Abstract
Big data is a term used for any collection of datasets whose size and complexity exceeds the capabilities of traditional data processing applications. Big data repositories, including molecular, clinical, and epidemiology data, offer unprecedented research opportunities to help guide scientific advancement. Advantages of big data can include ease and low cost of collection, ability to approach prospectively and retrospectively, utility for hypothesis generation in addition to hypothesis testing, and the promise of precision medicine. Limitations include cost and difficulty of storing and processing data, need for advanced techniques for formatting and analysis, and concerns about accuracy, reliability, and security. We discuss sources of big data and tools for its analysis to help inform the treatment and management of dermatologic diseases.
What is big data?
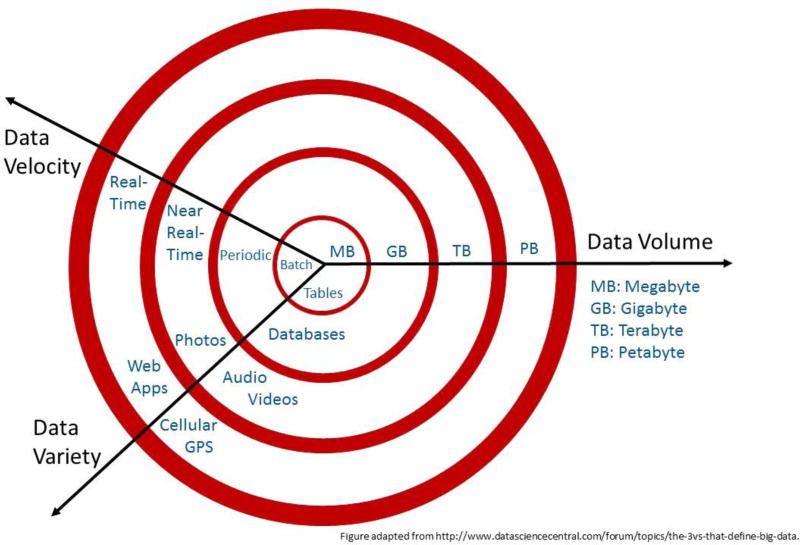
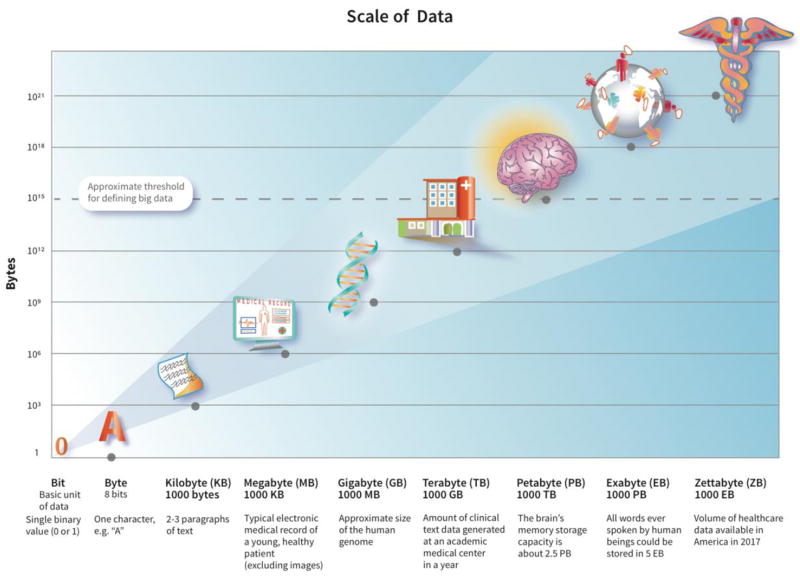
Big data is commonly defined as data so large or complex that traditional data processing and analytic approaches are inadequate. The ‘3 V’s’ that characterize big data are volume (amount of data), velocity (speed at which data is generated and processed), and variety (types of data) (Laney, 2001), all of which have been growing rapidly (Figure 1). While there is no predefined threshold for volume, in general, anything ≥1 petabyte is considered big data (1015 bytes, or the approximate size of 1 million human genomes, Figure 2). The ability to monitor, record, and store information from large populations from sources including electronic medical records, insurance claims, surveys, disease registries, biospecimens, apps and social media, the internet, and personal monitoring devices has shepherded the era of big data into use in healthcare. The volume of healthcare data in the United States in 2017 is rapidly approaching zettabyte levels (iHT2, 2013). This wealth of structured and unstructured data has the potential to substantially impact healthcare delivery through improved risk assessment, surveillance, diagnosis, and treatment methods.
Figure 1.
The 3 V’s of big data: volume (amount of data), velocity (speed at which data is generated), and variety (number of types of data), all of which have been growing rapidly. After “The 3Vs that define Big Data,” Diya Soubra, Data Science Central, http://www.datasciencecentral.com/forum/topics/the-3vs-that-define-big-data.
Figure 2.
Logarithmic scale depicting volume of big data.
What are some big data sources in healthcare?
There are many big data sources in healthcare. OptumLabs (https://www.optumlabs.com), an open collaborative research center, provides de-identified clinical data from electronic health records (EHR) and claims data for over 100 million insured members (Borah, 2016). Sentinel (https://www.sentinelinitiative.org), an FDA initiative, utilizes data from EHR, insurance claims, and registries to monitor post-marketing, real-world safety of medicines. Sentinel data were used to estimate the validity of ICD-9 codes for ascertaining Stevens-Johnson syndrome and toxic epidermal necrolysis in 12 collaborating research units, covering almost 60 million people (Davis et al., 2015). UK Biobank and Kaiser Permanente Biobank are examples of medical data and tissue samples collected for research purposes. UK Biobank (www.ukbiobank.ac.uk) is a cohort of 500,000 participants in the UK who have provided baseline information, blood, urine, and saliva samples, and are being followed prospectively through their regular care. The Kaiser Permanente Research Biobank (https://www.dor.kaiser.org/external/DORExternal/rpgeh) is comprised of 220,000 health plan members who have contributed genetic and EHR data. This was recently utilized in a large genome-wide association study of cutaneous squamous cell carcinoma (cSCC), which identified 10 single-nucleotide polymorphisms associated with cSCC at genome-wide significance and provided new insights into the genetics of heritable cSCC risks (Asgari et al., 2016). For genomic data, such as that found in biobanks, the National Center for Biotechnology Information has developed the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo), which acts as a public archive and repository of microarray, next-generation sequencing, and high-throughput functional genomic data. Geographic information systems, such as the National Cancer Institute Geographic Information Systems and Science for Cancer Control (https://gis.cancer.gov), capture geographic data that allow for mapping of disease trends. Solar ultraviolet (UV) radiation data is available through this system, and the association between cutaneous melanoma incidence rates and county-level UV exposure has been examined (Richards et al., 2011). Computer-based geographic information systems, web-based geospatial technologies, such as global positioning systems in smartphones, and geospatial modeling can be used to follow disease trends and to examine mobility and social networks and their impact on disease (Birch, 2016; Ray et al., 2016).
To enhance the utility of biomedical big data from these diverse sources, the NIH established Big Data to Knowledge (BD2K, https://datascience.nih.gov/bd2k). It aims to make digital data “Findable, Accessible, Interoperable, and Reusable (FAIR),” with the following specific goals: 1) improve the ability to find and use big data, 2) develop analysis tools for big data, 3) increase training in data science, and 4) establish centers of excellence in data science (Margolis et al., 2014). BD2K has funding opportunities in many areas, including curating, coordinating, and organizing big data, developing big data educational curricula, and improving big data standards (https://www.nlm.nih.gov/ep/BD2KGrants.html).
How do analytic techniques for big data differ from traditional data?
Although big data can be used for traditional hypothesis testing, and can be especially valuable for research on rare diseases or exposures, big data analyses are often hypothesis generating. Rather than test a hypothesis, they can provide evidence for new hypotheses that can later be tested with traditional techniques. Big data analyses often center around identifying patterns. Unlike traditional predictive modeling based on a small number of covariates, big data predictive modeling often involves variables that are not pre-selected. Thus, compared to traditional data analysis, big data has the potential to be more exploratory. Given the multiplicity inherent in the many potential patterns evaluated, such big data analyses benefit from special statistical methods that account for this multiple testing using p-value adjustments or false discovery rates.
Analytic techniques for big data
There are many computational and statistical methods used to analyze big data. Data mining is a process through which data are analyzed from different perspectives to identify unsuspected patterns. Using insurance claims, data mining with TreeScan software was used to explore unsuspected adverse reactions associated with antifungal drug exposure (Kulldorff et al., 2013). TreeScan is free data mining software available for download online (https://www.treescan.org). Cluster analysis focuses on grouping similar patients or observations by demographics, medical history, genetics, or geography. For example, the spatial scan statistic was used to detect geographic clusters of basal cell carcinomas in a Northern California population with the goal of targeting screening and prevention efforts (Ray et al., 2016). Another example is cluster analysis of different quality of life scoring systems in psoriasis patients, which showed lack of correlation of disease severity with psychological distress instruments (Sampogna et al., 2004).
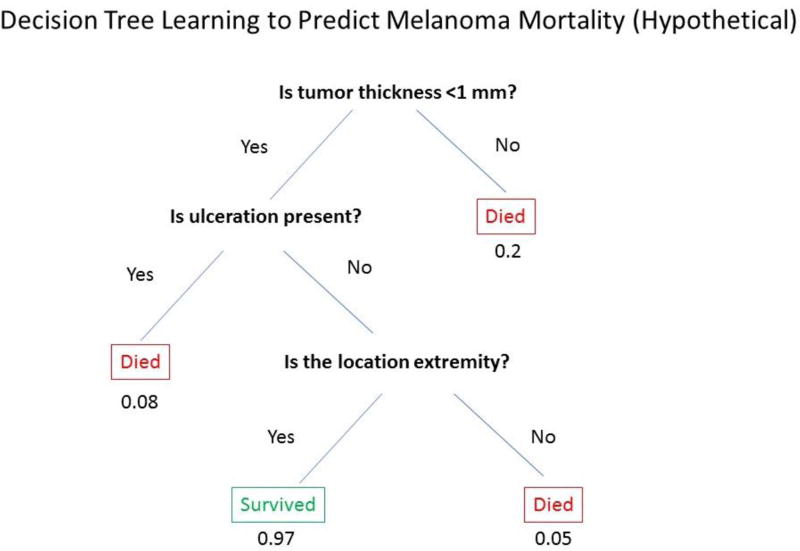
Machine learning allows algorithms to learn from a training dataset to make predictive models without specifying the model in advance. Machine learning is currently being explored to track pigmented lesions over time and identify lesions at higher risk for malignancy (Li et al., 2016). Machine learning was recently used to develop a diagnosis algorithm for skin cancer based on clinical images (Esteva et al., 2017). The algorithm, which uses only pixels and disease labels as inputs, matches the performance of dermatologists in identifying cancerous and non-cancerous lesions (Esteva et al., 2017). Deployable on mobile devices, machine learning algorithms that train computers to make reliable diagnoses directly from clinical images hold the potential to make a significant clinical impact by extending the reach of dermatologists beyond the clinic (Esteva et al., 2017). Decision-tree learning is a type of machine learning in which the independent variables are used to create a hierarchical tree structure with leaves and branches, which can predict an outcome (example, Figure 3). There are two main types of decision tree analyses: classification tree analysis, where the predicted outcome is dichotomous such as melanoma mortality, and regression tree analysis, where the predicted outcome is a continuous variable such as age at melanoma diagnosis. Both classification and regression tree analysis were used to identify histological features of melanoma associated with CDKN2A germline mutations (Sargen et al., 2015). Bayesian networks are another type of machine learning that use probabilistic graphs to explore relationships between for example symptoms and disease, to be used in clinical decision making or diagnosis. Cognitive computing is a type of machine learning that tries to mimic the functioning of the human brain. Natural language processing (NLP) algorithms allow computers to extract useful information from text, such as electronic health records, well enough to yield meaningful data. Such algorithms can identify mentions of a risk factor or of an outcome disease in clinic notes, recognizing that the same exposure or diagnosis can be expressed in many different ways and with potential misspellings, and distinguishing a positive diagnosis from a rule-out diagnosis. NLP has been used in dermatology research to find non-melanoma skin cancer diagnoses in electronic pathology reports (Eide et al., 2012).
Figure 3.
Hypothetical example illustrating the utility of decision-tree learning for melanoma mortality prediction showing “leaves” (independent variables) such as tumor thickness, ulceration, and tumor location, and probability of survival (outcome).
Analytic platforms for big data
There are two approaches to analytic platforms for big data: 1) a divide and conquer approach (distributed data) and 2) a centralized approach utilizing a platform that provides both database storage and analytics in a centralized fashion, such as SAP HANA (http://www.sap.com/product/technology-platform/hana.html). SAP HANA is a computing platform that offers tools for storing, managing, and analyzing big data. When big data is in different physical locations, distributed data analysis can be used with some of the analysis conducted locally on the complete data while the final analysis occurs centrally using summary data from each site. The advantage of distributed data for medical information is that data remains at their local site, minimizing storage costs and maximizing data integrity and patient privacy.
Summary and future directions in dermatology
Big data is more than just very large data or a large number of data sources, but encompasses a new approach to complex data. It offers a new, hypothesis generating framework to conduct research and requires novel analysis methods. It has significant advantages but also has limitations (Table 1), and traditional data analytics are still crucially important. In dermatology, big data can be used to improve risk prediction models, support targeted screening for high-risk individuals (e.g. targeted skin cancer screening), optimize management of a variety of skin diseases, and offer clinical decision support (e.g. assistance in deciding whether to biopsy a pigmented lesion). We can further investigate the genetics of skin disease (e.g. genome wide association studies) (Asgari et al., 2016; Frelinger, 2015) and examine distinct disease phenotypes within heterogeneous diseases that could benefit from tailored therapies (e.g. in psoriasis or eczema). Big data may be an excellent way to perform surveillance and evaluate safety of medications and devices, especially for rarer outcomes. Big data in dermatology presents spectacular opportunities, allowing researchers to maximize the potential of existing data sources and opening up new, efficient, and powerful methods for future research.
Table 1.
Advantages and limitations of big data
| Advantages | Limitations |
|---|---|
|
|
Acknowledgments
We would like to acknowledge Susan Gruber PhD for her assistance with reviewing the content of this manuscript.
Funding/Support: This research was supported by NIH grant R01CA166672 (MA).
Appendix
Multiple Choice Questions
-
What are the “3 V’s” that characterize big data?
- Value, viability, and variety
- Volume, velocity, and viability
- Volume, velocity, and variety
- Volume, value, and variety
Explanation: The correct answer is c. The “3 V’s” that characterize big data are volume (amount of data); velocity (speed at which data is generated); and variety (number of types of data).
-
What distinguishes big data analyses from traditional data analyses?
- Can be used to both test and generate hypotheses
- Variables are often not pre-selected for prediction modeling
- Often center around identifying and evaluating patterns
- All of the above
Explanation: The correct answer is d. Big data can be used not only to test hypotheses, but also to generate new hypotheses that can later be tested with traditional data approaches. In big data predictive modeling, variables are often not pre-selected, which allows big data to be more exploratory and have a broader scope compared to traditional data. Big data analyses often center around identifying and evaluating patterns.
-
What analytic technique focuses on grouping similar patients by characteristics such as demographics, genetics, or geography and can be used to inform geographically targeted screening and prevention efforts?
- Cluster analysis
- Decision-tree learning
- Bayesian networks
- Cognitive computing
Explanation: The correct answer is a. Cluster analysis focuses on grouping similar patients or observations by variables such as demographics, medical history, genetics, or geography. Cluster analysis was used to detect statistically significant geographic clusters of basal cell carcinomas in a Northern California population with the goal of informing geographically targeting screening and prevention efforts. Decision-tree learning is a type of machine learning in which the independent variables are used to create a hierarchical tree structure with leaves and branches, which can predict an outcome. Bayesian networks use probabilistic graphs to explore relationships between variables. Cognitive computing is a type of machine learning that tries to mimic the functioning of the human brain
-
Which of the following is NOT a limitation of big data?
- Storage may require considerable resources
- Formatting and analysis may require advanced computer science
- Can only be used for retrospective analyses
- More complex security and information privacy concerns than traditional datasets
Explanation: The correct answer is c. Big data has several significant limitations. Storage of big data may require considerable resources. Advanced computer science may be required to format and analyze big datasets. Big datasets often have more complex security and information privacy concerns than traditional datasets. Quality control can be difficult and often has to be done through small representative samples. The advantages of a big data approach are many, however. Big data provides a large sample size and can be inexpensive to acquire. Both prospective and retrospective approaches are often available. Multiple data points from different sources can be combined in ways never before possible, leveraging the advantages of different collection sources and smaller datasets.
-
Which of the following is NOT a potential application of big data?
- Improve risk prediction for very rare diseases
- Identify distinct disease phenotypes in heterogeneous diseases that may merit different therapies
- Identify causal associations
- Perform drug and medical device surveillance
Explanation: The correct answer is c. Big data is not used to identify causal associations. Big data, however, has many other potential applications that present powerful research opportunities in dermatology. Big data applications include improved risk prediction for rare diseases and diseases not included in traditional datasets; identification of distinct disease phenotypes in heterogeneous diseases that may merit different therapies; and surveillance of drug and medical device safety.
Footnotes
Conflict of Interest: Dr. Asgari has received research funding to her institution from Pfizer Inc. and Valeant Pharmaceuticals, but these associations have not influenced our work on this paper. The authors have no other potential conflicts of interest to disclose.
References
- Asgari MM, Wang W, Ioannidis NM, Itnyre J, Hoffmann T, Jorgenson E, et al. Identification of Susceptibility Loci for Cutaneous Squamous Cell Carcinoma. The Journal of investigative dermatology. 2016;136:930–7. doi: 10.1016/j.jid.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch P. Powering geospatial analysis: public geo datasets now on Google Cloud. 2016 < https://cloudplatform.googleblog.com/2016/10/powering-geospatial-analysis-public-geo-datasets-now-on-Google-Cloud.html> Accessed.
- Borah BJ. [Accessed December 14 2016];Optum Labs Overview. 2016 <https:// http://www.allianceforclinicaltrialsinoncology.org/main/cmsfile?cmsPath=/Public/AnnualMeeting/files/Prevention-OptumLabsOverview.pdf>.
- Davis RL, Gallagher MA, Asgari MM, Eide MJ, Margolis DJ, Macy E, et al. Identification of Stevens-Johnson syndrome and toxic epidermal necrolysis in electronic health record databases. Pharmacoepidemiology and drug safety. 2015;24:684–92. doi: 10.1002/pds.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide MJ, Tuthill JM, Krajenta RJ, Jacobsen GR, Levine M, Johnson CC. Validation of claims data algorithms to identify nonmelanoma skin cancer. The Journal of investigative dermatology. 2012;132:2005–9. doi: 10.1038/jid.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–8. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frelinger JA. Big Data, Big Opportunities, and Big Challenges. The journal of investigative dermatology Symposium proceedings. 2015;17:33–5. doi: 10.1038/jidsymp.2015.38. [DOI] [PubMed] [Google Scholar]
- iHT2. [Accessed December 14 2016];Transforming Health Care Through Big Data. 2013 < http://c4fd63cb482ce6861463-bc6183f1c18e748a49b87a25911a0555.r93.cf2.rackcdn.com/iHT2_BigData_2013.pdf>.
- Kulldorff M, Dashevsky I, Avery TR, Chan AK, Davis RL, Graham D, et al. Drug safety data mining with a tree-based scan statistic. Pharmacoepidemiology and drug safety. 2013;22:517–23. doi: 10.1002/pds.3423. [DOI] [PubMed] [Google Scholar]
- Laney D. 3D data management: Controlling data volume, variety and velocity. Application Delivery Strategies 2001 [Google Scholar]
- Li Y, Esteva A, Kuprel B, Novoa R, Ko J, Thrun S. Skin Cancer Detection and Tracking using Data Synthesis and Deep Learning. arXiv preprint arXiv: 161201074 2016 [Google Scholar]
- Margolis R, Derr L, Dunn M, Huerta M, Larkin J, Sheehan J, et al. The National Institutes of Health's Big Data to Knowledge (BD2K) initiative: capitalizing on biomedical big data. Journal of the American Medical Informatics Association : JAMIA. 2014;21:957–8. doi: 10.1136/amiajnl-2014-002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray GT, Kulldorff M, Asgari MM. Geographic Clusters of Basal Cell Carcinoma in a Northern California Health Plan Population. JAMA dermatology. 2016;152:1218–24. doi: 10.1001/jamadermatol.2016.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards TB, Johnson CJ, Tatalovich Z, Cockburn M, Eide MJ, Henry KA, et al. Association between cutaneous melanoma incidence rates among white US residents and county-level estimates of solar ultraviolet exposure. Journal of the American Academy of Dermatology. 2011;65:S50–7. doi: 10.1016/j.jaad.2011.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampogna F, Sera F, Abeni D IDI Multipurpose Psoriasis Research on Vital Experiences (IMPROVE) Investigators. Measures of clinical severity, quality of life, and psychological distress in patients with psoriasis: a cluster analysis. The Journal of investigative dermatology. 2004;122:602–7. doi: 10.1046/j.0022-202X.2003.09101.x. [DOI] [PubMed] [Google Scholar]
- Sargen MR, Kanetsky PA, Newton-Bishop J, Hayward NK, Mann GJ, Gruis NA, et al. Histologic features of melanoma associated with CDKN2A genotype. Journal of the American Academy of Dermatology. 2015;72:496–507. doi: 10.1016/j.jaad.2014.11.014. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]