Abstract
BACKGROUND
Limited information is available regarding primary care clinicians’ response to pharmacogenomic Clinical Decision Support (PGx-CDS) alerts integrated in the electronic health record.
METHODS
In February 2015, 159 clinicians in the Mayo Clinic primary care practice were sent e-mail surveys to understand their perspectives on the implementation and use of pharmacogenomic testing in their clinical practice. Surveys assessed how the clinicians felt about pharmacogenomics and whether they thought electronic PGx-CDS alerts were useful. Information was abstracted on the number of CDS alerts the clinicians received between October, 2013 and the date their survey was returned. CDS alerts were grouped into two categories: alert recommended caution using the prescription or the alert recommended an alternate prescription. Finally, data were abstracted regarding whether the clinician changed their prescription in response to the alert recommendation.
RESULTS
The survey response rate was 57% (n=90). Overall, 52% of the clinicians did not expect to use or did not know whether they would use pharmacogenomic information in their future prescribing practices. Additionally, 53% of the clinicians felt that the alerts were confusing, irritating, frustrating, or that it was difficult to find additional information. Finally, only 30% of the clinicians that received a CDS alert changed their prescription to an alternative medication.
CONCLUSIONS
Our results suggest a lack of clinician comfort with integration of pharmacogenomic data into primary care. Further efforts to refine PGx-CDS alerts to make them as useful and user-friendly as possible are needed to improve clinician satisfaction with these new tools.
Keywords: pharmacogenetics, genomics, practice patterns, clinical practice patterns
INTRODUCTION
Pharmacogenomics (PGx) is the study of the role of genetic variation in drug response phenotypes.(1–4) The effects of genetic variation can range from serious, potentially life-threatening adverse drug reactions, to lack of therapeutic efficacy. As a result, the clinical implementation of pharmacogenomics at the point-of-care may make it possible to avoid adverse drug reactions, maximize drug efficacy, reduce drug-drug interactions, and select medications based on the genetic profile of individual patients. Over the past decade, a number of genetic variants with demonstrated clinical utility have been identified and incorporated into drug labels by the US Food and Drug Administration.(5) There is now general agreement that pharmacogenomics represents an area within genomic science that could positively impact clinical medicine. However, the incorporation of pharmacogenomics into clinical practice has been slow and challenging, even at major academic medical centers.(6)
Multiple barriers to implementing pharmacogenomics into clinical practice exist, including availability of testing, evidence-based recommendations for prescribing, and integration of results into electronic health records (EHR). Several studies have demonstrated that clinicians feel unprepared to deal with pharmacogenomics information for their patients, and are unsure about the impact that such information will have on their clinical practices.(7–11) Thus, one of the largest barriers has been the education of providers about pharmacogenomics, both generally, and at the time of drug prescribing using clinical decision support (CDS) tools.
While multiple studies have demonstrated concerns among clinicians regarding use of pharmacogenomics data in clinical practice, previous studies of the impact of PGx-CDS on clinical practice have shown mixed results, but have focused predominantly on management of patients with a family history of disease or specific cancers.(12) Evidence is therefore limited regarding the actual impact of available pharmacogenomics data on primary care practice.
Beginning in January 2013, Mayo Clinic implemented CDS alerts in the electronic drug prescribing system that provided patient-specific pharmacogenomics information to guide treatment.(13) In October 2013, primary care clinicians were provided additional educational materials about pharmacogenomics and online resources to use with their patients who had undergone pharmacogenomic testing. Clinicians who received the informational materials were surveyed regarding their perspectives on pharmacogenomics, and EHRs were abstracted to assess the impact of the alerts on prescribing practices. In this report, we summarize and describe early clinician experience with pharmacogenomics in the clinical setting.
METHODS
Setting and Study Participants
Mayo Clinic is a tertiary medical center located in Rochester, Minnesota, which also supports a large primary care practice for local residents, employees, and their families. Our study population consisted of 159 primary care clinicians that cared for 1,013 patients participating in the Right Drug, Right Dose, Right Time Protocol (RIGHT Protocol).(13) Participants in the RIGHT Protocol research study were sequenced for variants in the CYP2C19, CYP2C9, VKORC1, SLCO1B1, and CYP2D6 genes. In addition, the primary care clinicians for the RIGHT patients also cared for other Mayo Clinic patients that did not participate in the RIGHT Protocol, but had received pharmacogenomic testing for clinical indications to guide dosing of clopidogrel, tamoxifen, or thiopurines. Pharmacogenomic information is integrated into the Mayo Clinic EHR, and CDS was implemented in 2013 to guide clinicians in their prescribing choices. In October 2014, the primary care clinicians comprising our study population were sent a printed packet of materials containing a letter that described the RIGHT Protocol, a list of their patients that were participating in the study, and copies of the pharmacogenomic educational materials that their patients received.
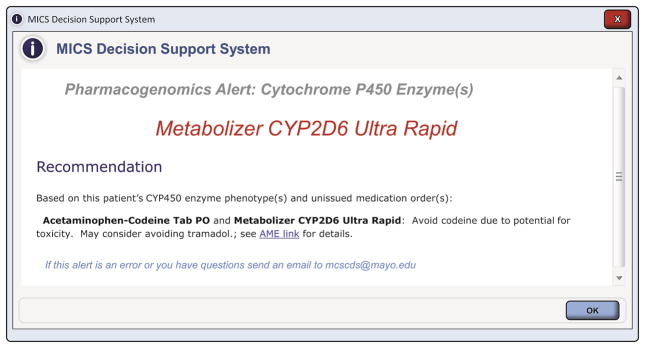
The Mayo Clinic CDS Program consists of a multidisciplinary team that includes clinical experts, pharmacists, clinical informaticians, and information technology analysts that develop evidenced-based CDS alerts. The PGx CDS interventions (alerts, reminders, screen changes, problem list documentation, etc.) were designed to deliver the necessary information and to facilitate further actions including cancelling a medication order, entering a problem or allergy, and connecting to additional related information in a web-based educational tool called “AskMayoExpert”.(14) When a clinician enters a prescription order (either new or renewal) and the patient has relevant genetic test results in the EHR, an electronic alert lists the patient’s phenotype, along with recommendations to adjust dose or consider an alternative medication. Links to additional information and educational materials are also provided (Figure 1).(13) At the time of the survey, 12 CDS alerts had been implemented in clinical practice (Supplemental Table 1).
Figure 1.
Example of a PGx-CDS alert.
Data Collection
Surveys
In February, 2015, the primary care physicians were sent e-mail surveys to understand their perspectives on the implementation and use of pharmacogenomic testing in their clinical practice. Specifically, surveys were designed to assess how these clinicians felt about pharmacogenomic in general, and whether they thought the corresponding CDS alerts were useful. Following the initial survey invitation, clinicians received two additional e-mail reminders at 2 and 4 weeks after the initial contact. Responses to survey questions were summarized using descriptive statistics.
The patients participating in the RIGHT Protocol(13) were mailed surveys to assess their satisfaction with and understanding of their pharmacogenomic test results. Details regarding this survey and data summarizing the patient responses to the survey are summarized elsewhere.(15) However, in this report, we summarize the RIGHT Protocol patient responses to the survey question “I am confident that my health care provider will use my pharmacogenomic information when prescribing medication for me.” Overall, 819 (81%) RIGHT Protocol patients responded to this question, and responses were collected on a 5-point scale ranging from “strongly agree” to “strongly disagree”.
Alert data
Following receipt of the surveys, we extracted information on the number of PGx-CDS alerts the participating clinicians received between October 2013 and the date their survey was returned for both RIGHT Protocol participants and other patients with pharmacogenomic information. Two of the authors (JLS and EJB) independently abstracted final prescription information and information on the pharmacogenomic phenotype of the affected patients from their EHRs. Differences in abstraction were resolved via consensus between the reviewers. CDS alerts were grouped into two categories: alert recommended caution with the prescription or the alert recommended an alternate prescription. If the final patient prescription differed from the initial prescription that triggered the alert, we concluded that the clinician had changed the initial prescription in response to the alert recommendation. Data were summarized using descriptive statistics.
RESULTS
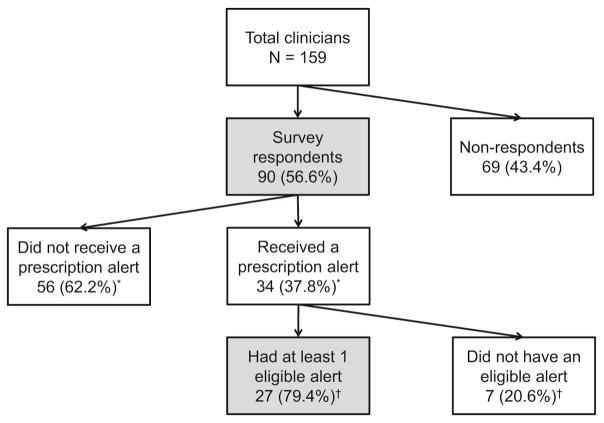
Overall, 90 (57% of 159) of the clinicians returned a survey (Figure 2). Clinician survey responses are summarized in Table 1. Twenty-six (30% of 87) clinicians noted that pharmacogenomics was part of their formal training or medical education, and 8 (9% of 90) had already talked to a patient about their pharmacogenomic results. However, 45 (52% of 86) clinicians did not plan to use, or did not know whether they would use, pharmacogenomic results in the future, and only 6 (7% of 87) expected to order or recommend a pharmacogenomic test for their patients in the next 6 months. In contrast to these results, 614 (75% of 819) patients responding to the RIGHT Protocol survey question reported that they “somewhat agreed” or “strongly agreed” that their health care providers would use their pharmacogenomic information when prescribing a medication for them.
Figure 2.
Flow chart of the study participants. Data are reported for the participants in the dark grey boxes (survey respondents and survey respondents who received at least one eligible alert).
Table 1.
Summary of Clinician Responses to the Survey
| All respondents N=90 |
||
|---|---|---|
|
| ||
| Question | N* | % |
| Pharmacogenomics was part of training/medical education | 26/87 | 30 |
| Talked to a patient about pharmacogenomics results | 8/90 | 9 |
| Plan to use pharmacogenomics results in the future to:† | ||
| Guide future prescribing selection when appropriate | 32/86 | 37 |
| Guide future dosing when appropriate | 27/86 | 31 |
| Call for pharmacy consultation | 17/86 | 20 |
| Do not plan to use | 6/86 | 7 |
| Do not know at this time | 39/86 | 45 |
| Expect to order or recommend a pharmacogenomic test in next 6 months | 6/87 | 7 |
| Remembered receiving the informational packet | 27/90 | 30 |
| Informational packet was at least somewhat helpful‡ | 19/27 | 70 |
| Remembered seeing a drug-gene alert | 37/87 | 43 |
| Sometimes, often, or always changed the prescription due to alert§ | 15/37 | 41 |
| Perception of the PGx-CDS alert§,† | ||
| Positive response (alert was helpful, convenient, or easy to understand) | 12/36 | 33 |
| Negative response (alert was confusing, irritating, frustrating, or it was difficult to find additional information) | 19/36 | 53 |
| Both positive and negative responses | 5/36 | 14 |
Non-missing values reported; responders not shown, and not included in the percentages based on non-missing values.
Respondents could check all that applied.
Denominator is the 27 participants who remembered receiving the informational packet.
Denominator is the 37 participants who remembered seeing a drug-gene alert.
Overall, 37 (43% of 87) clinicians remembered seeing a pharmacogenomic alert for at least one of their patients. Of those that remembered receiving an alert, 12 (33% of 36) reported only positive responses (thought the alert was helpful, convenient, or easy to understand), and 19 (53% of 36) selected only negative responses (thought that it was difficult to find additional information or that the alert was confusing, irritating, or frustrating). An additional 5 (14% of 36) providers reported both positive and negative responses. When asked how to improve the alerts, suggestions included providing specific guidance with alternative medications, specific recommendations for dose adjustments, and developing web-based algorithms or care process models.
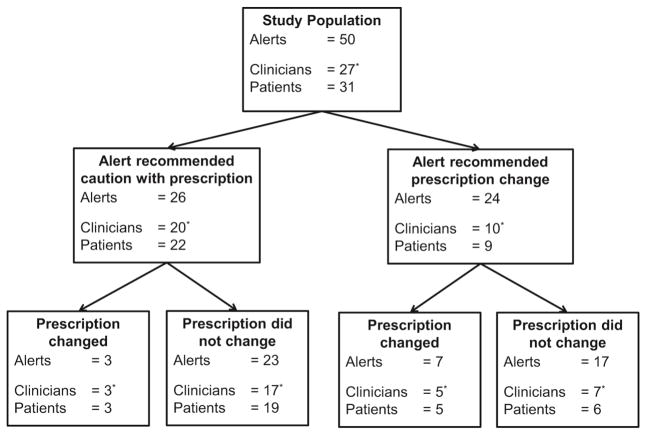
Among 34 clinicians that received pharmacogenomic alerts, 7 clinicians were excluded because none of their non-RIGHT patients with alerts gave permission for their EHR to be used for research or because they had only received alerts that were not related to an eligible prescription (Figure 2). Figure 3 shows the number of clinicians that received at least one eligible PGx-CDS alert, and how they responded following the alert. Overall, 50 CDS alerts were activated for 31 unique patients. Most (47; 94% of 50) of the alerts related to a potential codeine or tramadol interaction with a CYP2D6 phenotype, 2 (4%) related to a potential interaction between clopidogrel and a CYP2C19 phenotype, and 1 (2%) related to a potential interaction with a TPMT phenotype. Twenty-six (52% of 50) alerts cautioned the clinician that the prescribed medications might be less effective in their patients, and the clinicians changed their prescriptions in 3 (12% of 26) of the cases (Figure 3). Twenty-four (48% of 50) of the alerts recommended an alternate prescription, and the clinicians changed their prescriptions in 7 (29% of 24) cases (Figure 3). The proportion of prescriptions that changed was greater when clinicians received an alert recommending an alternate prescription (29% vs 12%), but the difference was not statistically significant (Fisher’s exact p value = 0.16). Overall, 8 (30% of 27) of the clinicians changed at least one prescription in response to a pharmacogenomic alert. In the 40 cases where the prescription did not change, EHR documentation was available in 6 (15% of 40) cases that indicated the clinician had reviewed the pharmacogenomic information, but, based on clinical judgment, felt that changing the prescription was not warranted (e.g. patient had used medication in the past with no concerns). Finally, most of the patients (24; 77% of 31) had received the prescription at least once prior to the alert.
Figure 3.
Flow chart of the number and types of eligible alerts received during the study period, the number of clinicians and patients affected by each alert, and the clinician response to the alert.
DISCUSSION
In this study, we examined the clinician experience with PGx-CDS integrated into the EHR. Overall, we found that over half of the clinicians that completed surveys did not expect to use pharmacogenomic information or did not know whether they would use this information in their future prescribing practices. Additionally, over half of the clinicians that received an alert felt that the alerts were confusing, irritating, frustrating, or that it was difficult to find additional information. Finally, when we examined the prescriptions that followed receipt of an alert, only 30% of the clinicians changed at least one prescription in response to a PGx-CDS alert.
Over half (53%) of the surveyed clinicians did not find the PGx-CDS alerts to be useful. These results are consistent with other studies reporting variable satisfaction with and utility of pharmacogenomic alerts.(16–19) Alert fatigue is a significant problem for clinicians, and a recent review noted that alerts must be highly sensitive and specific, must be clear and unambiguous, and must be seamlessly integrated into the work flow in order to be useful to clinicians and effective for patients.(20) Further research and efforts to ensure PGx-CDS alerts meet all of these goals could significantly improve clinician satisfaction.
We also found that about 45% of the survey respondents did not know whether they would use pharmacogenomic alerts in the future. Appropriate education for these clinicians could potentially improve satisfaction with the pharmacogenomic alerts. However, while some studies have found pharmacogenomics education useful,(21, 22) there are considerable challenges to developing effective educational strategies. In particular, previous studies have shown that providers have limited time available to pursue education related to pharmacogenomics.(18, 23) Clinicians desire education at point-of-care to guide prescribing, which can be costly to develop and to update.(24) Finally, pharmacogenomics is a relatively new field and broader evidence of the efficacy of pharmacogenomics on clinical outcomes is needed. If such evidence accumulates, use of pharmacogenomic information will become standard of care, and may make clinicians more comfortable with the use of such information in their daily practice.
Importantly, our data suggest a disconnect between patient expectations and actual clinical practice. Most of the RIGHT Protocol patients from a previous survey believed that their health care providers would use their pharmacogenomic information when prescribing medications; however, over half of the clinicians surveyed did not expect to use, or did not know whether they would use, pharmacogenomic information in the future. This disconnect is especially problematic for clinicians because patient perspectives on the use of genomic data for personalized care may be significantly influenced by the media and for-profit genetic testing companies, which can market directly to consumers. For example, recent articles in the New York Times highlight the “Promise of Genetic Testing in Medicine”,(25) while an additional on-line article highlights a new partnership between RiteAid pharmacies and Harmonyx, a genetic testing company, to offer a range of genetic tests directly to consumers.(26) Such stories highlight the enthusiasm for use of genetic testing in clinical practice, but clinician engagement will be key to ensuring that genetic information is actually translated into clinical care. Patients typically do not have the background knowledge to fully interpret their genetic data, and independent companies marketing these tests are not typically integrated with health care systems. Direct-to-consumer pharmacogenomic testing has been associated with increases in physician utilization, because patients will bring genetic testing results to the attention of their health care providers.(27) However, our data indicate that clinician concerns about use of pharmacogenomic information may pose one possible stumbling block toward integration and use of genomic testing into care, and could prevent pharmacogenomic data from being fully utilized in clinical practice.
Previous studies have indicated that computerized alerts often have positive effects on electronic prescribing practices,(28) but reports of overrides of such alerts have ranged from 49–96%.(29) Data on CDS alerts specific to pharmacogenomics are more limited, but Bell and colleagues reported that alerts were over-ridden only 5% of the time among pediatric patients with lymphoma or leukemia.(30) By contrast, Goldspiel and colleagues found that overrides of alerts for pediatric patients prescribed abacavir, allopurinol, and carbamazepine occurred in 90% of the prescriptions. In the majority of these cases, the clinician noted that the patient was already taking the medication with no adverse events.(31) Our data are in agreement with results seen for general computerized alerts, and results reported by Goldspiel et al. Overall, 70% of the clinicians in our study that received a PGx-CDS alert for their patients did not change their initial prescription to an alternative medication. It was not possible for us to ascertain the rationale for the final prescriptions from most of the EHR notes. However, if the patient had tolerated the prescription in the past with no adverse effects, the clinician may not have felt the need to change the prescription, regardless of the availability of the new pharmacogenomic information. In support of this hypothesis, most of the patients in this study (77%) had received the same prescription at a prior date. Pharmacogenomic information may therefore be most useful for patients that receive a new prescription for the first time. In addition, prescribing decisions are probably guided by the likely severity of the outcome. As noted above, Bell and colleagues found that PGx-CDS recommendations in their patient population were rarely over-ridden. However, the CDS alerts in that study were for pediatric cancer patients, and failure to change the prescription could result in severe patient outcomes (e.g. myelosuppression).(30) In our study, the PGx-CDS alerts noted that the patients would respond less well to the prescribed medications, but the medications were typically not completely contraindicated. Since pharmacogenomics is a relatively new field, the likely repercussions of failing to change a prescription are not known for all medications. In many cases, failing to change a prescription may not be life-threatening, and may not significantly change key health outcomes. Additional data are therefore urgently needed to assess the impact of pharmacogenomics on relevant clinical and health care outcomes.
Limitations of our study include the relatively low survey participation rate. Survey response rates among health care providers have been consistently declining over time, and response rates among physicians tend to be lower than response rates in the general population.(32) Our 57% response rate is similar to or greater than other survey responses observed among primary care providers, and is very close to the minimum 60% response rate required by some medical and public health journals.(32) However, it is still possible that clinicians that participated in our study may differ from those that did not participate. For example, those that were particularly satisfied or dissatisfied with the CDS alerts may have been more likely to respond to the survey, and actual rates of dissatisfaction may be higher or lower than reported. In addition, our results represent the experience of a single institution, and these results may not be generalizable to other settings. Mayo Clinic is a large, tertiary referral center with a well-integrated primary care practice. As such, primary care providers at this institution often have more resources available than smaller, independent primary care practices. The primary care physicians at this site might be expected to be more willing to accept new health care initiatives compared to physicians at other sites, and our results may therefore represent best case scenarios for integration of pharmacogenomics into clinical practice. Finally, the small number of actionable alerts received during the study period may make our estimates of clinician agreement with alert recommendations unstable.
CONCLUSION
Our results indicate that clinicians are not comfortable with the integration of pharmacogenomic data into their clinical practice. Since most patients expect that their pharmacogenomic data will help guide their care decisions, further efforts to educate clinicians about the utility of pharmacogenomic data for clinical practice, and efforts to refine PGx-CDS alerts to make them useful and user-friendly, may close the gap between the clinician’s approach and patient expectations.
Supplementary Material
Clinical Significance.
Implementation of pharmacogenomics (PGx) at the point-of-care may make it possible to optimally select medications based on a patient’s genetic profile.
Our study indicates that primary care providers are not comfortable with using PGx data in routine practice, and overrides of electronic PGx clinical decision support alerts are common.
Further efforts are needed to optimize the clinical use of PGx data.
Acknowledgments
The authors thank Robin Adams for her assistance with manuscript preparation and submission.
Funding: This research was funded by the Mayo Clinic Center for Individualized Medicine and the Robert D. and Patricia E. Kern Center for the Science of Health Care Delivery.
Footnotes
Conflict of Interest: The authors have no conflicts of interest to report.
Authorship: All authors had access to the data and a role in writing the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364(12):1144–53. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weinshilboum R. Inheritance and drug response. N Engl J Med. 2003;348(6):529–37. doi: 10.1056/NEJMra020021. [DOI] [PubMed] [Google Scholar]
- 3.Weinshilboum R, Wang L. Pharmacogenomics: bench to bedside. Nat Rev Drug Discov. 2004;3(9):739–48. doi: 10.1038/nrd1497. [DOI] [PubMed] [Google Scholar]
- 4.Weinshilboum RM, Wang L. Pharmacogenetics and pharmacogenomics: development, science, and translation. Annu Rev Genomics Hum Genet. 2006;7:223–45. doi: 10.1146/annurev.genom.6.080604.162315. [DOI] [PubMed] [Google Scholar]
- 5.United States Food and Drug Administration. Table of pharmacogenomic biomarkers in drug labeling. Silver Spring, MD: 2015. [accessed 03/25/2016]. updated 05/20/2015. Available from: http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.Htm. [Google Scholar]
- 6.McCarthy JJ, McLeod HL, Ginsburg GS. Genomic medicine: a decade of successes, challenges, and opportunities. Sci Transl Med. 2013;5(189):189sr4. doi: 10.1126/scitranslmed.3005785. [DOI] [PubMed] [Google Scholar]
- 7.Johansen Taber KA, Dickinson BD. Pharmacogenomic knowledge gaps and educational resource needs among physicians in selected specialties. Pharmgenomics Pers Med. 2014;7:145–62. doi: 10.2147/PGPM.S63715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanek EJ, Sanders CL, Taber KA, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91(3):450–8. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 9.Haga SB, Burke W, Ginsburg GS, et al. Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clin Genet. 2012;82(4):388–94. doi: 10.1111/j.1399-0004.2012.01908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unertl KM, Field JR, Price L, Peterson JF. Clinician perspectives on using pharmacogenomics in clinical practice. Pers Med. 2015;12(4):339–47. doi: 10.2217/pme.15.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel HN, Ursan ID, Zueger PM, et al. Stakeholder views on pharmacogenomic testing. Pharmacotherapy. 2014;34(2):151–65. doi: 10.1002/phar.1364. [DOI] [PubMed] [Google Scholar]
- 12.Welch BM, Kawamoto K. Clinical decision support for genetically guided personalized medicine: a systematic review. J Am Med Inform Assoc. 2013;20(2):388–400. doi: 10.1136/amiajnl-2012-000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89(1):25–33. doi: 10.1016/j.mayocp.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cook DA, Sorensen KJ, Nishimura RA, et al. A comprehensive information technology system to support physician learning at the point of care. Acad Med. 2015;90(1):33–9. doi: 10.1097/ACM.0000000000000551. [DOI] [PubMed] [Google Scholar]
- 15.Olson JE, Rohrer Vitek CR, Bell EJ, et al. Participant understanding of CYP2D6 genetic test results and attitudes toward pharmacogenomics: Mayo Clinic RIGHT Protocol; Abstract #1071. Presented at the 2016 American College of Medical Genetics Annual Clinical Genetics Meeting; March 8–12, 2016; Tampa, FL. [Google Scholar]
- 16.Devine EB, Lee CJ, Overby CL, et al. Usability evaluation of pharmacogenomics clinical decision support aids and clinical knowledge resources in a computerized provider order entry system: a mixed methods approach. Int J Med Inform. 2014;83(7):473–83. doi: 10.1016/j.ijmedinf.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Overby CL, Devine EB, Abernethy N, et al. Making pharmacogenomic-based prescribing alerts more effective: A scenario-based pilot study with physicians. J Biomed Inform. 2015;55:249–59. doi: 10.1016/j.jbi.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selkirk CG, Weissman SM, Anderson A, Hulick PJ. Physicians’ preparedness for integration of genomic and pharmacogenetic testing into practice within a major healthcare system. Genet Test Mol Bioma. 2013;17(3):219–25. doi: 10.1089/gtmb.2012.0165. [DOI] [PubMed] [Google Scholar]
- 19.Peterson JF, Field JR, Shi Y, et al. Attitudes of clinicians following large-scale pharmacogenomics implementation. Pharmacogenomics J. 2015 Aug 11; doi: 10.1038/tpj.2015.57. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Sijs H, Aarts J, Vulto A, Berg M. Overriding of drug safety alerts in computerized physician order entry. J Am Med Inform Assoc. 2006;13(2):138–47. doi: 10.1197/jamia.M1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohrer Vitek CR, Nicholson WT, Schultz C, Caraballo PJ. Evaluation of the use of clinical decision support and online resources for pharmacogenomics education. Pharmacogenomics. 2015;16(14):1595–603. doi: 10.2217/pgs.15.100. [DOI] [PubMed] [Google Scholar]
- 22.Reed EK, Johansen Taber KA, Ingram Nissen T, et al. What works in genomics education: outcomes of an evidenced-based instructional model for community-based physicians. Genet Med. 2015 Nov 19; doi: 10.1038/gim.2015.144. Advance online publication. [DOI] [PubMed] [Google Scholar]
- 23.Grimshaw JM, Eccles MP, Walker AE, Thomas RE. Changing physicians’ behavior: what works and thoughts on getting more things to work. J Contin Educ Health Prof. 2002;22(4):237–43. doi: 10.1002/chp.1340220408. [DOI] [PubMed] [Google Scholar]
- 24.Institute of Medicine. Improving genetics education in graduate and continuing health professional education: Workshop summary. Washington D. C: The National Academies Press, Washington D.C; 2015. [PubMed] [Google Scholar]
- 25.Vigliotti G. The Promise of Genetic Testing in Medicine. [Accessed 03/25/2016];New York Times [serial on the Internet] 2015 Dec 21;2015 Available from: http://www.nytimes.com/roomfordebate/2015/10/22/the-promise-of-genetic-testing-in-medicine. [Google Scholar]
- 26.PharmacoGeneticsTesting.com. Genetic testing partnership between RiteAid and Harmonyx. [Accessed 03/25/2016];Pharmacogenetic Information and News [serial on the Internet] 2015 Dec 21;2015 Available from: http://pharmacogeneticstesting.com/pharmacogenomic-companies/harmonyx-genetic-testing-kits-available-at-rite-aid-nationwide/ [Google Scholar]
- 27.Bloss CS, Schork NJ, Topol EJ. Direct-to-consumer pharmacogenomic testing is associated with increased physician utilisation. J Med Genet. 2014;51(2):83–9. doi: 10.1136/jmedgenet-2013-101909. [DOI] [PubMed] [Google Scholar]
- 28.Schedlbauer A, Prasad V, Mulvaney C, et al. What evidence supports the use of computerized alerts and prompts to improve clinicians’ prescribing behavior? J Am Med Inform Assoc. 2009;16(4):531–8. doi: 10.1197/jamia.M2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCoy AB, Thomas EJ, Krousel-Wood M, Sittig DF. Clinical decision support alert appropriateness: a review and proposal for improvement. Ochsner J. 2014;14(2):195–202. [PMC free article] [PubMed] [Google Scholar]
- 30.Bell GC, Crews KR, Wilkinson MR, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc. 2014;21(e1):e93–9. doi: 10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldspiel BR, Flegel WA, DiPatrizio G, et al. Integrating pharmacogenetic information and clinical decision support into the electronic health record. J Am Med Inform Assoc. 2014;21(3):522–8. doi: 10.1136/amiajnl-2013-001873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLeod CC, Klabunde CN, Willis GB, Stark D. Health care provider surveys in the United States, 2000–2010: a review. Eval Health Prof. 2013;36(1):106–26. doi: 10.1177/0163278712474001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.