Abstract
Objectives
To evaluate diagnostic image quality of 3D late gadolinium enhancement (LGE) with high isotropic spatial resolution (~1.4mm3) images reconstructed from randomly under-sampled k-space using LOw-dimensional-structure Self-learning and Thresholding (LOST).
Materials and Methods
We prospectively enrolled 270 patients (181 men; 55±14 years) referred for myocardial viability assessment. 3D LGE with isotropic spatial resolution of 1.4±0.1 mm3 was acquired at 1.5T using LOST acceleration rate of 3 to 5. In subset of 121 patients, 3D LGE or phase-sensitive LGE were acquired with parallel imaging with acceleration rate of 2 for comparison. Two readers evaluated image quality using a scale of 1 (poor) to 4 (excellent) and assessed for scar presence. McNemar test statistic was used to compare proportion of detected scar between the two sequences. We assessed association between image quality and characteristics (age, gender, torso dimension, weight, heart rate), using generalized linear models. Overall LGE detection proportions for 3D LGE with LOST were similar between readers 1 and 2 (16.30% versus 18.15%). For image quality, readers gave 85.9% and 80.0% respectively for images categorized as good or excellent. Overall proportion of scar presence was not statistically different from conventional 3D LGE (28% vs. 33% (P= 0.17) for reader 1 and 26% vs. 31% (P= 0.37) for reader 2). Increasing subject heart rate was associated with lower image quality (estimated slope of =-0.009 (P= 0.001)).
Conclusions
High-resolution 3D LGE with LOST yields good to excellent image quality in >80% of patients and identifies patients with LV scar at the same rate as conventional 3D LGE.
Keywords: compressed sensing, late gadolinium enhancement, myocardial viability
Introduction
Myocardial scar and fibrosis can be imaged using late gadolinium enhancement (LGE) cardiac magnetic resonance (MR) (1). LGE has been utilized in identifying ventricular tachycardia (VT) substrates, predicting risk of sudden cardiac death (2–4). Furthermore, the presence and extent of LGE or heterogeneous left ventricular (LV) scar strongly predict adverse cardiac events including appropriate implantable cardioverter defibrillator therapy (5–8). However, challenges in LGE imaging remain. The right ventricle (RV) can also develop fibrosis, but this is difficult to detect due to limited spatial resolution and its thin wall. Similarly, LGE enables imaging of left atrial (LA) scar and fibrosis in patients with atrial fibrillation (9–12), however, the low spatial resolution of current LGE imaging sequences makes it challenging to reliability image LA scar. Therefore, there is an unmet clinical need to further improve LGE spatial resolution in a clinically feasible scan time.
LV LGE imaging is commonly performed using two-dimensional (2D) breath-hold imaging with a slice thickness of 8–10 mm (13). Three-dimensional (3D) LGE acquisition is an alternative approach (14–19), which enables imaging with higher spatial resolution for better assessment of LV scar heterogeneity, RV or LA scar. However, 3D high-resolution LGE requires long scan times, which has limited its clinical adoption. For example, in a recent study by Andreu et. al. (20), the reported 3D LGE scan time was 16.4 ± 7.2 minutes for a spatial resolution of 1.4×1.4×1.4 mm3. Besides patient’s comfort, a long scan time also creates additional artifacts in 3D LGE, such as difficulty in nulling of healthy myocardium due to temporal changes in contrast agent concentration. Parallel imaging can reduce the scan time; however, acceleration rate is often limited to a factor of 2 because of the LGE low signal-to-noise ratio. Compressed sensing (CS) has been previously used to further reduce the 3D LGE scan time (18,21). LOw-dimensional-structure Self-learning and Thresholding (LOST) reconstruction technique has been shown to enable 3D LGE acceleration rates up to rate 3 (21,22). The combination of parallel imaging and LOST has also been shown to further accelerate imaging up to rate 6 in coronary MRI (32). However, CS-based image reconstructions (e.g. LOST) are not yet commercially available. Image reconstruction therefore needs to be performed manually off-line, which is not ideal in clinical settings. Furthermore, prior literature assessing diagnostic image quality of high-resolution 3D LGE with accelerated data acquisition and CS reconstruction is lacking.
Materials and Methods
All imaging sequences were implemented on a 1.5-T Philips Achieva (Philips Healthcare, Best, The Netherlands) system with a 32-channel cardiac phased-array receiver coil. The research protocol was approved by our institutional review board, and written informed consent was obtained from all participants in a HIPAA-compliant manner.
Patient Study
In this prospective study, we recruited patients referred for clinical cardiac MR exam with LGE assessment. Indications for imaging included known or suspected coronary disease, non-ischemic cardiomyopathy or atrial fibrillation. LGE images were acquired 10 to 20 minutes after a bolus (2 ml/s) infusion of 0.2 mmol/kg of Gd-DTPA (Magnevist, @Bayer Schering Pharma AG, Germany), or 0.1 to 0.2 mmol/kg of Gd-BOPTA (MultiHance, @Bracco, Rome, Italy). All patients with eGFR between 30 to 60mL/min/1.73m2 received 0.1 mmol/kg of Gd-BOPTA.
A free-breathing electrocardiogram (ECG)-triggered navigator-gated inversion-recovery gradient echo imaging sequence was used for all acquisitions. Prior to each scan, a Look-Locker acquisition was used to select the inversion time to null the LV myocardial signal. To simplify the clinical protocol, the ECG trigger delay was selected to be the longest trigger time, thereby accommodating the LGE sequence acquisition window. For patients with heart rates >80 bpm (n=29), imaging was performed in systole. A respiratory navigator (2D spiral pencil-beam) placed on the dome of the right hemidiaphragm was used for respiratory motion compensation, utilizing prospective real-time correction. A navigator with fixed scan time was used to adaptively change the acquisition window throughout the scan to achieve a fixed navigator efficiency of 60% (23). A gradient echo imaging sequence with the following parameters was used for 3D LGE: TR/TE= 5.2/2.6 ms, field of view=320×320×100−120 mm3, flip angle=25°. Imaging was performed axially, covering the entire heart. Saturation bands along the phase-encode direction were used to reduce fold-over artifacts. A right-left phase-encoding direction was used to reduce respiratory artifacts from the chest wall. The spatial resolution varied from 1.0 to 1.5 mm3 (except one patient who was imaged with spatial resolution of 2 mm3 to accommodate for large patient size) with mean ± standard deviation of 1.4 ± 0.1 mm3.
In a subset of patients, we also acquired 3D LGE or 3D PSIR images. These images were acquired with a uniform undersampling pattern and were reconstructed on the scanner using Philips SENSE reconstruction implementation. The ordering of acquisition was randomly changed in different patients. The typical imaging parameters for 3D LGE were: spatial resolution = 1.5×1.5×3 mm3, FOV = 320×320×100−120 mm3, flip angle = 25°, TR/TE= 5.3/2.5 ms, acceleration rate of 2, and for 3D PSIR LGE: spatial resolution = 1.5×1.5×5 mm3, FOV = 320×320×100−120 mm3, TR/TE=5.7/2.7 ms, flip angle = 15°, and acceleration rate of 2. The imaging parameters for 3D LGE and 3D PSIR LGE were our standard clinical imaging protocol and were not modified in our study.
LOST Data Acqusition and Reconstruction
3D random undersampling was implemented for the accelerated acquisitions (21,24). A pseudo-random k-space undersampling pattern was generated, by fully sampling a part of the central k-space (corresponding to 15–20% of ky lines and 25% of kz lines) and randomly discarding the edges. This pattern was stored as a lookup table, and a profile reordering was performed to sort the selected ky-kz lines based on their ky and kz location in a radial fashion (25). The acceleration rate was 3 in 150 patients, 4 in 25 patients, 5 in 92 patients, and 6 in 3 patients. The acceleration rate was selected by the technologists. In patients with longer imaging protocols, a higher acceleration rate was chosen. The total acquisition times were approximately 6:25 minutes for 3-fold accelerated acquisitions and approximately 4 minutes for 5-fold accelerated acquisitions at a heart rate of 70 bpm. The exact scan time varied between different subjects to accommodate for changes in imaging field of view, spatial resolution and heart rate.
The detailed explanation for the reconstruction algorithm and parameters has been previously described (21,24). The randomly undersampled k-space data is reconstructed using an iterative B1-weighted LOST algorithm (26). LOST reconstruction uses an initial image estimate to adaptively identify 2D image blocks of similar signal content, and to generate an adaptive sparse representation for the data. These blocks are declared to be similar if this distance is less than a pre-defined fixed threshold. Such similar blocks were then grouped into the similarity cluster of that voxel. The adaptive sparse representation was generated by applying a 3D fast Fourier transform (FFT) to each similarity cluster. De-aliasing of the data was performed via shrinkage of the 3D FFT coefficients of the similarity clusters. The iterative B1-weighted reconstruction approach uses the coil sensitivity information for data consistency during the reconstruction. For faster implementation, coil compression (27) is applied to these 3D coil sensitivity maps to compress the 32-channel coil data to 10 “virtual” channels. The coil compression was performed only once, prior to the iterative portion of the algorithm. At every iteration of the B1-weighted LOST algorithm: 1) the current combined-coil image estimate is thresholded using LOST, 2) individual coil images are generated by the voxel-wise multiplication of the coil sensitivity map of that coil and the combined image, 3) data consistency is enforced by replacing the acquired k-space locations with the acquired lines for each of these individual coil images, and 4) a new combined-coil image estimate is generated by summing the voxel-wise product of the data-consistent coil images and the complex conjugate of the coil sensitivity maps across the coil dimension. LOST reconstruction was implemented in Matlab (The MathWorks, Natick, MA), with the adaptive learning and nonlinear shrinkage portions implemented in C++. The same reconstruction parameters were used in all cases and acceleration rates, allowing for fully automated reconstructions (21,24).
Automated Reconstruction Framework
A software platform was developed to automate the communication and data transfer between the scanner and the remote processing units. Figure 1 shows a schematic diagram of these connections illustrating the sequence of the program workflow. After a scan is completed, the operator uses the program to request the reconstruction of the scanned data, which initiates the following fully automated workflow: 1) the program inquires the scanner database for the scan information including the raw data and the imaging parameters, 2) the database returns the desired information, 3) raw data are packaged along with the imaging parameters and reconstruction options, and are then sent to a remote processing unit for image reconstruction, 4) during the processing, the program periodically connects with the remote processing unit and updates the progress of the operation, and 5) upon reconstruction completion, the reconstructed images are automatically pulled from the remote processing unit, packaged into a Digital Imaging and Communications in Medicine (DICOM) format and then pushed to the scanner database, from where it can also be sent to the hospital Picture Archiving and Communication System (PACS). In this workflow, multiple datasets can be sent at the same time or sequentially. All workflow processes are implemented such that they are performed in the background and do not block operator interaction with the scanner console. Before sending the data, the software generates and attaches a random unique identifier (UID) for the specific data, and then uses this UID to inquire about progress, retrieve the results when ready, and push them into the scanner database with the proper patient and series information to comply with HIPAA, especially if the remote system is located outside the imaging facility or using available web-based cloud computational system. The operator can choose to have the remote processing occur on the scanner machine or at a different remote station on the network, or even on a CPU cluster or GPU server on the same network. In our implementation, a CPU cluster was used for performing the reconstruction. Once the reconstructed images are available in the scanner database or in the PACS, they can be viewed and stored similar to images reconstructed by the vendor reconstruction system. The software program was developed in Matlab (The MathWorks, Natick, MA).
Figure 1.
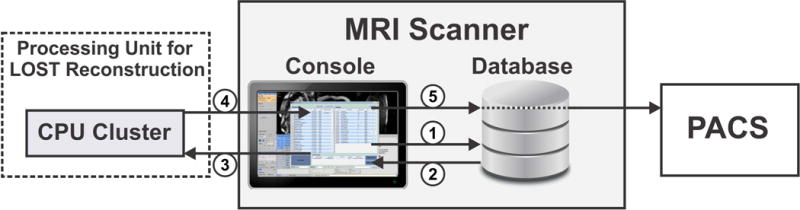
Integration of the automated reconstruction framework into the clinical workflow: (1) the scanner database is queried for scan raw data and imaging parameters, (2) the database returns the requested information, (3) raw data are packed and sent to the Central Processing Unit (CPU) cluster for LOST reconstruction, (4) progress and results are updated upon request from the operator, (5) reconstructed images are pushed to the scanner database, from where they can also be sent to the hospital PACS.
Image Analysis
LOST reconstructed 3D LGE images were written into DICOM format and were used for subjective assessment. In patients with available clinical 3D LGE or PSIR images, reconstructed images from the scanner vendor software were available and were used for assessment. Subjective qualitative assessment was performed by two independent readers (each with 10 years of experience in interpreting clinical cardiac MR exams (CT, FD), blinded to patient history. These readers were not aware of the specifics of the reconstructions, however because of variations in contrast and resolution, they could differentiate between LOST reconstructed images vs. conventional SENSE reconstruction. To minimize this impact, image assessment for LOST and SENSE reconstruction were performed in two separate sessions. Readers reviewed each dataset in Osirix (Pixmeo SARL, Geneva, Switzerland) and were free to adjust the window leveling. Both original and multi-planar reformatted images were used for subjective assessment.
For each dataset, the presence of LGE was assessed using a 3-point scale (absent with confidence = 0, present with confidence = 1, unable to interpret/inconclusive = 2). Furthermore, each dataset was qualitatively assessed using a four-point ordinal system based on overall image quality to assess for viability/scar: 1, poor (large artifacts/no confidence in interpreting LGE in > 50% of the myocardium/); 2, fair (moderate artifacts/LGE interpretable in 50–70% of the myocardium); 3, good (small artifacts/LGE interpretable in 75–90% of the myocardium); 4, excellent (no artifacts/LGE interpretable in >90% of the myocardium).
To assess the potential impact of various patient characteristics on image quality, we extracted the following parameters for each patient: age, gender, heart rate, weight, torso dimension (i.e. posteroanterior (back to chest wall) and transverse (right to left)).
Statistical Analysis
We first analyzed data for all subjects with 3D LGE with LOST. The proportion of the detected LGE presence (“Present with Confidence”) was reported for each reader, as well as the agreement Kappa statistic and 95% confidence interval. Stratification by acceleration rate was carried out to yield these estimates within each rate stratum (acceleration rate 3, 4, and 5). We performed similar analysis for image quality by grouping “good” and “excellent” image quality in one category and estimating the proportion of “good or excellent” for overall image quality and by stratification of rate. Kappa statistic was computed. Second, we analyzed data for the sample of 121 subjects with data from both sequences (3D LGE with LOST, and conventional 3D LGE). We also reported the same statistics as in the sample of 270 subjects. Additionally, we compared the “Present with Confidence” of LGE between the two sequences using McNemar test statistic to account for dependency in the sample, (since subjects in this group were imaged with both sequences). We used SAS software version 9.4 for data management and statistical analysis.
Results
We recruited 270 patients (181 men; 54.9 ± 14.1 years) who were imaged using high-resolution 3D LGE with LOST. We also acquired 3D LGE or 3D PSIR images in subset of 121 patients. The reconstruction framework allowed successful automation of image reconstruction in all cases without the need for any interaction with research scientists on the investigator’s team. Figure 2 shows an example of 3D LGE with high isotropic resolution, as well as 3D PSIR LGE image acquired in a 65-year-old-man with history of coronary artery disease and ventricular tachycardia. Improved spatial resolution allows better depiction of complex scar geometry in the patient. In high-resolution images, several additional small focal enhancements can be seen that were not seen in 3D PSIR images due to limited spatial resolution. The isotropic spatial resolution also allows reformatting in any desired orientation (as shown) with identical image quality as the original axial images. Because of non-isotopic spatial resolution of PSIR LGE, there is additional partial voluming and lower image quality in reformatted images. Viability images from a patient with hypertrophic cardiomyopathy (HCM) are shown in Figure 3. The isotropic spatial resolution of accelerated 3D LGE shows the complex nature of the scar in this patient including papillary muscle. While hyperenhancement can be assessed with 3D PSIR LGE, the visualization of complex scar and scar patchiness are better seen with high-resolution 3D LGE with isotropic spatial resolution. Figure 4 shows viability images in a 62 years-old male patient with sarcoidosis and LV aneurysms. Higher spatial resolution images improve detection of hyperenhancement region, as well as confirm the involvement of right ventricle.
Figure 2.
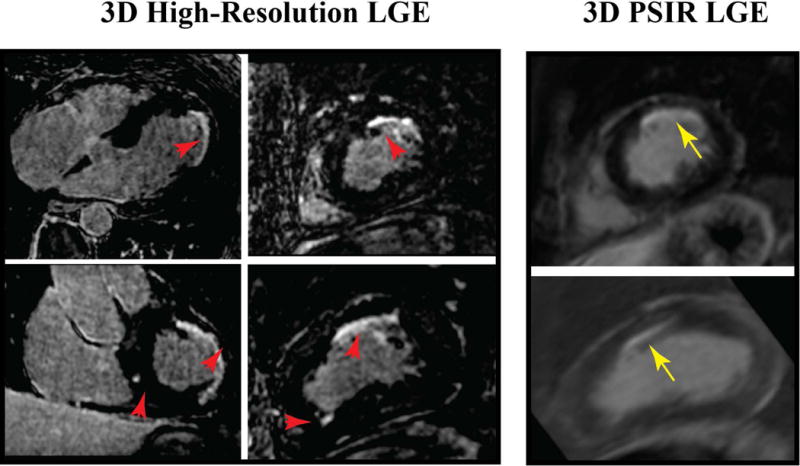
Reformatted 3D Late Gadolinium Enhancement (LGE) images (3D high-resolution LGE with isotropic spatial resolution with LOST rate 5 and 3D Phase Sensitive Inversion recovery (PSIR) LGE with non-isotropic spatial resolution) in a 65 year-old man with coronary artery disease, symptomatic nonsustained ventricular tachycardia and presyncope referred for evaluation of LV scar. Subendocardial LGE in the left anterior descending coronary distribution can be seen in all images. Additionally, smaller scar in remote areas is visible at different slice locations with significantly better depiction of scar in 3D high-resolution LGE with isotropic spatial resolution. Smaller areas of scar are not visible in 3D PSIR LGE with non-isotropic spatial resolution.
Figure 3.
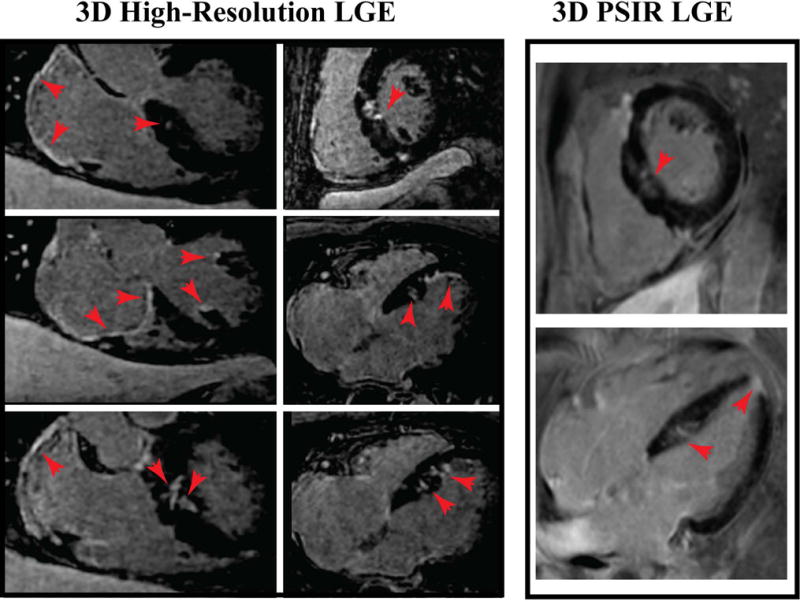
Reformatted 3D LGE of a 67 year-old man with hypertrophic cardiomyopathy. Focal hypertrophy of the mid inferior left ventricular septum. High resolution 3D high-resolution LGE images acquired with LOST rate 3 demonstrate a more detailed depiction of scar compared with 3D PSIR LGE.
Figure 4.
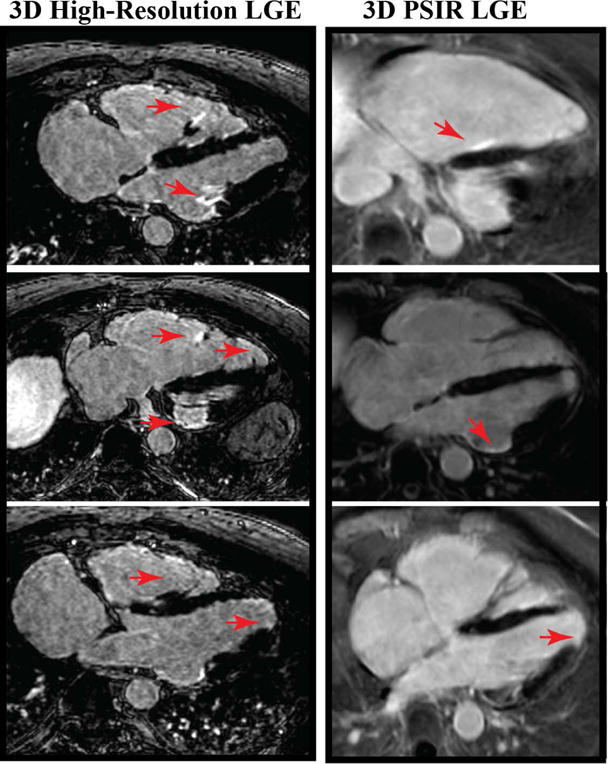
Viability images from a 62 year-old man with cardiac sarcoidosis, comparing high-resolution 3D LGE with 1.2 mm3 isotropic resolution with LOST rate 4 with 3D PSIR LGE with non-isotropic resolution. 3D high-resolution LGE significantly improves visualization of scar and shows involvement of the right ventricular (RV) free wall as well as signal enhancement in the RV papillary muscles and apical LV aneurysm.
Table 1 shows the proportions of “Present with Confidence” for the presence of detected LGE in 3D LGE with LOST; 16.30% and 18.15% for reader 1 and reader 2 respectively. These proportions agree well with a Kappa statistic of 0.78 (95% CI: 0.68–0.88). These proportions are also similar when the sample was stratified by acceleration rate (3, 4, 5). For acceleration rate 4, the sample size is small, and compared to rate 3 and 5, the proportions are smaller. Similar analysis was carried out for image quality. The proportion of “Good or Excellent” is high, 85.93% for reader 1, and 80.00% for reader 2, with a fair Kappa statistic of 0.43 (95% CI 0.22–0.57). Reader 1 scored 4, 34, 88, and 144 as poor, fair, good and excellent, respectively, while reader 2 scored 7, 47, 135, and 81 as poor, fair, good and excellent, respectively. Stratification by rate also yielded similar proportions.
Table 1.
Subjective assessment of 3D LGE with LOST reconstruction (N = 270) for two readers and inter-reader agreement for images acquired via different acceleration rates.
| HYPER-ENHANCEMENT ON LGE | ||||
|---|---|---|---|---|
| Present with Confidence for All Rates (N=270) |
Present with Confidence for Rate=3 (N=150) |
Present with Confidence for Rate=4 (N=25) |
Present with Confidence for Rate=5 (N=92) |
|
| Reader 1 3D LGE with LOST | 16.30% | 15.33% | 8.00% | 20.65% |
| Reader 2 - 3D LGE with LOST | 18.15% | 16.00% | 12.00% | 22.83% |
| IMAGE QUALITY | ||||
|
Good or Excellent for All Rates (N=270) |
Good or Excellent for Rate=3 (N=150) |
Good or Excellent for Rate=4 (N=25) |
Good or Excellent for Rate=5 (N=92) |
|
| Reader 1 - 3D LGE with LOST | 85.93% | 88.00% | 96.00% | 79.35% |
| Reader 2 - 3D LGE with LOST | 80.00% | 79.33% | 96.00% | 76.09% |
| INTER-READER AGREEMENT | ||||
| Kappa | 95% Confidence Interval | |||
| 3D LGE with LOST Score | 0.78 | 0.68–0.88 | ||
| 3D LGE with LOST Image Quality | 0.43 | 0.29–0.57 | ||
Table 2 shows the analysis for the 121 subjects with data for both 3D LGE with LOST, and conventional 3D LGE. For reader 1, the overall proportion of “Present with Confidence” is 28.93% for 3D LGE with LOST, and is not statistically different from 33.88% for conventional 3D LGE (p-value=0.157, McNemar test). We stratified this sample by acceleration rate 3, 4, 5, and found no significant statistical difference between the two proportions. Numerically, 3D LGE with LOST estimates are slightly less than those of the conventional 3D LGE overall, and within each stratum of acceleration rate. The agreement between the two readers via Kappa statistic is high at 0.90 (95% CI: 0.81–0.99) for “Present with Confidence” for LGE detection. Conventional 3D LGE also yields a high Kappa of 0.87 (95% CI: 0.77–0.96). For image quality, “Good or Excellent” is 85.95% for reader 1 for 3D LGE with LOST, and 84.30% for conventional 3D LGE (p-vale=0.637). For reader 2, there is also no statistical difference between the 2 approaches (78.51% versus 85.95%, p-value=0.083). The agreement is fair with Kappa of 0.36 (95% CI: 0.15–0.56) for 3D LGE with LOST. For Conventional 3D LGE, the Kappa is high 0.80 (95% CI: 0.65–0.96).
Table 2.
Subjective assessment of the subset of patients with both 3D LGE with LOST reconstruction and conventional 3D LGE (N = 121) for two readers and inter-reader agreement for images acquired via different acceleration rates (3 to 5).
| Hyper-enhancement on LGE | ||||
|---|---|---|---|---|
| Present with Confidence for All Rates (N=121) |
Present with Confidence for Rate=3 (N=48) |
Present with Confidence for Rate=4 (N=13) |
Present with Confidence for Rate=5 (N=60) |
|
| Reader 1 - 3D LGE with LOST | 28.93% | 33.33% | 7.69% | 30.00% |
| Reader 1 - Conventional 3D LGE | 33.88% | 35.42% | 30.77% | 33.33% |
| P-value Comparison of LOST and Conventional LGE for Reader 1* | 0.157 | 0.739 | 0.083 | 0.414 |
| Reader 2 - 3D LGE with LOST | 26.45% | 29.17% | 7.69% | 28.33% |
| Reader 2 - Conventional 3D LGE | 31.40% | 35.42% | 23.08% | 30.00% |
| P-value Comparison of LOST and Conventional LGE for Reader 2* | 0.134 | 0.317 | 0.157 | 0.655 |
| IMAGE QUALITY | ||||
|
Good or Excellent for All Rates (N=121) |
Good or Excellent for Rate=3 (N=48) |
Good or Excellent for Rate=4 (N=13) |
Good or Excellent for Rate=5 (N=60) |
|
| Reader 1 - 3D LGE with LOST | 85.95% | 85.42% | 100.00% | 83.33% |
| Reader 1 - Conventional 3D LGE | 84.30% | 85.42% | 100.00% | 80.00% |
| P-value Comparison of LOST and Conventional Image Quality for Reader 1* | 0.637 | 1.000 | 1.000 | 0.564 |
| Reader 2 - 3D LGE with LOST | 78.51% | 75.00% | 92.71% | 78.33% |
| Reader 2 - Conventional 3D LGE | 85.95% | 87.50% | 100.00% | 81.67% |
| P-value Comparison of LOST and Conventional Image Quality for Reader 2* | 0.083 | 0.109 | 1.000 | 0.564 |
| INTER-READER AGREEMENT | ||||
| Kappa# | 95% Confidence Interval | |||
| 3D LGE with LOST Score | 0.90 | 0.81–0.99 | ||
| Conventional 3D LGE Score | 0.87 | 0.77–0.96 | ||
| 3D LGE with LOST Image Quality | 0.36 | 0.15–0.56 | ||
| Conventional 3D LGE Image Quality | 0.80 | 0.65–0.96 | ||
McNemar test;
Kappa based on 2 by 2 table Present with Confidence versus otherwise, Good or Excellent
The baseline characteristics of the study population and torso size measurements are shown in Table 3. Increasing subject heart rate, weight, and torso dimension are associated negatively with subjective image quality (Table 4), but only heart rate reached statistical significance.
Table 3.
Baseline characteristics of the study population (n = 270).
| Baseline Patient characteristics | N=270 |
|---|---|
| Age years (range) | 55 (18–85) |
| Male subjects | 181 (66.7%) |
| Heart rate (/min) | 66 ± 13 |
| Weight (kg) | 83 ± 15 |
| Chest posteroanterior dimension (cm) | 23.1 ± 3.4 |
| Chest transverse dimension (cm) | 35.3 ± 3.6 |
Table 4.
Relationship between subjective average qualitative image assessment scores and patient factors.
| Patient Factors | Change in mean of average image score per unit change in patient factor* | P-value |
|---|---|---|
| Heart Rate | −0.009 (0.003) | 0.001 |
| Weight | −0.003 (0.003) | 0.323 |
| Chest AP dimension | −0.012 (0.015) | 0.421 |
| Chest transverse dimension | −0.027 (0.015) | 0.064 |
Slope estimate and standard error from multivariable generalized linear model.
Discussion
We have developed and implemented a clinically feasible image reconstruction framework for accelerated data acquisition and reconstruction using the LOST technique. Image quality of 3D LOST-accelerated LGE was evaluated in 270 patients with known or suspected cardiovascular disease referred for clinical viability assessment. We demonstrated that using this approach, 3D LGE with high isotropic spatial resolution is clinically feasible with acceptable image quality in most patients.
Despite the growth in development of CS-based image reconstruction techniques, there have been very limited efforts in clinical translation and evaluation (28–35). Sharma et. al. (30) evaluated CS based reconstruction for routine neuro-imaging sequences and found that CS was able to moderately accelerate certain neuroimaging sequences without severe loss of clinically relevant information. For those sequences with coarser spatial resolution and/or at a higher acceleration factor, artifacts degraded the quality of the reconstructed image to the point where they are of minimal/no clinical value. Vasanawala et. al. (31) showed the feasibility of improved pediatric imaging to achieve higher resolution and/or faster imaging in 34 patients in a clinical setting. Hsiao et. al. (32) assessed the potential of CS in four-dimensional (4D) phase-contrast MR imaging for the evaluation of valvular insufficiency and intra-cardiac shunts in 34 patients with congenital heart disease and demonstrated that the CS 4D phase-contrast sequence can augment conventional cardiac MR imaging by improving sensitivity for and depiction of hemodynamically significant shunts and valvular regurgitation. Roujol et. al. evaluated the performance of CS-accelerated ECG gated pulmonary vein MRA in 19 patients with atrial fibrillation (29). Mann et. al. assessed the utility of CS in quantification of the fat fraction in 11 patients with type 2 diabetes and concluded that acceptable image quality can be achieved with acceleration up to a factor of 3.8 (28). These exploratory studies demonstrated the potential and clinical feasibility of the CS-based approach for accelerated imaging; however, evaluations were only performed in a small and selected number of patients (i.e. between 20–40).
Despite excellent image quality, the long reconstruction time remains one of the main limitations of this technique, with a typical reconstruction time of 1 hour for high-resolution 3D LGE. In our implementation, a 3D volume reconstructed by zero filling of the under sampled data will be available immediately after imaging. Because of fully sampled center of k-space, we found these images to be helpful in predicting the image quality of the CS reconstructed images and often scar can be seen in these images. While this workflow does not work on cases where immediate access to reconstructed data is needed (e.g. real-time MRI), immediate reconstruction is not necessary in our specific application of 3D LGE. In our clinical practice, images from each patient are typically evaluated the next day, allowing sufficient time for image reconstruction for clinical evaluation. Further research to reduce the reconstruction time and development of alternative reconstruction techniques is warranted.
Our study only evaluated the image quality of isotropic 3D LGE with high resolution in our tertiary care medical center’s referral population. The clinical impact of high spatial resolution imaging for different diseases should be carefully assessed in future studies. Not all patients may need imaging with high spatial resolution, however for some cardiovascular diseases, this may be clinically warranted. For example, a higher spatial resolution may allow better visualization of the LV scar morphology, which may facilitate assessment for ventricular arrhythmia. In addition, a volumetric isotropic 3D LGE allows for image reconstruction in any orientation and multi-planar visualization of the data, which further improves confidence in detection of scar by confirming the scar in different views. Further studies are needed to establish the clinical impact of high-resolution isotropic 3D LGE. We also found that there were differences in scoring between the two readers for the LOST accelerated imaging dataset. While the proportion of good to excellent quality was similar between the two readers, one of our reader’s scores were more positive and majority of images were scored as excellent.
Image reconstruction for CS is commonly performed offline, which has hindered evaluation of its various reconstruction algorithms in a clinical environment. Other stand-alone reconstruction frameworks have been described. A flexible real-time MRI reconstruction platform (i.e. RTHawks) has been widely applied in real-time MRI (36). A real-time display and reconstruction system was also developed at the National Institutes of Health and is widely used for interventional procedures (37). Recently, a flexible stand-alone MRI image reconstruction framework (38,39) has been developed which provides additional flexibility for reconstruction and use of cloud computing to facilitate image reconstruction. These reconstruction platforms are designed mainly for situations where real-time or immediate reconstruction of MRI data is needed, such as image guidance or real-time imaging.
Our study has several limitations. We did not perform a direct comparison between 3D and 2D LGE imaging, where the latter is the current clinical standard at many medical centers. Currently, all patients imaged for LV viability at our medical center are imaged using 3D LGE. The spatial resolution varied between different patients to accommodate patient size and heart rate. Due to the inherent de-noising in CS, we cannot directly perform SNR or CNR measurements. We used fixed image reconstruction parameters for all studies and did not optimize the parameters based on acceleration rate.
In conclusion, 3D LGE with LOST yields good to excellent image quality in >80% of patients and identifies patients with LV scar at the same rate as conventional 3D LGE. High isotropic spatial resolution (1–1.4 mm3) detects smaller scar and enables multi-planar reconstruction of 3D LGE to facilitate scar visualization.
Acknowledgments
We thank Sophie Berg, Kraig V. Kissinger, and Beth Goddu for patient recruitment and scanning.
Grant Support: The project described was supported by NIH: R01EB008743, R01HL129185-01, R01HL127015, R21HL127650, AHA: 15EIA22710040. Dr. Akcakaya was supported by NIH K99HL111410 and R00HL111410.
References
- 1.Kim RJ, Fieno DS, Parrish TB, et al. Relationship of MRI delayed contrast enhancement to irreversible injury, infarct age, and contractile function. Circulation. 1999;100(19):1992–2002. doi: 10.1161/01.cir.100.19.1992. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt A, Azevedo CF, Cheng A, et al. Infarct Tissue Heterogeneity by Magnetic Resonance Imaging Identifies Enhanced Cardiac Arrhythmia Susceptibility in Patients With Left Ventricular Dysfunction. Circulation. 2007;115(15):2006–2014. doi: 10.1161/CIRCULATIONAHA.106.653568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan AT, Shayne AJ, Brown KA, et al. Characterization of the peri-infarct zone by contrast-enhanced cardiac magnetic resonance imaging is a powerful predictor of post-myocardial infarction mortality. Circulation. 2006;114(1):32–39. doi: 10.1161/CIRCULATIONAHA.106.613414. [DOI] [PubMed] [Google Scholar]
- 4.Roes SD, Borleffs CJ, van der Geest RJ, et al. Infarct tissue heterogeneity assessed with contrast-enhanced MRI predicts spontaneous ventricular arrhythmia in patients with ischemic cardiomyopathy and implantable cardioverter-defibrillator. Circulation Cardiovascular imaging. 2009;2(3):183–190. doi: 10.1161/CIRCIMAGING.108.826529. [DOI] [PubMed] [Google Scholar]
- 5.Assomull RG, Prasad SK, Lyne J, et al. Cardiovascular magnetic resonance, fibrosis, and prognosis in dilated cardiomyopathy. J Am Coll Cardiol. 2006;48(10):1977–1985. doi: 10.1016/j.jacc.2006.07.049. [DOI] [PubMed] [Google Scholar]
- 6.Wu KC, Weiss RG, Thiemann DR, et al. Late gadolinium enhancement by cardiovascular magnetic resonance heralds an adverse prognosis in nonischemic cardiomyopathy. J Am Coll Cardiol. 2008;51(25):2414–2421. doi: 10.1016/j.jacc.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iles L, Pfluger H, Lefkovits L, et al. Myocardial fibrosis predicts appropriate device therapy in patients with implantable cardioverter-defibrillators for primary prevention of sudden cardiac death. Journal of the American College of Cardiology. 2011;57(7):821–828. doi: 10.1016/j.jacc.2010.06.062. [DOI] [PubMed] [Google Scholar]
- 8.Rayatzadeh H, Tan A, Chan RH, et al. Scar heterogeneity on cardiovascular magnetic resonance as a predictor of appropriate implantable cardioverter defibrillator therapy. J Cardiovasc Magn Reson. 2013;15(1):31. doi: 10.1186/1532-429X-15-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGann CJ, Kholmovski EG, Oakes RS, et al. New magnetic resonance imaging-based method for defining the extent of left atrial wall injury after the ablation of atrial fibrillation. J Am Coll Cardiol. 2008;52(15):1263–1271. doi: 10.1016/j.jacc.2008.05.062. [DOI] [PubMed] [Google Scholar]
- 10.Peters DC, Wylie JV, Hauser TH, et al. Recurrence of atrial fibrillation correlates with the extent of post-procedural late gadolinium enhancement: a pilot study. JACC Cardiovasc Imaging. 2009;2(3):308–316. doi: 10.1016/j.jcmg.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marrouche NF, Wilber D, Hindricks G, et al. Association of atrial tissue fibrosis identified by delayed enhancement MRI and atrial fibrillation catheter ablation: the DECAAF study. JAMA. 2014;311(5):498–506. doi: 10.1001/jama.2014.3. [DOI] [PubMed] [Google Scholar]
- 12.Moghari MH, Peters DC, Smink J, et al. Pulmonary vein inflow artifact reduction for free-breathing left atrium late gadolinium enhancement. Magnetic Resonance in Medicine. 2011;66(1):180–186. doi: 10.1002/mrm.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kramer C, Barkhausen J, Flamm S, et al. Standardized cardiovascular magnetic resonance (CMR) protocols 2013 update. Journal of Cardiovascular Magnetic Resonance. 2013;15(1):91. doi: 10.1186/1532-429X-15-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen TD, Spincemaille P, Weinsaft JW, et al. A fast navigator-gated 3D sequence for delayed enhancement MRI of the myocardium: comparison with breathhold 2D imaging. J Magn Reson Imaging. 2008;27(4):802–808. doi: 10.1002/jmri.21296. [DOI] [PubMed] [Google Scholar]
- 15.Peukert D, Laule M, Taupitz M, Kaufels N, Hamm B, Dewey M. 3D and 2D delayed-enhancement magnetic resonance imaging for detection of myocardial infarction: preclinical and clinical results. Acad Radiol. 2007;14(7):788–794. doi: 10.1016/j.acra.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Peters DC, Appelbaum EA, Nezafat R, et al. Left ventricular infarct size, peri-infarct zone, and papillary scar measurements: A comparison of high-resolution 3D and conventional 2D late gadolinium enhancement cardiac MR. J Magn Reson Imaging. 2009;30(4):794–800. doi: 10.1002/jmri.21897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moghari MH, Chan RH, Hong SN, et al. Free-breathing cardiac MR with a fixed navigator efficiency using adaptive gating window size. Magnetic Resonance in Medicine. 2012 doi: 10.1002/mrm.24210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roujol S, Basha TA, Akcakaya M, et al. 3D late gadolinium enhancement in a single prolonged breath-hold using supplemental oxygenation and hyperventilation. Magn Reson Med. 2014;72(3):850–857. doi: 10.1002/mrm.24969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saranathan M, Rochitte CE, Foo TK. Fast, three-dimensional free-breathing MR imaging of myocardial infarction: a feasibility study. Magn Reson Med. 2004;51(5):1055–1060. doi: 10.1002/mrm.20061. [DOI] [PubMed] [Google Scholar]
- 20.Andreu D, Ortiz-Pérez JT, Fernández-Armenta J, et al. 3D delayed-enhanced magnetic resonance sequences improve conducting channel delineation prior to ventricular tachycardia ablation. Europace. 2015;17(6):938–945. doi: 10.1093/europace/euu310. [DOI] [PubMed] [Google Scholar]
- 21.Akcakaya M, Rayatzadeh H, Basha TA, et al. Accelerated late gadolinium enhancement cardiac MR imaging with isotropic spatial resolution using compressed sensing: initial experience. Radiology. 2012;264(3):691–699. doi: 10.1148/radiol.12112489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akcakaya M, Basha TA, Goddu B, et al. Low-dimensional-structure self-learning and thresholding: Regularization beyond compressed sensing for MRI Reconstruction. Magn Reson Med. 2011;66(3):756–767. doi: 10.1002/mrm.22841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moghari MH, Chan RH, Hong SN, et al. Free-breathing cardiac MR with a fixed navigator efficiency using adaptive gating window size. Magn Reson Med. 2012;68(6):1866–1875. doi: 10.1002/mrm.24210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akcakaya M, Basha TA, Goddu B, et al. Low-dimensional-structure self-learning and thresholding: Regularization beyond compressed sensing for MRI Reconstruction. Magnetic Resonance in Medicine. 2011;66(3):756–767. doi: 10.1002/mrm.22841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akcakaya M, Basha TA, Chan RH, et al. Accelerated contrast-enhanced whole-heart coronary MRI using low-dimensional-structure self-learning and thresholding. Magnetic Resonance in Medicine. 2012;67(5):1434–1443. doi: 10.1002/mrm.24242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akcakaya M, Basha TA, Chan RH, Manning WJ, Nezafat R. Accelerated isotropic sub-millimeter whole-heart coronary MRI: compressed sensing versus parallel imaging. Magn Reson Med. 2014;71(2):815–822. doi: 10.1002/mrm.24683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang F, Vijayakumar S, Li Y, Hertel S, Duensing GR. A software channel compression technique for faster reconstruction with many channels. Magn Reson Imaging. 2008;26(1):133–141. doi: 10.1016/j.mri.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 28.Mann LW, Higgins DM, Peters CN, et al. Accelerating MR Imaging Liver Steatosis Measurement Using Combined Compressed Sensing and Parallel Imaging: A Quantitative Evaluation. Radiology. 2016;278(1):247–256. doi: 10.1148/radiol.2015150320. [DOI] [PubMed] [Google Scholar]
- 29.Roujol S, Foppa M, Basha TA, et al. Accelerated free breathing ECG triggered contrast enhanced pulmonary vein magnetic resonance angiography using compressed sensing. J Cardiovasc Magn Reson. 2014;16:91. doi: 10.1186/s12968-014-0091-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma SD, Fong CL, Tzung BS, Law M, Nayak KS. Clinical image quality assessment of accelerated magnetic resonance neuroimaging using compressed sensing. Invest Radiol. 2013;48(9):638–645. doi: 10.1097/RLI.0b013e31828a012d. [DOI] [PubMed] [Google Scholar]
- 31.Vasanawala SS, Alley MT, Hargreaves BA, Barth RA, Pauly JM, Lustig M. Improved pediatric MR imaging with compressed sensing. Radiology. 2010;256(2):607–616. doi: 10.1148/radiol.10091218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsiao A, Lustig M, Alley MT, Murphy MJ, Vasanawala SS. Evaluation of valvular insufficiency and shunts with parallel-imaging compressed-sensing 4D phase-contrast MR imaging with stereoscopic 3D velocity-fusion volume-rendered visualization. Radiology. 2012;265(1):87–95. doi: 10.1148/radiol.12120055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaspan ON, Fleysher R, Lipton ML. Compressed sensing MRI: a review of the clinical literature. Br J Radiol. 2015;88(1056):20150487. doi: 10.1259/bjr.20150487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang T, Chowdhury S, Lustig M, et al. Clinical performance of contrast enhanced abdominal pediatric MRI with fast combined parallel imaging compressed sensing reconstruction. J Magn Reson Imaging. 2014;40(1):13–25. doi: 10.1002/jmri.24333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akcakaya M, Hu P, Chuang ML, et al. Accelerated noncontrast-enhanced pulmonary vein MRA with distributed compressed sensing. J Magn Reson Imaging. 2011;33(5):1248–1255. doi: 10.1002/jmri.22559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos JM, Wright GA, Pauly JM. Flexible real-time magnetic resonance imaging framework. Conf Proc IEEE Eng Med Biol Soc. 2004;2:1048–1051. doi: 10.1109/IEMBS.2004.1403343. [DOI] [PubMed] [Google Scholar]
- 37.Guttman MA, Kellman P, Dick AJ, Lederman RJ, McVeigh ER. Real-time accelerated interactive MRI with adaptive TSENSE and UNFOLD. Magn Reson Med. 2003;50(2):315–321. doi: 10.1002/mrm.10504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen MS, Sorensen TS. Gadgetron: an open source framework for medical image reconstruction. Magn Reson Med. 2013;69(6):1768–1776. doi: 10.1002/mrm.24389. [DOI] [PubMed] [Google Scholar]
- 39.Xue H, Inati S, Sorensen TS, Kellman P, Hansen MS. Distributed MRI reconstruction using Gadgetron-based cloud computing. Magn Reson Med. 2015;73(3):1015–1025. doi: 10.1002/mrm.25213. [DOI] [PMC free article] [PubMed] [Google Scholar]


