rs11045828 | 12 | 21338231 | A | 0.307 | −0.35 | ↓ | 0.052 | 5.33E –11 | SLCO1B1 | intron | – | SLCO1B3 |
| E1C/E1 | rs10841755 | 12 | 21338889 | A | 0.306 | −0.35 | ↓ | 0.052 | 8.13E–11 | SLCO1B1 | Intron | – | SLCO1B3 |
| E1C/E1 | rs11045834 | 12 | 21232363 | A | 0.302 | −0.34 | ↓ | 0.053 | 3.08E –10 | SLCO1B1 | intron | – | SLCO1B3 |
| E1C/E1 | rs4149056 | 12 | 21222816 | C | 0.136 | 0.42 | ↑ | 0.071 | 8.67E–09 | SLCO1B1 | Exon | Missense | – |
| E1C/E1 | rs11045819 | 12 | 21329813 | A | 0.158 | −0.38 | ↓ | 0.068 | 3.87E–08 | SLCO1B1 | Exon | Missense | – |
| E1/Andr | rs10953024 | 7 | 90504067 | C | 0.088 | −0.49 | ↓ | 0.100 | 1.45E–06 | CDK14 | Intron | – | STEAP1 |
| E1/Andr | rs1453308 | 2 | 237186325 | A | 0.198 | −0.33 | ↓ | 0.069 | 2.06E–06 | ASB18 | 5′upstream | – | – |
| E1/Andr | rs16952550 | 18 | 7807948 | C | 0.171 | −0.33 | ↓ | 0.069 | 2.09E–06 | PTPRM | Intron | – | – |
| E1/Andr | rs10224911 | 7 | 90539427 | A | 0.083 | −0.49 | ↓ | 0.102 | 2.11E–06 | CDK14 | Intron | – | – |
| E1/Andr | rs17601876 | 15 | 51553909 | A | 0.500 | 0.24 | ↑ | 0.052 | 3.50E–06 | CYP19A1 | Intron | – | CYP19A1 |
| E1 | rs12586722 | 14 | 70890140 | A | 0.155 | 0.30 | ↑ | 0.063 | 1.48E–06 | LOC105370555 | 5′ upstream | – | ADAM20P1, AL357153.1 |
| E1 | rs12436808 | 14 | 70904607 | C | 0.156 | 0.30 | ↑ | 0.063 | 2.19E–06 | LOC105370555 | Intron | – | ADAM20P1, AL357153.1 |
| E1 | rs3764182 | 14 | 70919161 | A | 0.156 | 0.30 | ↑ | 0.063 | 2.21E–06 | ADAM21 | Intron | – | ADAM20P1, AL357153.1 |
| E1 | rs114864695 | 14 | 70924955 | A | 0.162 | 0.31 | ↑ | 0.064 | 2.55E–06 | ADAM21 | Exon | Missense | ADAM20P1, AL357153.1 |
| Andr | rs57712673 | 19 | 14885800 | A | 0.349 | 0.36 | ↑ | 0.067 | 1.39E–07 | EMR2/ADGRE2 | Intron | – | EMR2 |
| Andr | rs1108015 | 19 | 14886784 | C | 0.354 | 0.35 | ↑ | 0.067 | 2.10E–07 | EMR2/ADGRE2 | Intron | – | EMR2 |
| Andr | rs59964204 | 7 | 36939322 | A | 0.096 | −0.45 | ↓ | 0.087 | 3.19E–07 | ELMO1 | Intron | – | – |
| Andr | rs34346910 | 10 | 88427209 | C | 0.111 | 0.49 | ↑ | 0.097 | 5.67E–07 | LDB3 | 5′upstream | – | – |
| Andr | rs6988985 | 8 | 144007104 | C | 0.461 | 0.25 | ↑ | 0.050 | 6.65E–07 | CYP11B2 | 5′ upstream | – | LYNX1, LY6E |
| Andr | rs3802230 | 8 | 143992864 | A | 0.433 | 0.24 | ↑ | 0.047 | 6.88E–07 | CYP11B2 | 3′UTR | – | LYNX1, LY6E |
Expression quantitative trait loci (eQTL) information was obtained from the GTEx database. An eQTL is a variant or SNP that is associated with the mRNA expression of a given gene
Direction of effect = (↑) indicates that the minor allele of the SNP is associated with an increase in the hormone or hormone ratio. Direction of effect = (↓) indicates that the minor allele of the SNP is associated with a decreased in the hormone or hormone ratio
Fig. 3.
Mean plasma E1C concentrations for each genotype of rs4149056 (with 95% confidence intervals shown) in postmenopausal women with ER+ breast cancer. Red points with red horizontal bars indicate the mean values for each genotype
As anticipated, there was a strong association between plasma concentrations of E1C and those of its precursor, E1 (Pearson’s r = 0.63, p = 5.15E–83). However, regression analyses showed that there was no direct effect of the rs4149056 nsSNP on E1 concentrations, but the positive relationship between E1 and EIC concentrations was modified by rs4149056 (Supplementary Table 2). Specifically, individuals homozygous for the variant allele had, on average, a smaller increase in E1 per unit increase in E1C concentrations, whereas those homozygous for the wildtype genotype had, on average, a greater increase in E1 per unit increase in E1 (Supplementary Fig. 2)—compatible with the functional effect of the rs4149056 C allele. This suggests that even when there was a similar quantitative increase in E1C among our subjects, the subsequent increase in E1 was smaller in women who were homozygous variant for the SLCO1B1 missense SNP. Perhaps, this might be due to fewer EIC molecules entering the liver for desulfation in subjects with decreased OATP1B1 transport activity.
E1C/E1 GWAS and the SLCO1B1 SNP signal
The Manhattan plot for the GWAS of the E1C/E1 ratio is shown in Fig. 1B (Q–Q plots in Supplementary Fig. 3). Numerous SNPs in this GWAS achieved genome-wide significance (Supplementary Table 3) and they all mapped to SLCO1B1 (Fig. 2). The linkage disequilibrium (LD) structure among the top SNPs suggested the existence of two independent signals in the SLCO1B1 gene. The rs4149056 nsSNP that was the top GWAS signal for E1C concentration also displayed a genome-wide significant association with E1C/E1 (p = 8.67E–09) (Table 3). However, this SNP was not in LD (r2 = 0.013) with the top SNP for the E1C/E1 GWAS, rs10841753 (p = 2.95E–12), (see Fig. 2B) an intronic SNP with a minor allele C that was associated with decreased E1C/E1. This SNP was not an expression quantitative trait locus (eQTL) for SLCO1B1, but it was in perfect LD (r2 = 1) with another SLCO1B1 missense variant, rs11045819, that was also associated with E1C/E1 (p = 3.87E–08) (Table 3). This second missense variant, rs11045819 (463C>A), resulted in a proline to threonine (Pro155Thr) change in the protein encoded by SLCO1B1 and had a minor allele (A) frequency of 15% in our sample of women who were primarily of European ancestry. This variant was absent in populations of East Asian ancestry and had a MAF of 6% in populations of African ancestry based on 1000 Genomes data [38]. Unlike rs4149056, rs11045819 does not appear to alter the activity of the OATP1B1 transporter [36].
Three of the top SNPs from this GWAS, rs11045828 (p = 5.33E–11), rs10841755 (p = 8.13–11), and rs11045834 (p = 3.08E–11) (Table 3), mapped to an SLCO1B1 intron and were significant eQTLs for the nearby SLCO1B3 gene, but not for SLCO1B1 (GTEx) [39]. For each of these SNPs, the variant allele was associated with decreased E1C/E1 and with decreased expression of SLCO1B3 (rs11045828: effect size for expression = −0.4, p = 1.6E–06; rs10841755: effect size = −0.4, p = 1.7E–06; 11045834: effect size = −0.38 p = 4.7E–06). SLCO1B3 also encodes a hepatic influx transporter, maps 200 kb upstream of SLCO1B1, and transports E1Cs [37]. All three of the SNPs that were eQTLs for SLCO1B3 were in high LD [r2 = 1.0] with each other. In addition to being associated with E1C/E1, these SNPs were also significantly associated with decreased E1C concentrations (rs11045828: p = 5.68E–06, rs10841755: p = 8.68E–06, 11045834: p = 1.61E–06) but not with E1 concentrations. Since SLCO1B3 encodes an influx transporter one might anticipate that SNPs associated with decreased expression of SLCO1B3 might be associated with increased E1C concentrations, but our data clearly demonstrate the opposite.
E1 GWAS
We also performed a GWAS for E1 concentrations (Fig. 1C, Supplementary Fig. 4) and the top SNP was rs12586722 which mapped to LOC105370555 on chromosome 14 (Table 3, Supplementary Table 4), but this SNP did not achieve genome-wide significance. The minor allele A (MAF = 0.16) for this SNP was associated with increased E1 concentrations (p = 1.48E–06). This SNP was a cis-eQTL for ADAM20P1, a nearby processed pseudogene [39]. The minor allele for the SNP was associated with decreased expression. Another SNP of possible interest in this region was rs114864695 (p = 2.55E–06), a missense variant in exon 2 of ADAM21 (Table 3). It resulted in a 739A>C change in the genomic sequence and a Lys247Gln change in the encoded amino acid sequence. The functional consequences of this missense variant are unknown. According to GTEx, this SNP is also a cis-eQTL for ADAM21P1, and the C allele is associated with increased expression of this putative pseudogene (effect size = −0.49, p = 1.8E–09).
E1/androstenedione ratio GWAS
Although there were no SNPs that reached genome-wide significance in the E1/androstenedione GWAS (Supplementary Fig. 5), there were several SNPs with suggestive significance. Those polymorphisms included SNPs in or near CDK14, ASB18, PTPRM, and CYP19A1 (Table 3, Supplementary Table 5). CYP19A1 was of interest since it encodes aromatase, the enzyme that catalyzes the conversion of androstenedione to E1 and of testosterone to E2. The minor allele (G) for the top SNP in this gene, rs17601876 (p = 3.50E–06), was associated with an increased E1/androstenedione (Table 3). Several of the SNPs from this GWAS were cis-eQTLs for CYP19A1, including rs7175531 (MAF = 0.34, p = 6.18E–05) and rs2414095 (MAF = 0.34, p = 7.02E–05). Minor alleles for both SNPs were associated with decreased E1/androstenedione among our patients and with decreased CYP19A1 expression [39]. Since decreased CYP19A1 would result in decreased conversion of androstenedione to E1, the directional effects for both relationships were consistent.
Androstenedione GWAS
In our androstenedione GWAS (Supplementary Fig. 6c), the main signal included intronic SNPs for EMR2/ADGRE2, with rs57712673 having the lowest p value (p = 1.39E–07) (Table 3). The minor allele for this SNP (A) was associated with increased androstenedione concentrations as well as decreased expression of EMR2 (effect size = −0.2, p = 8.0E–07). Another signal of interest involved a cluster of SNPs on chromosome 8 within and flanking CYP11B1 and CYP11B2 (Supplementary Fig. 6c, Supplementary Table 6), genes encoding enzymes in the steroid hormone biosynthesis pathway. Within this region, the minor allele (T) of the SNP with the lowest p value, rs6988985, was associated with increased androstenedione (p = 6.65E–07) (Table 3). CYP11B1 expression is regulated by adrenocorticotrophic hormone (ACTH) which regulates the quantity of 17-hydroxyprogesterone available for conversion to androstenedione in the adrenal glands.
Discussion
The major signal observed in our E1C GWAS was a missense variant in SLCO1B1, rs4149056. GWAS for the ratio of E1C/E1 yielded the same signal, as well as an additional independent signal that included another SLCO1B1 missense variant, rs11045819. These two nsSNPs are approximately 1700 bp apart and map to adjacent SLCO1B1 exons, 5 and 6, but they are not in LD with each other in European, African, or Asian populations. The variant allele for rs4149056 (521T>C) results in a Val174Ala amino acid change in the fourth transmembrane segment of the OATP1B1 protein. This variant is a well-studied pharmacogenomic marker identified initially during a GWAS for statin-induced myopathy [40]. This variant has also been associated with concentrations of bilirubin [41], sex hormone-binding globulin [42], and various metabolites [43]. OATP1B1, the transporter encoded by SLCO1B1, is expressed primarily in hepatic tissue, and transports numerous endogenous compounds, including E1Cs (E1S), from the systemic circulation into hepatocytes [37]. Functional studies have shown that the rs4149056 variant allele is associated with decreased membrane expression of the OATP1B1 transporter, and thus, reduced transport activity [36]. Reduced transport would increase the plasma concentration and systemic exposure to OATP1B1 substrates, which is compatible with our observations. In our patients, those with one or more copies of the rs4149056 C variant allele had higher mean plasma concentrations of E1C than patients homozygous for the major allele (CC: 873.6 pg/mL, TT: 285.6 pg/mL) (Fig. 3). Thus, women who carried the minor allele had increased systemic exposure to E1Cs.
The second SLCO1B1 missense variant, rs11045819 (463C>A), resulted in a Pro155Thr change in the extra-cellular loop region of OATP1B1. However, in vitro studies failed to show that 463C>A affects either protein expression for the encoded transporter or its uptake activity [32]. In our patients, the variant allele (A) for this SNP was associated with decreased E1C/E1, and thus a lower exposure to circulating E1C. There were no patients in our sample who were homozygous for variants for both mis-sense SNPs. The 463C>A variant might be significantly associated with E1C/E1 because it is in modest LD (r2 = 0.517) with 3 intronic SNPs that displayed statistically significant associations with the E1C/E1 ratio, and which were also cis-eQTLs for SLCO1B3. That gene encodes OATP1B3, a solute carrier transporter that is also capable of transporting E1Cs, but with lower transport efficiency than OATP1B1 [44].
Both the non-synonymous and other variants in SLCO1B1 associated with elevated plasma E1C concentrations may be clinically important since elevated E1C concentrations have previously been linked to increased breast cancer risk in postmenopausal women [15, 16]. In addition, ten SNPs in SLCO1B1 were previously shown to be associated with increased risk for breast cancer in postmenopausal women in the California Teachers Study (CTS) [45]. Although none of those SNPs showed genome-wide significant associations with E1C concentrations in our data, seven were nominally associated, and one SNP (rs4149058) exhibited suggestive statistical significance (p = 5.02E–06). That SNP was associated with a 23% increase in breast cancer risk in the CTS and with increased E1C concentrations in our study. Rs4149058 was in modest LD (r2 = 0.59) with the rs4149056 missense variant that was the top SNP signal in our E1C GWAS. Rs4149056 was not genotyped in the CTS, nor were most of our variants that displayed genome-wide significant associations with E1C or E1C/E1.
The increase in systemic exposure to E1C by ER+ breast cancer patients who were homozygous variant for rs4149056 may mean that their tumors take up more E1C, a precursor for E1. Since SLCO1B1 is not expressed in either normal or cancerous breast tissue according to the GTEx (39) and TCGA databases (46–47), one can assume that this missense SNP will not affect E1C transport into breast tumor tissue. Other OATPs that have the capacity to transport E1S (33), such as OATP2B1, OATP3A1, and OATP4A1, are highly expressed in breast tumors. Those OATPs tend to be ubiquitously expressed, but have lower hepatic expression than does OATP1B1 (35). Furthermore, TCGA data indicate that the breast tumor expression of these genes is comparable to key breast cancer genes such as ESR1 and CCND1 [46, 47]. Hypothetically, elevated circulating E1Cs could indirectly augment the proliferation of breast tumors that have the capacity to take up and hydrolyze E1Cs to form E1. In kinetic studies, ER+ cell lines have much higher rates of E1C uptake than do normal breast cell lines [48, 49]. Furthermore, ER+ cell lines had lower Km values for OATP-mediated transport of E1Cs compared to other breast cancer cell lines, indicating that ER+ cell lines may have greater transport efficiency [49]. Higuchi et al. [50] demonstrated up-regulation of the expression of the OATP1B1, OATP1A2, OATP4A2, and OATP5A1 transporters in a letrozole-resistant MCF7 breast cancer cell line. Those cell lines were shown to proliferate preferentially in E1C supplemented media, a situation analogous to elevated circulating plasma concentrations. They also showed that treatment with an STS inhibitor reduced proliferation of the letrozole-resistant cell lines, results that support the possible clinical application of STS inhibitors. Further highlighting the biological and potential clinical importance of the STS-mediated pathway for estrogen production, Utsumi et al. [51] demonstrated that patients with high steroid sulfatase [STS] expression in breast tumors had a higher risk of recurrence than those with low expression. This implies that higher plasma E1C exposure, higher tumor E1C uptake, and higher STS expression might contribute to risk for tumor recurrence. Therefore, STS inhibitors might prove beneficial for ER+ breast cancer patients with high circulating E1C during adjuvant AI therapy. Selection of patients for STS inhibitor trials has been a challenge, and it is possible that rs4149056 could help address this issue. We assume that patients with high circulating concentrations of E1C might benefit from STS inhibition. Thus, patients who are homozygous variant for rs4149056 and are on an aromatase inhibitor could be selected for clinical trials of STS inhibitor as add-on therapy.
In summary, our GWAS of plasma E1 and its major precursors in postmenopausal women with ER+ breast cancer identified SNP signals across SLCO1B1 that point to a mechanism that might contribute to increased tumor exposure to estrogen. The SLCO1B1-encoded hepatic influx transporter regulates circulating concentrations of E1C, a “reservoir” for E1 that can increase the exposure of patients to active estrogens after hydrolysis of the conjugates. This pathway remains active in patients who are on aromatase inhibitor therapy. We found that two independent missense variants in SLCO1B1 were associated with either increased or decreased circulating E1C concentrations and/or E1C/E1 ratio—results that require replication in future studies. These variants may provide a means by which to identify ER+ breast cancer patients on AI therapy who are at increased risk for tumor recurrence because of elevated tumor E1C exposure, and who might benefit from additional therapy such as STS inhibition.
Supplementary Material
Acknowledgments
This work was supported, in part, by National Institutes of Health Grants U19 GM61388 (The Pharmacogenomics Research Network), R01 GM28157, U01 HG005137, R01 CA138461, R01 CA133049, and P50 CA166201 (Mayo Clinic Breast Cancer Specialized Program of Research Excellence), The RIKEN Center for Integrative Medical Sciences, the Biobank Japan Project funded by the Ministry of Education, Culture, Sports, Science and Technology (Japan), and a generous gift from the Prospect Creek Foundation, ClinicalTrials.gov study number NCT00283608. Tanda Dudenkov was supported by the National Institute of General Medical Sciences (T32 GM 65841).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-017-4243-3) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest The authors declare no potential conflicts of interest.
References
- 1.Silberstein GB, Van Horn K, Shyamala G, Daniel CW. Essential role of endogenous estrogen in directly stimulating mammary growth demonstrated by implants containing pure antiestrogens. Endocrinology. 1994;134(1):84–90. doi: 10.1210/endo.134.1.8275973. [DOI] [PubMed] [Google Scholar]
- 2.Laidlaw IJ, Clarke RB, Howell A, Owen AW, Potten CS, Anderson E. The proliferation of normal human breast tissue implanted into athymic nude mice is stimulated by estrogen but not progesterone. Endocrinology. 1995;136(1):164–171. doi: 10.1210/endo.136.1.7828527. [DOI] [PubMed] [Google Scholar]
- 3.Hurd C, Khattree N, Dinda S, Alban P, Moudgil VK. Regulation of tumor suppressor proteins, p53 and retinoblastoma, by estrogen and antiestrogens in breast cancer cells. Oncogene. 1997;15(8):991–995. doi: 10.1038/sj.onc.1201233. [DOI] [PubMed] [Google Scholar]
- 4.Kohler BA, Sherman RL, Howlader N, Jemal A, Ryerson AB, Henry KA, et al. Annual report to the nation on the status of cancer, 1975–2011, featuring incidence of breast cancer subtypes by race/ethnicity, poverty, and state. J Natl Cancer Inst. 2015 doi: 10.1093/jnci/djv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rich RL, Hoth LR, Geoghegan KF, Brown TA, LeMotte PK, Simons SP, Hensley P, Myszka DG. Kinetic analysis of estrogen receptor/ligand interactions. Proc Natl Acad Sci. 2002;99:8562–8567. doi: 10.1073/pnas.142288199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasqualini JR, Gelly C, Nguyen BL, Vella C. Importance of estrogen sulfates in breast cancer. J Steroid Biochem Mol Biol. 1989;34(1–6):155–163. doi: 10.1016/0022-4731(89)90077-0. [DOI] [PubMed] [Google Scholar]
- 7.Ruder HJ, Loriaux L, Lipsett MB. Estrone sulfate: production rate and metabolism in man. J Clin Investig. 1972;51(4):1020–1033. doi: 10.1172/JCI106862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noel CT, Reed MJ, Jacobs HS, James VH. The plasma concentration of oestrone sulphate in postmenopausal women: lack of diurnal variation, effect of ovariectomy, age and weight. J Steroid Biochem Mol Biol. 1981;14(11):1101–1105. doi: 10.1016/0022-4731(81)90039-x. [DOI] [PubMed] [Google Scholar]
- 9.Vermeulen A, Deslypere JP, Paridaens R, Leclercq G, Roy F, Heuson JC. Aromatase, 17 beta-hydroxysteroid dehydrogenase and intratissular sex hormone concentrations in cancerous and normal glandular breast tissue in postmenopausal women. Eur J Cancer Clin Oncol. 1986;22(4):515–525. doi: 10.1016/0277-5379(86)90121-5. [DOI] [PubMed] [Google Scholar]
- 10.Pasqualini JR, Chetrite G, Blacker C, Feinstein MC, Delalonde L, Talbi M, Maloche C. Concentrations of estrone, estradiol, and estrone sulfate and evaluation of sulfatase and aromatase activities in pre- and postmenopausal breast cancer patients. J Clin Endocrinol Metab. 1996;81(4):1460–1464. doi: 10.1210/jcem.81.4.8636351. [DOI] [PubMed] [Google Scholar]
- 11.Roberts KD, Rochefort JG, Bleau G, Chapdelaine A. Plasma estrone sulfate levels in postmenopausal women. Steroids. 1980;35:179–187. doi: 10.1016/0039-128x(80)90101-4. [DOI] [PubMed] [Google Scholar]
- 12.Raftogianis R, Creveling C, Weinshilboum R, Weisz J. Estrogen metabolism by conjugation. J Natl Cancer Inst Monogr. 2000;27(113):24. doi: 10.1093/oxfordjournals.jncimonographs.a024234. [DOI] [PubMed] [Google Scholar]
- 13.Iwamori M. Estrogen Sulfatase. In: B-M, editor. Enzymology. Academic Press; 2005. pp. 293–302. [DOI] [PubMed] [Google Scholar]
- 14.Key T, Appleby P, Barnes I, Reeves G, Endogenous H Breast Cancer Collaborative Group. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J Natl Cancer Inst. 2002;94(8):606–616. doi: 10.1093/jnci/94.8.606. [DOI] [PubMed] [Google Scholar]
- 15.Missmer SA, Eliassen AH, Barbieri RL, Hankinson SE. Endogenous estrogen, androgen, and progesterone concentrations and breast cancer risk among postmenopausal women. J Natl Cancer Inst. 2004;96(24):1856–1865. doi: 10.1093/jnci/djh336. [DOI] [PubMed] [Google Scholar]
- 16.Tworoger SS, Rosner BA, Willett WC, Hankinson SE. The combined influence of multiple sex and growth hormones on risk of postmenopausal breast cancer: a nested case-control study. Breast Cancer Res. 2011;13(5):R99. doi: 10.1186/bcr3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyoshi Y, Tanji Y, Taguchi T, Tamaki Y, Noguchi S. Association of serum estrone levels with estrogen receptor-positive breast cancer risk in postmenopausal Japanese women. Clin Cancer Res. 2003;9(6):2229–2233. [PubMed] [Google Scholar]
- 18.Kaaks R, Rinaldi S, Key TJ, Berrino F, Peeters PHM, Biessy C, et al. Postmenopausal serum androgens, oestrogens and breast cancer risk: the European prospective investigation into cancer and nutrition. Endocr Relat Cancer. 2005;12(4):1071–1082. doi: 10.1677/erc.1.01038. [DOI] [PubMed] [Google Scholar]
- 19.Bonney RC, Reed MJ, Davidson K, Beranek PA, James VH. The relationship between 17 beta-hydroxysteroid dehydrogenase activity and oestrogen concentrations in human breast tumours and in normal breast tissue. Clin Endocrinol. 1983;19(6):727–739. doi: 10.1111/j.1365-2265.1983.tb00051.x. [DOI] [PubMed] [Google Scholar]
- 20.Honma N, Saji S, Hirose M, Horiguchi S, Kuroi K, Hayashi S, et al. Sex steroid hormones in pairs of tumor and serum from breast cancer patients and pathobiological role of androstene-3beta, 17beta-diol. Cancer Sci. 2011;102(10):1848–1854. doi: 10.1111/j.1349-7006.2011.02018.x. [DOI] [PubMed] [Google Scholar]
- 21.Dowsett M, Forbes JF, Bradley R, Ingle J, Aihara T, Bliss J, et al. Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet. 2015;386(10001):1341–1352. doi: 10.1016/S0140-6736(15)61074-1. [DOI] [PubMed] [Google Scholar]
- 22.Purohit A, Woo LW, Singh A, Winterborn CJ, Potter BV, Reed MJ. In vivo activity of 4-methylcoumarin-7-O-sulfamate, a nonsteroidal, nonestrogenic steroid sulfatase inhibitor. Cancer Res. 1996;56(21):4950–4955. [PubMed] [Google Scholar]
- 23.Stanway SJ, Purohit A, Woo LW, Sufi S, Vigushin D, Ward R, et al. Phase I study of STX 64 [667 Coumate] in breast cancer patients: the first study of a steroid sulfatase inhibitor. Clin Cancer Res. 2006;12(5):1585–1592. doi: 10.1158/1078-0432.CCR-05-1996. [DOI] [PubMed] [Google Scholar]
- 24.Ishida H, Nakata T, Suzuki M, Shiotsu Y, Tanaka H, Sato N, et al. A novel steroidal selective steroid sulfatase inhibitor KW-2581 inhibits sulfated-estrogen dependent growth of breast cancer cells in vitro and in animal models. Breast Cancer Res Treat. 2007;106(2):215–227. doi: 10.1007/s10549-007-9495-x. [DOI] [PubMed] [Google Scholar]
- 25.Palmieri C, Januszewski A, Stanway S, Coombes RC. Irosustat: a first-generation steroid sulfatase inhibitor in breast cancer. Expert Rev Anticancer Ther. 2011;11(2):179–183. doi: 10.1586/era.10.201. [DOI] [PubMed] [Google Scholar]
- 26.Dunning AM, Dowsett M, Healey CS, Tee L, Luben RN, Folkerd E, et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst. 2004;96(12):936–945. doi: 10.1093/jnci/djh167. [DOI] [PubMed] [Google Scholar]
- 27.Haiman CA, Dossus L, Setiawan VW, Stram DO, Dunning AM, Thomas G, et al. Genetic variation at the CYP19A1 locus predicts circulating estrogen levels but not breast cancer risk in postmenopausal women. Cancer Res. 2007;67(5):1893–1897. doi: 10.1158/0008-5472.CAN-06-4123. [DOI] [PubMed] [Google Scholar]
- 28.Beckmann L, Husing A, Setiawan VW, Amiano P, Clavel-Chapelon F, Chanock SJ, et al. Comprehensive analysis of hormone and genetic variation in 36 genes related to steroid hormone metabolism in pre- and postmenopausal women from the breast and prostate cancer cohort consortium [BPC3] J Clin Endocrinol Metab. 2011;96(2):E360–E367. doi: 10.1210/jc.2010-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M, Ingle JN, Fridley BL, Buzdar AU, Robson ME, Kubo M, et al. TSPYL5 SNPs: association with plasma estradiol concentrations and aromatase expression. Mol Endocrinol. 2013;27(4):657–670. doi: 10.1210/me.2012-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prescott J, Thompson DJ, Kraft P, Chanock SJ, Audley T, Brown J, et al. Genome-wide association study of circulating estradiol, testosterone, and sex hormone-binding globulin in postmenopausal women. PLoS One. 2012;7(6):e37815. doi: 10.1371/journal.pone.0037815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingle JN, Buzdar AU, Schaid DJ, Goetz MP, Batzler A, Robson ME, et al. Variation in anastrozole metabolism and pharmacodynamics in women with early breast cancer. Cancer Res. 2010;70(8):3278–3286. doi: 10.1158/0008-5472.CAN-09-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ingle JN, Kalari KR, Buzdar AU, Robson ME, Goetz MP, Desta Z, et al. Estrogens and their precursors in postmenopausal women with early breast cancer receiving anastrozole. Steroids. 2015;99:32–38. doi: 10.1016/j.steroids.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van der Waerden BL. Order tests for the two-sample problem and their power. Indag Math. 1952;14:453–458. [Google Scholar]
- 35.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tirona RG, Leake BF, Merino G, Kim RB. Polymorphisms in OATP-C: identification of multiple allelic variants associated with altered transport activity among European- and African-Americans. J Biol Chem. 2001;276(38):35669–35675. doi: 10.1074/jbc.M103792200. [DOI] [PubMed] [Google Scholar]
- 37.Obaidat A, Roth M, Hagenbuch B. The expression and function of organic anion transporting polypeptides in normal tissues and in cancer. Annu Rev Pharmacol Toxicol. 2012;52:135–151. doi: 10.1146/annurev-pharmtox-010510-100556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genomes Project. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.The GTEx Consortium. Human genomics. The genotype-tissue expression [GTEx] pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Search Collaborative Group. SLCO1B1 variants and statin-induced myopathy–a genomewide study. N Engl J Med. 2008;359(8):789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 41.Johnson AD, Kavousi M, Smith AV, Chen MH, Dehghan A, Aspelund T, et al. Genome-wide association meta-analysis for total serum bilirubin levels. Hum Mol Genet. 2009;18(14):2700–2710. doi: 10.1093/hmg/ddp202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coviello AD, Haring R, Wellons M, Vaidya D, Lehtimaki T, Keildson S, et al. A genome-wide association meta-analysis of circulating sex hormone-binding globulin reveals multiple loci implicated in sex steroid hormone regulation. Plos Genet. 2012;8(7):e1002805. doi: 10.1371/journal.pgen.1002805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gui C, Miao Y, Thompson L, Wahlgren B, Mock M, Stieger B, Hagenbuch B. Effect of pregnane X receptor ligands on transport mediated by human OATP1B1 and OATP1B3. Eur J Pharmacol. 2008;584(1):57–65. doi: 10.1016/j.ejphar.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee E, Schumacher F, Lewinger JP, Neuhausen SL, Anton-Culver H, Horn-Ross PL, et al. The association of polymorphisms in hormone metabolism pathway genes, menopausal hormone therapy, and breast cancer risk: a nested case-control study in the California teachers study cohort. Breast Cancer Res. 2011;13(2):R37. doi: 10.1186/bcr2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cbio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci Signal. 2013;6(269):l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nozawa T, Suzuki M, Takahashi K, Yabuuchi H, Maeda T, Tsuji A, Tamai I. Involvement of estrone-3-sulfate transporters in proliferation of hormone-dependent breast cancer cells. J Pharmacol Exp Ther. 2004;311(3):1032–1037. doi: 10.1124/jpet.104.071522. [DOI] [PubMed] [Google Scholar]
- 49.Banerjee N, Allen C, Bendayan R. Differential role of organic anion-transporting polypeptides in estrone-3-sulphate uptake by breast epithelial cells and breast cancer cells. J Pharmacol Exp Ther. 2012;342(2):510–519. doi: 10.1124/jpet.112.192344. [DOI] [PubMed] [Google Scholar]
- 50.Higuchi T, Endo M, Hanamura T, Gohno T, Niwa T, Yamaguchi Y, et al. Contribution of estrone Sulfate to cell proliferation in aromatase inhibitor (AI) –resistant, hormone receptor-positive breast cancer. PLoS One. 2016;11(5):e0155844. doi: 10.1371/journal.pone.0155844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Utsumi T, Yoshimura N, Takeuchi S, Ando J, Maruta M, Maeda K, Harada N. Steroid sulfatase expression is an independent predictor of recurrence in human breast cancer. Cancer Res. 1999;59(2):377–381. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.