Abstract
Inactivating mutations in the melanocortin 3 receptor (Mc3r) have been described as causing obesity in mice, but the physiologic effects of MC3R mutations in humans have been less clear. Here we review the MC3R polymorphisms and mutations identified in humans, and the in vitro, murine, and human cohort studies examining their putative effects. Some, but not all, studies suggest that the common human MC3R variant T6K+V81I, as well as several other rare, function-altering mutations, are associated with greater adiposity and hyperleptinemia with altered energy partitioning. In vitro, the T6K+V81I variant appears to decrease MC3R expression and therefore cAMP generation in response to ligand binding. Knockin mouse studies confirm the T6K+V81I variant increases feeding efficiency and the avidity with which adipocytes derived from bone or adipose tissue stem cells store triglycerides. Other MC3R mutations occur too infrequently in the human population to make definitive conclusions regarding their clinical effects.
Introduction
Human body weight is a complex trait determined by the interactions between environmental influences and genetic risk factors [1]. Twin and adoption studies suggest heritable characteristics explain 60–80% of the observed variance of human body weight [2]. It is also well established that the hypothalamus has a critical role in maintaining energy balance by regulating appetite and metabolism, and perturbations of hypothalamic function can lead to obesity [3]. Therefore, significant research has recently focused on the genes and proteins involved in hypothalamic control of body weight, including the leptin-melanocortin pathway. One hypothalamic protein, the melanocortin 3 receptor (MC3R), is believed to be one of the many factors important for regulating energy homeostasis; however its role is not completely understood. Moreover, recent evidence suggests that MC3R may regulate metabolism not only via central but also via peripheral actions [4–6]. Herein we briefly review MC3R structure and function, discuss the polymorphisms and mutations identified in humans, and summarize the in vitro, murine, and human studies examining their putative effects.
MC3R and its downstream signaling pathways
MC3R is a member of the melanocortin receptor family. Other members of the family include MC1R, important for skin and hair pigmentation; MC2R or the adrenocorticotropin (ACTH) receptor, critical for proper cortisol regulation; MC4R, integral for proper energy homeostasis and metabolism; and MC5R, important for piloerection [7, 8]. The human MC3R was first cloned in 1993 as a single exon gene located in chromosome 20q13.2. A sequence with high homology was then later found in mice on chromosome 2q [9–11]. The MC3R gene is highly conserved, with orthologs in 171 organisms including monkey, dog, mouse and rat (https://www.ncbi.nlm.nih.gov/gene/4159). MC3R and MC4R have 58% identity and 76% similarity [12].
Studies have evaluated the transcript structure of murine Mc3r and human MC3R by examining its untranslated regions (UTRs) [13, 14], which can play important roles in gene expression, mRNA stability, and translational efficiency. Research from our group [14] demonstrated that the murine Mc3r 5′ UTR has two transcription start sites: one 368 bases upstream of the translation start site and another 440 bases upstream, with putative initiator sequences associated with each start site. The murine Mc3r 3′ UTR terminates 1286 bases after the translational stop codon, but with a 787 base splice between consensus splice donor and acceptor sites from 171 bases to 958 bases downstream of the stop codon. The human MC3R 5′ UTR, on the other hand, has a transcription start site 527–544 bases upstream of the classical translation start site, with most transcripts starting 533 bases upstream, near a putative initiator sequence. There is also a 248 base splice from 140 to 388 bases upstream of the start codon between consensus splice donor and acceptor sites [13, 14]. The human MC3R 3′ UTR was found to end only 115–160 bases after the translational stop codon [13, 14]. The human MC3R has more than one potential start site for translation, with additional in-frame ATGs located 37 and 109 codons downstream. The first ATG is found only in humans and nonhuman primates, while the second ATG appears widely conserved among vertebrates [13]. Park et al. reported that the full-length 5’ UTR directs utilization of the second in-frame ATG as the primary translation start site instead of the canonically-assumed first ATG [13]. This leads to a protein transcript which is 37 codons shorter than the classical MC3R transcript [15]; although other research suggests that translation can occur from the first ATG site as well [16]. These studies suggested the possibility of transcript heterogeneity for both human MC3R and murine Mc3r that could potentially enable tissue-specific gene regulation.
MC3R is a G-protein-coupled receptor (GPCR), consisting of seven transmembrane helices with and extracellular N-terminus and an intracellular C-terminus [Figure 1]. While some GPCRs, such as MC4R, demonstrate substantial basal activity, MC3R likely has little or no basal activity [17]. Upon ligand binding, MC3R’s effects are believed primarily to be mediated via the Gsα protein, which activates adenylyl cyclase and produces cAMP. This secondary messenger, in turn, initiates further downstream signaling. MC3R has also been shown to activate ERK1/2 signaling, although somewhat inconsistent data have been reported [18–20]. Further studies are needed to elucidate how the two signaling pathways behave in different tissues.
Figure 1.
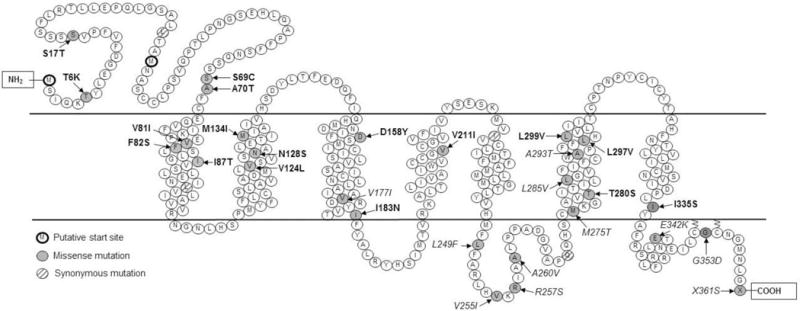
MC3R mutations and variants identified in humans. Mutations in bold font represent possibly deleterious mutations. Mutations in italics represent presumed benign sequence variants. Adapted from [54, 88].
MC3R and metabolic function
MC3R was initially found to be expressed in areas of the central nervous system associated with complex behaviors related to food ingestion and autonomic function [21]. Relatively high levels of expression are observed in hypothalamic and limbic structures; however expression has also been reported in peripheral tissues including kidney, heart, liver, gut, leukocytes, and adipose tissue [9, 22–24].
MC3R’s effects are governed by the binding of endogenous melanocortin ligands, which in turn are regulated and released by signals reflecting metabolic conditions. The endogenous agonists α-melanocyte-stimulating hormone (α-MSH) and γ-MSH are neuropeptides, cleaved from the pro-hormone proopiomelanocortin (POMC) that can activate MC3R. α-MSH is a non-selective melanocortin receptors agonist, capable of binding to MC1R, MC3R, MC4R, and MC5R [25]. γ-MSH, on the other hand, is more selective for MC3R [26, 27], suggesting that MC3R-specific regulatory pathways could exist.
Roles of MC3R in energy homeostasis
The MC3R appears to regulate metabolism in multiple distinct fashions in the CNS and at peripheral sites, such that its impact on energy homeostasis is not straightforward. Within the arcuate nucleus, the MC3R appears to function as an inhibitory autoreceptor for POMC neurons. Peripheral injection of D-Trp8-γ-MSH, a very selective and potent MC3R agonist [28], stimulates short-term feeding behavior, providing physiological evidence that, at least acutely, MC3R activation is orexigenic [26]. In the rostral arcuate nucleus, MC3R mRNA is expressed in about 40% of POMC neurons and >50% of NPY/AGRP neurons [29, 30]. Arcuate MC3R activation is believed to lead to release of GABA from NPY/AGRP neurons and thus inhibition of POMC neuronal activity. In rat arcuate brain slices, D-Trp8-γ-MSH increases the frequency of GABA-mediated mini-inhibitory postsynaptic currents in POMC neurons, causes hyperpolarization, and decreases action potential generation [31]. Roger Cone’s group has reported that mice with inactivation of MC3R have greater anorexia and loss of lean tissues in response to lipopolysaccharide injection or tumor growth [32], as well as impaired feeding response to fasting [33], all consistent with a role for MC3R activation as stimulatory for energy intake and gain of lean mass. Data also suggest activation of MC3R is essential for proper alignment of metabolic responses during entrainment to restricted feeding regimens [34] and that MC3R in the limbic system is important for motivating feeding after energy restriction [35].
Nevertheless, inactivation of Mc3r in mice is associated with an obesity phenotype that becomes more evident with high-fat diet feeding [36–38]. The obesity is moderate compared to Mc4r deficiency and total body mass is less affected than adipose tissue triglyceride content. Mc3r−/− mice exhibit increased fat mass but reduced fat-free mass, decreased linear growth and femur length [36, 37], and reduced adipose tissue inflammation [39] along with some evidence of reduced effects of leptin administration on food intake [40]. More consistently, however, Mc3r−/− mice show greater feeding efficiency (the ratio of weight gain to energy intake) [36]. They have also been reported to have decreased locomotor activity particularly during the dark phase [41] and altered circadian rhythmicity in constant dark conditions [42]. They are, under most conditions hypophagic or normophagic relative to controls and appear to maintain normal metabolic rate and metabolic profiles despite their obese phenotype. Thus, the obesity phenotype of Mc3r deficiency differs dramatically from that of murine Mc4r deficiency, where marked hyperphagia, increased lean mass, substantial insulin resistance, and adipose tissue inflammation are seen. It is clear that MC3R and MC4R play distinct roles in body weight and body composition regulation that are not redundant, because mice with simultaneous knockout of the Mc3r and Mc4r are more obese than mice with either knockout alone [37]. MC3R does not appear to act solely by altering the activation state of POMC neurons or of MC4R; it also regulates body weight at other central nervous system and peripheral sites.
The precise mechanisms through which MC3R alters metabolism to cause an obese state have not been fully identified. Several lines of evidence point to a role for MC3R in nutrient partitioning, namely towards adipose tissue and away from lean tissues [33]. Mc3r deletion has been found to result in slightly increased nadir corticosterone, but a somewhat suppressed glucocorticoid response to fasting. Several phenotypic features of Mc3r deficiency are reminiscent of a Cushing-like syndrome, including hypercorticosteronemia and increased visceral adiposity [33], although lipolysis and hormone-sensitive lipase expression in response to fasting is reported to be reduced in mice with Mc3r deficiency [33] and is usually well maintained in states of glucocorticoid excess [43]. Hypercorticosteronemia is also known to result in increased bone resorption and decreased bone deposition [44], and so the elevated corticosterone could potentially explain decreased linear growth of Mc3r-deficient mouse models. A study from Moller et al demonstrated mRNA expression of MC3R in human subcutaneous adipocytes, albiet to a lesser extent than MC1R, MC2R, or MC5R [23]. Additionally You et al showed mRNA expression of Mc3r in rat brain, liver and adipose tissue [45]. Taken together, the reduced capacity for lipolysis described in Mc3r−/− mice [33] along with the evidence above suggest adipose tissue might be an important site for MC3R-related body weight regulation.
Begriche et al., investigated the central versus peripheral roles of MC3R using a lox-stop-lox (LoxTB) sequence that globally blocked Mc3r transcription and could be reactivated in tissues of interest with tissue-specific Cre recombinase [6]. Homozygous carriers of the inactivating LoxTB Mc3r alleles (Mc3rTB/TB) displayed an obese phenotype similar to that observed in Mc3r−/− mice. Mc3rTB/TB mice displayed reduced lean mass, increased fat mass, and accelerated diet-induced obesity. However, after rescuing Mc3r expression only in the nervous system using a Nestin-Cre transgene, there was only a partial reduction of obesity in chow-fed conditions and little effect on high fat diet-induced obesity [6]. These data suggest that MC3R does not solely act in the CNS; rather, peripheral MC3R has a distinct pathway for body weight regulation that may be of importance for whole body homeostasis beyond the effects of the central receptors that have been the subject of much prior investigation.
To this point, liver microarray data suggest that Mc3r−/− mice exhibit increased expression of enzymes involved in hepatic lipogenesis and triglyceride synthesis with suppression of gluconeogenic enzymes [5]. There is also evidence for activation of a “cellular stress response” in the liver of Mc3r−/− mice, with increased expression of heat shock proteins and autophagy-related genes [5]. Further research is needed to investigate MC3R’s peripheral pathways of action.
Human Studies
Epidemiology of MC3R variants in Humans
In order to elucidate the molecular underpinnings of obesity in humans, linkage studies examining genotype-obesity phenotype correlations in different populations have been performed. A linkage analysis in French Canadians discovered a locus in 20q, homologous to an obesity quantitative trait locus found in mice, which was associated with increased adiposity and fasting insulin among humans [11]. A subsequent study described that obesity and fat mass percentage could be linked to markers in 20q13, within which is located the MC3R gene [46]. This locus does not seem to be represented, however, as a site with genome-wide significance in large genome-wide association studies. Based on the early linkage analyses and the Mc3r knockout obesity phenotype, sequencing of the MC3R gene and association studies with body weight in human cohorts have been performed in numerous populations.
The most common human variants encountered are C17A and G241A, which respectively change two amino acids in MC3R: Thr6Lys and Val81Ile (T6K+V81I). These variants occur in almost complete linkage disequilibrium. This common variant (T6K+V81I) has significant racial/ethnic differences in haplotype frequency. Results from published cohort studies show that homozygosity for this haplotype is rare in most races (<5%), except for non-Hispanic Blacks [Table 1] [47, 48]. Data from the 1000 Genomes Project confirm these findings, with the frequency of the minor allele varying greatly among African (44%), Asian (23%), Kuwaiti (14%), American (11%), and European (8%) samples [49]. The substitution of lysine for threonine is in the extracellular domain of the protein and potentially creates a novel site for ubiquitination. The substitution of isoleucine for valine (each a hydrophobic amino acid) is in the first transmembrane domain.
Table 1.
Distribution of T6K+V81I Haplotypes among different cohorts
| Year | Country | Cohort | No. of Subjects | HOM (%) | HET (%) | WT (%) | Reference |
|---|---|---|---|---|---|---|---|
| 2000 | US | Adult Females BMI≥40 or BMI<27 | 209 | 3.3 | 19.1 | 77.5 | [50] |
| Black | 51 | 11.8 | 43.1 | 45.1 | |||
| 2001 | France | Adults | 526 | 1.7 | 14.3 | 84.0 | [58] |
| 2002 | Canada | Adults | 222 | 0.9 | 19.8 | 79.3 | [59] |
| 2002 | NZ | Maori | 12 | 0 | 66.7 | 33.3 | [60] |
| 2003 | Finland1 | Adults BMI≥40 | 244 | 1.2 | 15.2 | 83.6 | [67] |
| 2004 | Greece2 | Women | 116 | 1.7 | 19.8 | 78.4 | [68] |
| 2005 | US | Children | 355 | 8.2 | 34.9 | 56.9 | [47] |
| Black | 152 | 15.8 | 52.6 | 31.6 | |||
| Caucasian | 176 | 1.7 | 18.2 | 80.1 | |||
| Other Race | 27 | 7.4 | 44.4 | 48.2 | |||
| 2007 | Singapore | Severely obese Asian children | 198 | 3.5 | 35.4 | 61.1 | [52] |
| Chinese | 105 | 2.9 | 36.2 | 61.0 | |||
| Malay | 68 | 0 | 35.3 | 64.7 | |||
| Indian | 19 | 21.1 | 36.8 | 42.1 | |||
| 2007 | Finland1 | Adults without T2D | 214 | 2.3 | 16.4 | 81.3 | [69] |
| 2007 | Italy | Obese children | 184 | 0 | 10.9 | 89.1 | [72] |
| 2009 | North America | Adults BMI≥40 or BMI≤25 | 1035 | 2.3 | 19.9 | 77.8 | |
| 2009 | US | Children | 416 | 7.7 | 30.3 | 62.0 | [64] |
| Black | 176 | 15.3 | 50.0 | 34.7 | |||
| 2010 | Chile | Obese children | 229 | 0.4 | 10.9 | 88.6 | [51] |
| 2010 | Belgium | Adults | 1321 | 0.6 | 13.6 | 85.8 | |
| 2011 | South Africa | Children | 431 | 17.4 | 49.0 | 33.6 | [48] |
| Black | 209 | 23.0 | 56.5 | 20.6 | |||
| 2013 | Poland | Obese children | 257 | 1.2 | 21.4 | 77.4 | [63] |
| Never-obese adults | 94 | 0 | 19 | 75 | |||
| 2015 | Thailand | Adults+children | 188 | 5.3 | 40.0 | 61.7 | [83] |
| 2016 | Singapore | Infants | 1090 | 6.1 | 35.3 | 58.6 | [65] |
| Chinese | 617 | 5.3 | 32.3 | 62.4 | |||
| Malay | 276 | 6.5 | 34.1 | 54.3 | |||
| Indian | 197 | 7.6 | 39.6 | 52.8 |
Subgroups presented in italics. HOM = homozygous for the minor allele variant, HET = heterozygous at either T6K or V81I, WT = homozygous wildtype. In studies where only one codon was reported, complete linkage disequilibrium was assumed.
T6K and V81I reported separately; frequencies of T6K+V81I haplotype estimated from the data given.
Data for only one subgroup was given.
Certain variants found in the 5′ flanking region also demonstrate different allelic frequencies among races, with −762 (T/C) and −239 (A/G) occurring more frequently in African Americans and Hispanics and −201 (C/G) more commonly in Caucasians [50, 51]. Most variants, however, occur too rarely to make firm conclusions regarding racial associations [Tables 2 and 3].
Table 2.
Rare MC3R coding variants
| Variant/Mutation | Domain | No. of Cases | In Silico Prediction PolyPhen-2 | cAMP | pERK1/2 | Ligand Binding | Cell surface Expression | References | |
|---|---|---|---|---|---|---|---|---|---|
| Obese | Lean | ||||||||
| F12F | EC N-terminus | 0* | 0 | [55] | |||||
| S17T | EC N-terminus | 3 | 0 | − | +a | + | + | − | [19, 55] |
| L35L | EC N-terminus | 1 | 0 | [84] | |||||
| S69C | EC N-terminus | 0 | 1 | ++ | +a | − | + | ++ | [19, 56, 62] |
| S69S | EC N-terminus | 1* | 0 | [55, 84] | |||||
| A70T | EC N-terminus | 1 | 0 | − | +a | − | − | +a | [19, 52, 56] |
| F82S | TM1 | 4 | 4 | ++ | ++ | − | ++ | ++ | [19, 62, 85] |
| I87T | TM1 | 1 | 2 | + | − | + | + | ++ | [19, 55, 56, 62] |
| L95L | TM1 | 2* | 1 | [55, 85] | |||||
| V124L | TM2 | 0 | 1 | ++ | [85] | ||||
| N128S | TM2 | 2 | 0 | ++ | + | + | − | [19, 84, 85] | |
| M134I | TM2 | 1 | 0 | + | +a | ++ | − | +a | [19, 52, 56] |
| D158Y | TM3 | 1 | 0 | ++ | ++ | ++ | ++ | − | [19, 55] |
| V177I | TM3 | 1 | 0 | − | − | − | − | − | [19, 55] |
| I183N | TM3 | 2 | 0 | ++ | ++ | − | − | +a | [19, 52, 53, 86, 87] |
| V211I | TM3 | 1 | 0 | − | + | − | [84] | ||
| I226I | TM4 | 3* | 0 | [55, 84, 85] | |||||
| L249F | IC loop 3 | 2 | 1 | ++ | − | − | + | − | [19, 55, 56, 85] |
| V255I | IC loop 3 | 0 | 1 | − | [85] | ||||
| R257S | IC loop 3 | 4 | 9 | ++ | − | − | + | − | [19, 55, 62, 63, 85] |
| A260V | IC loop 3 | 1 | 0 | − | − | − | − | ++ | [19, 56, 62] |
| Q270Q | IC loop 3 | 0* | 0 | [55] | |||||
| M275T | TM6 | 1 | 0 | + | − | − | − | ++ | [19, 56, 62] |
| T280S | TM6 | 2 | 0 | ++ | ++ | ++ | ++ | ++ | [19, 55, 56, 62] |
| L285V | TM6 | 0 | 1 | ++ | − | − | + | − | [19, 55] |
| A293T | TM6 | 2 | 0 | ++ | − | − | − | − | [10, 17, 19, 55] |
| L297V | TM6 | 1 | 0 | ++ | − | + | − | ++ | [19, 56, 62] |
| L299V | TM6 | 1 | 0 | ++ | ++ | − | ++ | +a | [19, 84] |
| I335S | TM7 | 5 | 0 | ++ | ++ | − | ++ | ++ | [10, 17, 55, 57, 63, 84] |
| E342K | IC C-terminus | 0 | 1 | ++ | [85] | ||||
| G353D | IC C-terminus | 0 | 1 | − | [85] | ||||
| X361S | IC C-terminus | 2 | 2 | − | − | − | − | [10, 17, 19, 55] | |
| c.397_726delins228 | TM3-TM4 (?) | 1 | 0 | ++ | [62] | ||||
not described whether one of subjects was obese or lean, ++ probably deleterious, + possibly deleterious, − probably benign or not significantly different from wildtype, EC = extracellular, TM = transmembrane domain, IC = intracellular,
Conflicting results between studies.
Table 3.
Non-coding MC3R locus variants
| Codon Variant/Mutation | Country | HET No. (%) | HOM No. (%) | Putative Functional Effects | References |
|---|---|---|---|---|---|
| 5′ UTR | |||||
| −769(T/C) | US | 28 (13.4) | 0 | None | [50] |
| −762(A/T) | US | 44 (21.1) | 0 | None | [50] |
| −239(A/G) (rs11575886) | US | 5 (2.4) | 0 | None | [50] |
| Chile | 19 (8.3) | 1 (0.44) | Homozygous mutants associated with higher waist-to-hip ratio. | [51] | |
| Chile | (8.5) | (0.5) | None | [70] | |
| −201(C/G) | US | 31 (14.8) | 0 | None | [50] |
| −4(T/C) | Italy & France | 2 (<0.01) | 0 | Decreases translation | [55] |
| rs4627642(A/T) | Finland | 82 (38.0) | 21 (9.7) | None | [69] |
| rs6024728 | Europe | None | [73] | ||
| rs6024730(G/A) | Finland | 59 (27.3) | 12 (5.6) | None | [69] |
| Europe | None | [73] | |||
| rs6024731 | Europe | None | [73] | ||
| rs16979603(T/C) | Finland | 69 (31.9) | 19 (8.8) | None | [69] |
| rs6014646(A/T) | Europe | T carriers may have more difficulty losing weight | [73] | ||
| rs6014649(G/A) | Finland | 29 (13.4) | 5 (2.3) | Linkage Disequilibrium with T6K+V81I (r2 > 0.8). A carriers associated with higher fasting glucose oxidation and lower fasting FFA. | [69] |
| rs6127698(G/T) | Finland | 110 (50.9) | 45 (20.8) | TT associated with higher lipid oxidation in fasted state. | [69] |
| Belgium | 510 (50.6) | 240 (23.8) | None | [66] | |
| Europe | None | [73] | |||
| rs11697509 | Europe | None | [73] | ||
| 3′ UTR | |||||
| +2138InsCAGACC (rs74181042) | Canada | 126 (38.9) | 15 (4.6) | Associated with variations in fat mass, percent body fat, and total abdominal fat. | [59] |
| US | 167 (40.0) | 25 (6.0) | None | [64] | |
| Chile | 67 (29.0) | 7 (3.0) | No anthropometric effects. Female carriers had higher emotional eating scores. Males had lower scores in the enjoyment to food subscale. | [51] | |
| Chile | (29.7) | (2.5) | None | [70] | |
| rs2870730 (G/C) | 60 (27.8) | 11 (5.1) | None | [69] | |
| rs1543873(T/G) | Europe | None | [73] | ||
| rs6099058 | Europe | None | [73] |
Subjects represented as n (%). HET = heterozygous for the minor allele, HOM = homozygous for the minor allele.
Functional Studies of MC3R variants
Functional studies of human MC3R variants shed light on the mechanisms behind the purported phenotypic differences. Constructs transfected into HEK293 cells demonstrated that the T6K+V81I MC3R has decreased maximal cAMP generation from α-MSH stimulation as compared to the wildtype construct, as well as decreased ligand binding capacity despite preserved binding affinity. This is in part due to decreased MC3R protein expression, despite normal mRNA transcription, suggesting that the variant may either have decreased translation or increased degradation. Despite this, membrane localization appears unaffected [47, 52]. When T6K and V81I constructs were examined separately, no significant differences were seen in cAMP generation, lending further credence to the notion that both T6K and V81I are likely necessary for phenotypic effects [47, 53].
The findings from the functional studies of rare MC3R variants are summarized in Table 2. Interestingly, except for a few mutations, the in silico prediction, functional studies, and phenotype of the affected individuals rarely coincide. Yang and Tao have proposed a classification system for the deleterious effects of MC3R mutations [54]. Certain variants, such as A260V, have decreased protein synthesis or accelerated protein degradation (Class I). Others, such as T280S, demonstrate significantly reduced cell surface expression, which expectedly results in decreased ligand binding, and nearly abolished signaling (Class II) [19, 55, 56]. Mutations such as D158Y have preserved translocation to the cell surface, but inherent defects in ligand binding lead to poor signaling (Class III) [19, 55]. Lastly, mutations that have normal cell surface expression and ligand binding but impaired signaling ability are considered Class IV, while putative benign polymorphisms are Class V. Few functional studies have been performed for synonymous variants, so their potential effects are unclear.
Surprisingly, several mutations (e.g. S69C, F82S, L299V, I335S) which had profoundly decreased cell surface expression and/or ligand binding, still retained normal pERK1/2 signaling capacity. Also, every mutation examined in one paper [56], demonstrated significantly decreased cell surface expression, contradicting earlier research in several cases (e.g. A70T, M134I), whereas most other MC3R mutations examined in other publications by this same lab had normal cell surface expression [19, 52, 53, 56, 57]. Ultimately, these functional studies need to be replicated to ensure that accurate conclusions can be drawn.
Metabolic Effects of MC3R variants
T6K + V81I
There is much debate over whether the T6K+V81I variant has any phenotypic effects. Li et al first sequenced MC3R in a cohort of morbidly obese females (BMI ≥ 40 kg/m2) and non-obese (BMI ≤ 27 kg/m2) controls, and found that the missense mutation V81I did not vary in frequency between the two groups [50]. Similarly, in a cohort study of French Caucasian adults, the T6K+V81I variant was not associated with T2D or obesity. However, among the nondiabetic, nonobese healthy controls, T6K+V81I was associated with greater fasting plasma glucose and insulin, as well as greater insulin levels at the 30, 60, and 90 minute time points of an oral glucose tolerance test (OGTT) [58]. Several other cohort studies also found no metabolic differences in carriers of the common variant, leading some to conclude that the MC3R T6K+V81I haplotype has no discernable phenotypic effect [16, 59–63].
However, many of the negative studies involved populations with relatively few individuals homozygous for the variant haplotype. If, hypothetically, the phenotypic effect from T6K+V81I primarily occurs in the homozygous state (i.e. autosomal recessive), it would not be surprising to find a lack of genotype-phenotype association in cohorts with a low minor allele frequency.
Indeed, in a relatively large US pediatric cohort enriched for obesity and African Americans, where 8% of subjects and 15.8% of African Americans were homozygous for the double mutation, significant metabolic associations were seen. Subjects homozygous for T6K+V81I had higher BMI-z, fat mass (FM), percent fat mass (%FM), and waist circumference [47]. The anthropometric results (BMI, BMI-z, FM, %FM) were reproduced in a later study from our laboratory, including 262 children not studied previously. Additionally, decreased fat-free mass and fat-free mass percentage were seen among the homozygous individuals [64], consistent with the knockout Mc3r mouse models [37].
It has been suggested that the T6K+V81I variant may exert its effects more prominently earlier in life and have much more profound effects in the homozygous than heterozygous state. A recent study from Singapore [65] examined subjects from birth until 48 months of age. By 24 months of age homozygous carriers had greater BMI-z and surrogate markers of fat mass (triceps skinfold and subscapular skinfold) than did wildtype infants, and these differences continued to increase with age [65].
Although most other studies, primarily with few homozygous carriers [Table 1], have not replicated these anthropometric findings, a pediatric cohort from Singapore associated variant carriers with increased FM, while obese T6K+V81I carriers in a Belgian cohort demonstrated greater BMI and body weight [52, 66].
Consistent with possibly increased fat mass, multiple studies have demonstrated higher leptin concentrations among T6K+V81I carriers [47, 52, 67, 68]. T6K+V81I may also be associated with insulin resistance, although the results to date have been inconsistent. Among pediatric cohorts, US children homozygous for T6K+V81I were found to have higher fasting insulin and insulin resistance (as measured by HOMA-IR) [47], whereas in a Singapore cohort, carriers were associated with lower insulin resistance [52]. Some adult cohorts have also suggested greater insulin resistance in carriers [58, 67, 68], whereas others have not [16, 59, 61, 69].
Associations with other markers of the metabolic syndrome have been mostly negative. Among a Chilean cohort of obese children, T6K+V81I carriers were at higher risk for Metabolic Syndrome, hypertriglyceridemia, and low HDL, but these results did not remain significant after correction for multiple comparisons [70]. Most other studies showed no differences among genotype with respect to triglycerides, cholesterol, or blood pressure [16, 47, 48, 61, 69], although a Kuwaiti study did show association with increased systolic blood pressure among variant carriers [49].
Determinants of possible causes of adiposity/obesity associated with the T6K+V81I haplotype have also been investigated. Consistent with knockout mouse models [36], glucose oxidation was increased and lipid oxidation decreased in T6K+V81I carriers [69, 71]. T6K+V81I variant carriers may also have greater difficulty losing weight, consistent with the “metabolically thrifty” phenotype [72, 73]. Two studies found no differences in energy intake between genotypes [52, 59], and in a third, the association no longer was significant when the data were examined separately by race [64]. Additionally, no significant differences were seen in resting energy expenditure [59, 64, 69], total energy expenditure [64, 71], respiratory quotient [59, 64], or physical activity [52, 64, 71, 72].
Eating behavior patterns have also been investigated, with negligible differences reported between genotypes. No differences were seen in an Asian pediatric cohort or Hispanic adult cohort on the Three-Factor Eating Questionnaire, a validated tool that examines eating restraint, disinhibition/cognitive restraint, emotional eating, and hunger [52, 74, 75]. Lower emotional eating was seen among boys (but not girls) carrying the minor alleles in a Hispanic cohort [51]. Aside from increased “slowness in eating” in infants, no differences were seen on any subscale of the Child Eating Behavior Questionnaire in infants or children [51, 65].
A mouse model for MC3R T6K+V81I
To clarify the role the MC3R T6K+V81I variant plays in obesity and metabolism, our group generated two novel homozygous knock-in mouse models [4] where we replaced the murine Mc3r with either human wildtype (hWT) MC3R (MC3RhWT/hWT) or the human doublemutant (hDM) T6K+V81I MC3R (MC3RhDM/hDM). The MC3RhWT/hWT mice were phenotypically normal, while the MC3RhDM/hDM mice exhibited greater fat mass (Figure 2) and feeding efficiency with reduced fat-free mass – results similar to those found in the Mc3r knockout mouse.
Figure 2.
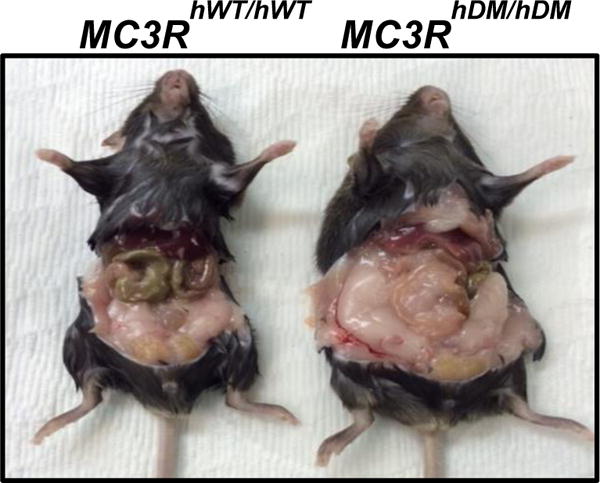
hMC3R knockin mice. The murine Mc3r was replaced with the human consensus “wildtype” sequence (hWT) or the C17A + G241A human “double-mutant” (hDM) variant, which changes two amino acids in MC3R: Thr6Lys and Val81Ile (T6K+V81I). Mice homozygous for MC3R hDM have greater adiposity than mice homozygous for hWT.
Despite their greater adiposity, MC3RhDM/hDM mice also showed reduced obesity-associated metabolic dysfunction [4]. MC3RhDM/hDM mice had well-maintained insulin sensitivity with similar serum glucose, triglycerides, cholesterol, free-fatty acid and corticosterone concentrations when compared to the non-obese MC3RhWT/hWT mice. MC3RhDM/hDM mice exhibited higher circulating leptin that appeared related to their increased adiposity rather than to the presence of leptin resistance, since suppression of food intake by leptin appeared normal [4]. Additionally, MC3RhDM/hDM mice demonstrated increased bone marrow fat, along with decreased crown-rump length and femur length. This was at least partially due to an alteration of mesenchymal stem cell fate, which seemed favored towards differentiation to triglyceride-accumulating adipocytes instead of osteoblasts. Additionally, unlike most other obese mouse models, MC3RhDM/hDM mice had greater circulating adiponectin, which we then also observed in samples from MC3RhDM/hDM humans [4]. It is possible that increased serum adiponectin may stimulate mesenchymal stem cell mobilization into adipose tissue, resulting in less bone mass [76] with shorter femur and reduced trabecular bone mineral density of MC3RhDM/hDM mice.
As noted above, in vitro studies find decreased MC3R protein expression and therefore partial inactivation of the MC3R by T6K+V81I [47]. Since T6K lies between the first and second in-frame translational start site, it might not necessarily be part of the translated protein [13]. We have proposed that T6K+V81I MC3R might affect the proportion of protein translated from the first and second in-frame ATGs and thus potentially interfere with MC3R actions, for instance by altering its membrane localization [13]. MC3RhDM/hDM mice do not appear to be phenocopies of MC3R−/− mice. For example, when compared with wildtype mice, MC3RhDM/hDM mice do not exhibit hyperglycemia under chow-fed conditions or increased inflammation in WAT under high-fat diet conditions. More studies are needed to understand better how T6K+V81I may affect receptor expression, stability, intracellular trafficking and signaling in vivo.
In summary, the metabolic effects of the T6K+V81I haplotype are not entirely straightforward given the conflicting results between studies. In humans, the most dramatic associations - namely increased BMI and fat mass - seem to occur in individuals homozygous for T6K+V81I and primarily in youth. In the heterozygous state or in adulthood, the haplotype’s effects are milder. Why this is the case needs further investigation. Studies of the MC3RhDM/hDM knockin mouse model, though, confirm this variant increases the predisposition for triglyceride accumulation, with what appear to be direct effects at peripheral sites including adipose tissue.
Other coding variants
To date, 33 rare coding variants have been identified [Table 2]. These mutations have been identified in too few individuals to make definitive conclusions regarding their phenotypic consequences, and no mouse models exist to date. However, examining the in silico prediction, functional studies, and characteristics of the affected subjects and their family members, we suggest that six are likely pathologic mutations, ten are likely benign polymorphisms, six are synonymous (insufficiently studied), and the effect is unclear for 11 other variants.
The variants D158Y, I183N, T280S, L299V, I335S, and c.397_726delins228 are likely pathologic. In all of these cases cAMP production is significantly decreased and all affected individuals are obese with no cases of lean subjects described. In most cases the obesity phenotype occurred early in age, which is to be expected from an inherited genetic condition. The D158 residue has been shown to be integral for ligand binding, while I335 is part of the highly-conserved N/DPxxY motif which plays important roles in ligand binding, G-protein coupling, and internalization [77–79].
Sequence variants S17T, S69C, A70T, F82S, I87T, V124L, N128S, M134I, V211I, L297V, and E342K, have been classified as “possibly pathologic”, because the physiologic consequences of these variants are unclear. For example, S17T was identified in obese cases only, but in silico prediction modelling defines this as a benign variant, and data suggest that cAMP and pERK1/2 signaling remain relatively preserved [19, 55]. In other cases, such as S69C, F82S, I87T, V124L, and E342K, the variant was found in lean individuals, even though in silico prediction or functional studies suggest that the mutation may be deleterious. The reason for this discordance may be that any or all of these mutations may only cause a weak predisposition to obesity, or significant clinical effects may only be realized in the homozygous state.
The other identified sequence variants (V177I, L249F, V255I, R257S, A260V, M275T, L285V, A293T, G353D, X361S) we presume are likely benign. For most of these variants, lean individuals were found carrying the variant (e.g. V255I, G353D) and/or functional studies demonstrated normal ligand binding and signal transduction. Interestingly, most of these variants are located in the intracellular regions of the protein or in transmembrane domain 6, which classically are important areas for G-protein binding and therefore cAMP production. However, alanine scanning studies did not identify any of the aforementioned residues as critical for functioning [80].
Lastly, the observed synonymous mutations are assumed to be benign, although recent studies have suggested that even synonymous mutations can have functional, consequences [81, 82].
Non-coding variants
14 sequence variants in the 5′ UTR and four in the 3′ UTR for MC3R have also been identified [Table 3]. The −4 codon is located in the promoter region of MC3R, and cell transfection studies suggest that the T-to-C mutation at this site suppresses gene transcription. This variant has only been identified in two obese subjects and no lean individuals [55].
The rs6014649(G/A) polymorphisms is in high linkage disequilibrium with the T6K+V81I variant (r2 > 0.8), and minor allele carriers were associated with higher fasting glucose oxidation and lower fasting FFA [69]. In the same cohort, rs6127698(G/T) was shown to be associated with higher lipid oxidation in the fasted state among individuals homozygous for the minor allele, but otherwise no anthropometric differences were seen [69].
Among 3′UTR variants, homozygosity for +2138InsCAGACC in normal weight subjects was reported to be associated with increased FM and %FM. Conversely, in overweight subjects with homozygosity for the insertion allele was associated with decreased FM and %FM, and in obese subjects lower total abdominal fat was seen [59]. However, in other cohorts, no anthropometric differences were seen between +2138InsCAGACC carriers and wildtype subjects [51, 64, 70]. Among obese girls, +2138InsCAGACC carriers were reported in one study to have significantly higher emotional eating than wildtype subjects, whereas obese boys had lower food enjoyment, although it is not clear if these measures would have retained statistical significance had the study corrected for their multiple-comparisons [51].
Conclusions
Except for the common variants T6K and V81I, most sequence variants in MC3R are rare in humans. Several rare MC3R mutations abolish MC3R function, apparently leading to an obese phenotype, but most mutations are partially inactivating with unclear consequences and mouse models are lacking. The T6K+V81I haplotype is by far the most well studied; its phenotypic effects are most prominent in the homozygous state, with milder clinical effects seen in heterozygous carriers. Studies also suggest that the haplotype’s effects may be more pronounced in infancy and childhood. The ability of T6K+V81I to alter energy homeostasis is demonstrated by knockin mouse studies. Further research is needed to elucidate MC3R’s differential functions between tissue types as well as its activity and regulation during different stages of life.
Highlights.
Both central and peripheral actions of MC3R appear important for weight regulation.
Certain human MC3R mutations are associated with greater adiposity and hyperleptinemia.
MC3R function is altered by homozygosity for both rare alleles of the human variant T6K+V81I.
T6K+V81I knockin mouse models suggest MC3R is involved in determining how mesenchymal stem cell differentiate.
Acknowledgments
Sources of funding: This research was supported by the Division of Intramural Research of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), NIH ZIAHD00641 (to JAY), with supplemental funding from NIMHD and the NIH Clinical Center Bench to Bedside Program. Dr. Yanovski reports receiving unrelated grant funding from Zafgen Inc. for a clinical trial of pharmacotherapy to treat obesity and hyperphagia in patients with the Prader-Willi syndrome and from Rhythm Pharmaceuticals Inc. for support of sequencing of genes related to obesity and for a trial of setmelanotide to treat patients with monogenic obesity syndromes in the leptin signaling pathway. No sponsors were involved in the collection, analysis, or interpretation of data; writing of the manuscript; or decision to submit the manuscript for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclaimers: JAY is a Commissioned Officer in the United States Public Health Service. The opinions and assertions expressed herein are those of the authors and are not to be construed as reflecting the views of the Public Health Service or the Department of Health and Human Services.
References
- 1.Wardle J, et al. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am J Clin Nutr. 2008;87(2):398–404. doi: 10.1093/ajcn/87.2.398. [DOI] [PubMed] [Google Scholar]
- 2.Silventoinen K, Kaprio J. Genetics of tracking of body mass index from birth to late middle age: evidence from twin and family studies. Obesity facts. 2009;2(3):196–202. doi: 10.1159/000219675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morton GJ, Meek TH, Schwartz MW. Neurobiology of food intake in health and disease. Nat Rev Neurosci. 2014;15(6):367–78. doi: 10.1038/nrn3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee B, et al. A mouse model for a partially inactive obesity-associated human MC3R variant. Nat Commun. 2016;7:10522. doi: 10.1038/ncomms10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girardet C, et al. Unravelling the mysterious roles of melanocortin-3 receptors in metabolic homeostasis and obesity using mouse genetics. Int J Obes Suppl. 2014;4(Suppl 1):S37–44. doi: 10.1038/ijosup.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begriche K, et al. Genetic dissection of the functions of the melanocortin-3 receptor, a seven-transmembrane G-protein-coupled receptor, suggests roles for central and peripheral receptors in energy homeostasis. J Biol Chem. 2011;286(47):40771–81. doi: 10.1074/jbc.M111.278374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gantz I, Fong TM. The melanocortin system. Am J Physiol Endocrinol Metab. 2003;284(3):E468–74. doi: 10.1152/ajpendo.00434.2002. [DOI] [PubMed] [Google Scholar]
- 8.Ni XP, et al. Central receptors mediating the cardiovascular actions of melanocyte stimulating hormones. J Hypertens. 2006;24(11):2239–46. doi: 10.1097/01.hjh.0000249702.49854.fa. [DOI] [PubMed] [Google Scholar]
- 9.Gantz I, et al. Molecular cloning of a novel melanocortin receptor. J Biol Chem. 1993;268(11):8246–50. [PubMed] [Google Scholar]
- 10.Magenis RE, et al. Mapping of the ACTH, MSH, and neural (MC3 and MC4) melanocortin receptors in the mouse and human. Mamm Genome. 1994;5(8):503–8. doi: 10.1007/BF00369320. [DOI] [PubMed] [Google Scholar]
- 11.Lembertas AV, et al. Identification of an obesity quantitative trait locus on mouse chromosome 2 and evidence of linkage to body fat and insulin on the human homologous region 20q. J Clin Invest. 1997;100(5):1240–7. doi: 10.1172/JCI119637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tao YX. The melanocortin-4 receptor: physiology, pharmacology, and pathophysiology. Endocr Rev. 2010;31(4):506–43. doi: 10.1210/er.2009-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J, Sharma N, Cutting GR. Melanocortin 3 receptor has a 5′ exon that directs translation of apically localized protein from the second in-frame ATG. Mol Endocrinol. 2014;28(9):1547–57. doi: 10.1210/me.2014-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor-Douglas DC, et al. Evaluation of hypothalamic murine and human melanocortin 3 receptor transcript structure. Biochem Biophys Res Commun. 2014;454(1):234–8. doi: 10.1016/j.bbrc.2014.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarnow P, et al. Identification of the translation start site of the human melanocortin 3 receptor. Obes Facts. 2012;5(1):45–51. doi: 10.1159/000336070. [DOI] [PubMed] [Google Scholar]
- 16.Schioth HB, et al. Alternative translation initiation codon for the human melanocortin MC3 receptor does not affect the ligand binding. Eur J Pharmacol. 1996;314(3):381–4. doi: 10.1016/s0014-2999(96)00566-3. [DOI] [PubMed] [Google Scholar]
- 17.Tao YX. Functional characterization of novel melanocortin-3 receptor mutations identified from obese subjects. Biochim Biophys Acta. 2007;1772(10):1167–74. doi: 10.1016/j.bbadis.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Begriche K, et al. Melanocortin-3 receptors are involved in adaptation to restricted feeding. Genes Brain Behav. 2012;11(3):291–302. doi: 10.1111/j.1601-183X.2012.00766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang F, Huang H, Tao YX. Biased signaling in naturally occurring mutations in human melanocortin-3 receptor gene. Int J Biol Sci. 2015;11(4):423–33. doi: 10.7150/ijbs.11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z, Tao YX. Biased signaling initiated by agouti-related peptide through human melanocortin-3 and -4 receptors. Biochim Biophys Acta. 2016;1862(9):1485–94. doi: 10.1016/j.bbadis.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Anderson EJ, et al. 60 YEARS OF POMC: Regulation of feeding and energy homeostasis by alpha-MSH. Journal of molecular endocrinology. 2016;56(4):T157–74. doi: 10.1530/JME-16-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chhajlani V. Distribution of cDNA for melanocortin receptor subtypes in human tissues. Biochem Mol Biol Int. 1996;38(1):73–80. [PubMed] [Google Scholar]
- 23.Moller CL, et al. Melanocortin agonists stimulate lipolysis in human adipose tissue explants but not in adipocytes. BMC research notes. 2015;8:559. doi: 10.1186/s13104-015-1539-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Getting SJ, et al. Redundancy of a functional melanocortin 1 receptor in the anti-inflammatory actions of melanocortin peptides: studies in the recessive yellow (e/e) mouse suggest an important role for melanocortin 3 receptor. J Immunol. 2003;170(6):3323–30. doi: 10.4049/jimmunol.170.6.3323. [DOI] [PubMed] [Google Scholar]
- 25.Schioth HB, et al. Major pharmacological distinction of the ACTH receptor from other melanocortin receptors. Life Sci. 1996;59(10):797–801. doi: 10.1016/0024-3205(96)00370-0. [DOI] [PubMed] [Google Scholar]
- 26.Marks DL, et al. The regulation of food intake by selective stimulation of the type 3 melanocortin receptor (MC3R) Peptides. 2006;27(2):259–64. doi: 10.1016/j.peptides.2005.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voisey J, Carroll L, Daal A van. Melanocortins and their receptors and antagonists. Curr Drug Targets. 2003;4(7):586–97. doi: 10.2174/1389450033490858. [DOI] [PubMed] [Google Scholar]
- 28.Grieco P, et al. D-Amino acid scan of gamma-melanocyte-stimulating hormone: importance of Trp(8) on human MC3 receptor selectivity. J Med Chem. 2000;43(26):4998–5002. doi: 10.1021/jm000211e. [DOI] [PubMed] [Google Scholar]
- 29.Jegou S, Boutelet I, Vaudry H. Melanocortin-3 receptor mRNA expression in pro-opiomelanocortin neurones of the rat arcuate nucleus. J Neuroendocrinol. 2000;12(6):501–5. doi: 10.1046/j.1365-2826.2000.00477.x. [DOI] [PubMed] [Google Scholar]
- 30.Bagnol D, et al. Anatomy of an endogenous antagonist: relationship between Agouti-related protein and proopiomelanocortin in brain. J Neurosci. 1999;19(18):RC26. doi: 10.1523/JNEUROSCI.19-18-j0004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowley MA, et al. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411(6836):480–4. doi: 10.1038/35078085. [DOI] [PubMed] [Google Scholar]
- 32.Marks DL, et al. Differential role of melanocortin receptor subtypes in cachexia. Endocrinology. 2003;144(4):1513–23. doi: 10.1210/en.2002-221099. [DOI] [PubMed] [Google Scholar]
- 33.Renquist BJ, et al. Melanocortin-3 receptor regulates the normal fasting response. Proc Natl Acad Sci U S A. 2012;109(23):E1489–98. doi: 10.1073/pnas.1201994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutton GM, et al. Central nervous system melanocortin-3 receptors are required for synchronizing metabolism during entrainment to restricted feeding during the light cycle. FASEB J. 2010;24(3):862–72. doi: 10.1096/fj.09-142000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mavrikaki M, et al. Melanocortin-3 receptors in the limbic system mediate feeding-related motivational responses during weight loss. Mol Metab. 2016;5(7):566–79. doi: 10.1016/j.molmet.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butler AA, et al. A unique metabolic syndrome causes obesity in the melanocortin-3 receptor-deficient mouse. Endocrinology. 2000;141(9):3518–21. doi: 10.1210/endo.141.9.7791. [DOI] [PubMed] [Google Scholar]
- 37.Chen AS, et al. Inactivation of the mouse melanocortin-3 receptor results in increased fat mass and reduced lean body mass. Nat Genet. 2000;26(1):97–102. doi: 10.1038/79254. [DOI] [PubMed] [Google Scholar]
- 38.Sutton GM, et al. Diet-genotype interactions in the development of the obese, insulin-resistant phenotype of C57BL/6J mice lacking melanocortin-3 or -4 receptors. Endocrinology. 2006;147(5):2183–96. doi: 10.1210/en.2005-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellacott KL, et al. Obesity-induced inflammation in white adipose tissue is attenuated by loss of melanocortin-3 receptor signaling. Endocrinology. 2007;148(12):6186–94. doi: 10.1210/en.2007-0699. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, et al. Targeted deletion of melanocortin receptor subtypes 3 and 4, but not CART, alters nutrient partitioning and compromises behavioral and metabolic responses to leptin. FASEB J. 2005;19(11):1482–91. doi: 10.1096/fj.05-3851com. [DOI] [PubMed] [Google Scholar]
- 41.Girardet C, Butler AA. Neural melanocortin receptors in obesity and related metabolic disorders. Biochim Biophys Acta. 2014;1842(3):482–94. doi: 10.1016/j.bbadis.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Begriche K, et al. The role of melanocortin neuronal pathways in circadian biology: a new homeostatic output involving melanocortin-3 receptors? Obes Rev. 2009;10(Suppl 2):14–24. doi: 10.1111/j.1467-789X.2009.00662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris C, et al. Large increases in adipose triacylglycerol flux in Cushingoid CRH-Tg mice are explained by futile cycling. Am J Physiol Endocrinol Metab. 2013;304(3):E282–93. doi: 10.1152/ajpendo.00154.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein RF, et al. Regulation of bone mass in mice by the lipoxygenase gene Alox15. Science. 2004;303(5655):229–32. doi: 10.1126/science.1090985. [DOI] [PubMed] [Google Scholar]
- 45.You P, et al. Effects of Melanocortin 3 and 4 Receptor Deficiency on Energy Homeostasis in Rats. Scientific reports. 2016;6:34938. doi: 10.1038/srep34938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JH, et al. Genome scan for human obesity and linkage to markers in 20q13. Am J Hum Genet. 1999;64(1):196–209. doi: 10.1086/302195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feng N, et al. Co-occurrence of two partially inactivating polymorphisms of MC3R is associated with pediatric-onset obesity. Diabetes. 2005;54(9):2663–7. doi: 10.2337/diabetes.54.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yako YY, et al. Negative association of MC3R variants with weight and blood pressure in Cape Town pupils aged 11–16 years. S Afr Med J. 2011;101(6):417–20. [PubMed] [Google Scholar]
- 49.Alsmadi O, et al. Leptin in association with common variants of MC3R mediates hypertension. Am J Hypertens. 2014;27(7):973–81. doi: 10.1093/ajh/hpt285. [DOI] [PubMed] [Google Scholar]
- 50.Li WD, et al. Melanocortin 3 receptor (MC3R) gene variants in extremely obese women. Int J Obes Relat Metab Disord. 2000;24(2):206–10. doi: 10.1038/sj.ijo.0801114. [DOI] [PubMed] [Google Scholar]
- 51.Obregon AM, et al. Melanocortin-3 receptor gene variants: association with childhood obesity and eating behavior in Chilean families. Nutrition. 2010;26(7–8):760–5. doi: 10.1016/j.nut.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 52.Lee YS, et al. The role of melanocortin 3 receptor gene in childhood obesity. Diabetes. 2007;56(10):2622–30. doi: 10.2337/db07-0225. [DOI] [PubMed] [Google Scholar]
- 53.Tao YX, Segaloff DL. Functional characterization of melanocortin-3 receptor variants identify a loss-of-function mutation involving an amino acid critical for G protein-coupled receptor activation. J Clin Endocrinol Metab. 2004;89(8):3936–42. doi: 10.1210/jc.2004-0367. [DOI] [PubMed] [Google Scholar]
- 54.Yang Z, Tao YX. Mutations in Melanocortin-3 Receptor Gene and Human Obesity. Prog Mol Biol Transl Sci. 2016;140:97–129. doi: 10.1016/bs.pmbts.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 55.Mencarelli M, et al. Rare melanocortin-3 receptor mutations with in vitro functional consequences are associated with human obesity. Hum Mol Genet. 2011;20(2):392–9. doi: 10.1093/hmg/ddq472. [DOI] [PubMed] [Google Scholar]
- 56.Yang F, Tao YX. Functional characterization of nine novel naturally occurring human melanocortin-3 receptor mutations. Biochim Biophys Acta. 2012;1822(11):1752–61. doi: 10.1016/j.bbadis.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z, Huang ZL, Tao YX. Functions of DPLIY motif and helix 8 of human melanocortin-3 receptor. J Mol Endocrinol. 2015;55(2):107–17. doi: 10.1530/JME-15-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hani EH, et al. Naturally occurring mutations in the melanocortin receptor 3 gene are not associated with type 2 diabetes mellitus in French Caucasians. J Clin Endocrinol Metab. 2001;86(6):2895–8. doi: 10.1210/jcem.86.6.7589. [DOI] [PubMed] [Google Scholar]
- 59.Boucher N, et al. A +2138InsCAGACC polymorphism of the melanocortin receptor 3 gene is associated in human with fat level and partitioning in interaction with body corpulence. Mol Med. 2002;8(3):158–65. [PMC free article] [PubMed] [Google Scholar]
- 60.Wong J, et al. Melanocortin-3 receptor gene variants in a Maori kindred with obesity and early onset type 2 diabetes. Diabetes Res Clin Pract. 2002;58(1):61–71. doi: 10.1016/s0168-8227(02)00126-2. [DOI] [PubMed] [Google Scholar]
- 61.Malczewska-Malec M, et al. Analysis of candidate genes in Polish families with obesity. Clin Chem Lab Med. 2004;42(5):487–93. doi: 10.1515/CCLM.2004.083. [DOI] [PubMed] [Google Scholar]
- 62.Calton MA, et al. Association of functionally significant Melanocortin-4 but not Melanocortin-3 receptor mutations with severe adult obesity in a large North American case-control study. Hum Mol Genet. 2009;18(6):1140–7. doi: 10.1093/hmg/ddn431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cieslak J, et al. Common polymorphism (81Val>Ile) and rare mutations (257Arg>Ser and 335Ile>Ser) of the MC3R gene in obese Polish children and adolescents. Mol Biol Rep. 2013;40(12):6893–8. doi: 10.1007/s11033-013-2808-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Savastano DM, et al. Energy intake and energy expenditure among children with polymorphisms of the melanocortin-3 receptor. Am J Clin Nutr. 2009;90(4):912–20. doi: 10.3945/ajcn.2009.27537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aris IM, et al. MC3R gene polymorphisms are associated with early childhood adiposity gain and infant appetite in an Asian population. Pediatr Obes. 2016;11(6):450–458. doi: 10.1111/ijpo.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zegers D, et al. Common melanocortin-3 receptor variants are not associated with obesity, although rs3746619 does influence weight in obese individuals. Endocrine. 2010;38(2):289–93. doi: 10.1007/s12020-010-9386-5. [DOI] [PubMed] [Google Scholar]
- 67.Schalin-Jantti C, et al. Melanocortin-3-receptor gene variants in morbid obesity. Int J Obes Relat Metab Disord. 2003;27(1):70–4. doi: 10.1038/sj.ijo.0802184. [DOI] [PubMed] [Google Scholar]
- 68.Yiannakouris N, et al. The Val81 missense mutation of the melanocortin 3 receptor gene, but not the 1908c/T nucleotide polymorphism in lamin A/C gene, is associated with hyperleptinemia and hyperinsulinemia in obese Greek caucasians. J Endocrinol Invest. 2004;27(8):714–20. doi: 10.1007/BF03347511. [DOI] [PubMed] [Google Scholar]
- 69.Rutanen J, et al. Single nucleotide polymorphisms of the melanocortin-3 receptor gene are associated with substrate oxidation and first-phase insulin secretion in offspring of type 2 diabetic subjects. J Clin Endocrinol Metab. 2007;92(3):1112–7. doi: 10.1210/jc.2006-1201. [DOI] [PubMed] [Google Scholar]
- 70.Suazo J, et al. Prevalence of metabolic syndrome in obese Chilean children and association with gene variants of the leptin-melanocortin system. J Pediatr Endocrinol Metab. 2013;26(11–12):1131–9. doi: 10.1515/jpem-2013-0084. [DOI] [PubMed] [Google Scholar]
- 71.Obregon AM, Diaz E, Santos JL. Effect of the melanocortin-3 receptor Thr6Lys and Val81Ile genetic variants on body composition and substrate oxidation in Chilean obese children. J Physiol Biochem. 2012;68(1):71–6. doi: 10.1007/s13105-011-0120-4. [DOI] [PubMed] [Google Scholar]
- 72.Santoro N, et al. Effect of the melanocortin-3 receptor C17A and G241A variants on weight loss in childhood obesity. Am J Clin Nutr. 2007;85(4):950–3. doi: 10.1093/ajcn/85.4.950. [DOI] [PubMed] [Google Scholar]
- 73.Santos JL, et al. Allelic variants of melanocortin 3 receptor gene (MC3R) and weight loss in obesity: a randomised trial of hypo-energetic high- versus low-fat diets. PLoS One. 2011;6(6):e19934. doi: 10.1371/journal.pone.0019934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vega JA, et al. Melanocortin-4 Receptor Gene Variation Is Associated with Eating Behavior in Chilean Adults. Ann Nutr Metab. 2016;68(1):35–41. doi: 10.1159/000439092. [DOI] [PubMed] [Google Scholar]
- 75.Stunkard AJ, Messick S. The three-factor eating questionnaire to measure dietary restraint, disinhibition and hunger. J Psychosom Res. 1985;29(1):71–83. doi: 10.1016/0022-3999(85)90010-8. [DOI] [PubMed] [Google Scholar]
- 76.Yu L, et al. Adiponectin regulates bone marrow mesenchymal stem cell niche through a unique signal transduction pathway: an approach for treating bone disease in diabetes. Stem cells. 2015;33(1):240–52. doi: 10.1002/stem.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang SX, Fan ZC, Tao YX. Functions of acidic transmembrane residues in human melanocortin-3 receptor binding and activation. Biochem Pharmacol. 2008;76(4):520–30. doi: 10.1016/j.bcp.2008.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen M, et al. Molecular characterization of human melanocortin-3 receptor ligand-receptor interaction. Biochemistry. 2006;45(4):1128–37. doi: 10.1021/bi0521792. [DOI] [PubMed] [Google Scholar]
- 79.Barak LS, et al. The conserved seven-transmembrane sequence NP(X)2,3Y of the G-protein-coupled receptor superfamily regulates multiple properties of the beta 2-adrenergic receptor. Biochemistry. 1995;34(47):15407–14. doi: 10.1021/bi00047a003. [DOI] [PubMed] [Google Scholar]
- 80.Wang ZQ, Tao YX. Functions of the third intracellular loop of the human melanocortin-3 receptor. Curr Pharm Des. 2013;19(27):4831–8. doi: 10.2174/1381612811319270005. [DOI] [PubMed] [Google Scholar]
- 81.Diederichs S, et al. The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol Med. 2016;8(5):442–57. doi: 10.15252/emmm.201506055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet. 2011;12(10):683–91. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 83.Wannaiampikul S, et al. Genetic variant screening of MC3R and MC4R genes in early-onset obese children and their relatives among a Thai population: family-based study. Genet Mol Res. 2015;14(4):18090–102. doi: 10.4238/2015.December.22.35. [DOI] [PubMed] [Google Scholar]
- 84.Zegers D, et al. Identification of three novel genetic variants in the melanocortin-3 receptor of obese children. Obesity (Silver Spring) 2011;19(1):152–9. doi: 10.1038/oby.2010.127. [DOI] [PubMed] [Google Scholar]
- 85.Zegers D, et al. Prevalence of rare MC3R variants in obese cases and lean controls. Endocrine. 2013;44(2):386–90. doi: 10.1007/s12020-012-9862-1. [DOI] [PubMed] [Google Scholar]
- 86.Lee YS, Poh LK, Loke KY. A novel melanocortin 3 receptor gene (MC3R) mutation associated with severe obesity. J Clin Endocrinol Metab. 2002;87(3):1423–6. doi: 10.1210/jcem.87.3.8461. [DOI] [PubMed] [Google Scholar]
- 87.Rached M, et al. Inactivation and intracellular retention of the human I183N mutated melanocortin 3 receptor associated with obesity. Biochim Biophys Acta. 2004;1689(3):229–34. doi: 10.1016/j.bbadis.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 88.melanocortin receptor 3 [Homo sapiens] 2016 December 27, 2016]; October 6, 2016 [Available from: https://www.ncbi.nlm.nih.gov/protein/170671732.


