Abstract
Study Objective
To illustrate the potential of metabolomics to identify novel biomarkers of illness severity in a child with fatal necrotizing pneumonia caused by methicillin-resistant Staphylococcus aureus (MRSA).
Design
Case report with two control groups and a metabolomics analysis.
Patients
An infant with fatal MRSA pneumonia, four children with influenza pneumonia (pneumonia control group), and seven healthy children with no known infections (healthy control group).
Measurements and Main Results
Urine samples were collected from all children. Metabolites were identified and quantified by using 1H-nuclear magnetic resonance spectrometry. Normalized metabolite concentration data from children with influenza pneumonia and healthy controls were compared by using an unpaired Student's t test. To identify differentiating metabolites of MRSA pneumonia, the fold change of each metabolite was calculated by dividing each urine metabolite concentration of the patient with fatal MRSA pneumonia by the median urine concentration values of the same metabolite of the patients with influenza pneumonia and healthy controls, respectively. Metscape (http://metscape.ncibi.org/), a bioinformatics tool, was used for data visualization and interpretation. Urine metabolite concentrations previously identified as associated with sepsis in children (e.g., 3-hydroxybutyrate, carnitine, and creatinine) were higher in the patient with fatal MRSA pneumonia compared with those of patients with influenza pneumonia and healthy controls. The concentrations of additional metabolites—acetone, acetoacetate, choline, fumarate, glucose and 3-aminoisobutyrate—were more than 25-fold higher in the patient with MRSA pneumonia than those of patients with influenza pneumonia and healthy controls.
Conclusion
These metabolic changes in the urine preceded the clinical severe sepsis phenotype, which suggests that detection of the extent of metabolic disruption can aid in the early identification of a sepsis phenotype in advance of clinical diagnosis. These data also support the utility of metabolomics for the development of clinical assays for the identification of patients with pediatric pneumonia at high risk for deterioration.
Keywords: metabolomics, pediatrics, septic shock, necrotizing pneumonia, nuclear magnetic resonance
Community-acquired pneumonia (CAP), a common cause of hospitalization among children, may be accompanied by complications within the pulmonary cavity, such as necrotizing pneumonia.1-3 Some children with CAP may also develop serious systemic complications such as sepsis. Identifying children with CAP who are at high risk for sepsis and subsequent clinical deterioration is challenging. For example, the first vital sign that is consistent with the criteria for systemic inflammatory response syndrome (SIRS)4 that needs to be met for a sepsis diagnosis occurs in >90% of pediatric patients with fever (>38.5°C) who visit the emergency department (ED). Of these patients, 82% are discharged from the ED without intravenous therapy and do not revisit the ED, whereas only 0.25% of these patients require critical care in the first 24 hours. Therefore, the SIRS criteria alone are insufficient to identify patients at risk of deterioration because these criteria have poor specificity in predicting the need for critical care in children.5 Although sepsis criteria have greater specificity, children may quickly deteriorate once these criteria are met, illustrating the great need for a tool that can predict the severity of the clinical course of disease before the patient deteriorates.4, 5
Metabolomics, the study of small molecules (<1 kDa), may provide a real-time snapshot of the biochemical impact of disease on the host. The objective of this study was to illustrate the use of metabolomics in identifying a patient at high risk for deterioration prior to meeting the clinical criteria of sepsis, prior to identifying the source of infection through conventional microbiologic techniques, and prior to the diagnosis of a rare complication of pneumonia. Specifically, our aim was to identify novel biomarkers of illness severity in a child with fatal necrotizing pneumonia caused by methicillin-resistant Staphylococcus aureus (MRSA).
Methods
Study Design and Patient Population
In this case report with a metabolomics analysis, two sets of controls were used for comparison with the patient with fatal MRSA pneumonia. The first set consisted of children diagnosed with influenza pneumonia (pneumonia controls), and the second set consisted of children who had no known infection (healthy controls). The children enrolled provided written assent (if applicable), and their parents or legal guardians provided written consent according to the study protocol approved by the institutional review board at Cincinnati Children's Hospital Medical Center (Cincinnati, OH).
Patient with Fatal MRSA Pneumonia
A previously healthy, fully immunized, 8-month-old infant presented to the ED of a tertiary-care children's hospital after experiencing 3 days of fever (maximum temperature 39.8°C), cough, difficulty breathing, emesis, diarrhea, rhinorrhea, and fatigue. Five hours after ED arrival, the infant was enrolled in a study entitled Catalyzing Ambulatory Research in Pneumonia Etiology and Diagnostic Innovations in Emergency Medicine (CARPE DIEM), a research study investigating the severity and etiology of pediatric pneumonia. The infant had no significant medical or family history. On ED arrival, the infant's vital signs were a heart rate of 208 beats/minute, rectal temperature 39.8°C, respiratory rate 50 breaths/minute, blood pressure 131/71 mm Hg, and room air oxygen saturation of 94% as measured by pulse oximetry (Figure 1). Physical examination revealed an ill, but not toxic-appearing, infant in moderate respiratory distress. Mild subcostal and intercostal retractions and diffuse rhonchi were present. The infant was well perfused, with a capillary refill time of <2 seconds. Initial diagnostic evaluation included a complete blood count significant for a white blood cell count of 1100 cells/mm3 and an absolute neutrophil count of 80 cells/mm3; a blood culture revealed no growth of an organism after 5 days of incubation (Table 1). The chest radiograph was concerning for multifocal pneumonia (Figure 2A). Serum electrolyte and blood urea nitrogen (BUN) levels were normal for the infant's age. A rapid venous blood gas was performed, which returned a mild base deficit (-4 mmol/L [normal range –2 to 2 mmol/L]).
Figure 1.
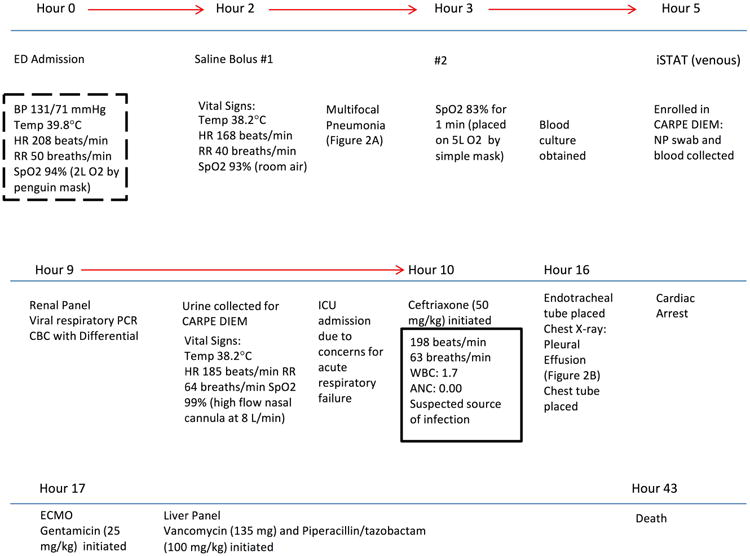
Timeline of the clinical course of the patient with fatal methicillin-resistant Staphylococcus aureus pneumonia. The dashed box indicates that systemic inflammatory response syndrome (SIRS) criteria were met, and the solid box indicates sepsis criteria were met. Information about antibiotic regimens can be found in the text. ED = emergency department; BP = blood pressure; HR = heart rate; RR = respiratory rate; SpO2 = oxygen saturation; CARPE DIEM = Catalyzing Ambulatory Research in Pneumonia Etiology and Diagnostic Innovations in Emergency Medicine; NP = nasopharyngeal; PCR = polymerase chain reaction; CBC= complete blood count; ICU = intensive care unit; WBC = white blood cell count; ANC = absolute neutrophil count; ECMO = extracorporeal membrane oxygenation. *Results from the istat venous point of care blood gas, renal panel, viral respiratory PCR panel, and the CBC with differential can be found in the text and Table 1.
Table 1. Demographic and Clinical Characteristics of the Patient with Fatal MRSA Pneumonia, Patients with Influenza Pneumonia, and Healthy Controls.
| Characteristic | Patient with Fatal MRSA Pneumoniaa | Patients with Influenza Pneumonia (n=4)a | Healthy Controls (n=7) |
|---|---|---|---|
| Age (yrs) | 0.67 | 9.5 (7.5-11.4) | 9.0 (7.5-11.0) |
| Male sex | 1 (100) | 2 (50) | 4 (57.1) |
| Caucasian | 1 (100) | 4 (100) | 5 (71.4) |
| Non-Hispanic ethnicity | 1 (100) | 4 (100) | 7 (100) |
| BMI (z-score)b | -0.98 | 0.30 (-0.79–1.89) | 0.55 (-1.47–2.10) |
| Vital signs | |||
| Heart rate (beats/minute) | 208 | 135 (116-184) | NA |
| Temperature (°C) | 39.8 | 37.3 (36.9- 38.1) | NA |
| Respiratory rate (breaths/minute) | 50 | 36 (24-48) | NA |
| Blood pressure (mm Hg) | 131/71 | 108/59 (93/55-117/58) | NA |
| Oxygen saturation (% on room air) | 94 | 96 (95-97) | NA |
| Laboratory results | |||
| White blood cell count (cells/mm3) | 1100 | 12,700 (9500-14,900) | NA |
| Absolute neutrophil count (cells/mm3) | 80 | 10,580 (6740-13,540) | NA |
| Blood culture | No growth after 5 days | No growth after 5 days | NA |
| Creatinine level (mg/dL) | 0.25 | NA | NA |
| Blood urea nitrogen level (mg/dL) | 7 | NA | NA |
| Rapid venous blood gas (mmol/L) | -4 | NA | NA |
| C-reactive protein level (mg/dL) | 4.34 | 5.7 (3.37-9.55) | NA |
| Procalcitonin level (ng/mL) | 14.1 | 0.3 (<0.1-0.4) | NA |
Data are mean (range) or no. (%) of patients in the influenza pneumonia and healthy control groups.
MRSA = methicillin-resistant Staphylococcus aureus; BMI = body mass index; NA = not applicable or available since these tests were not performed as part of clinical care in any of the patients with influenza pneumonia or in the assessment of the healthy controls.
At the time of presentation to the emergency department.
BMI is presented as z-score, which adjusts the height and weight of a child relative to their age. It is a more accurate comparison for BMI across ages.
Figure 2.
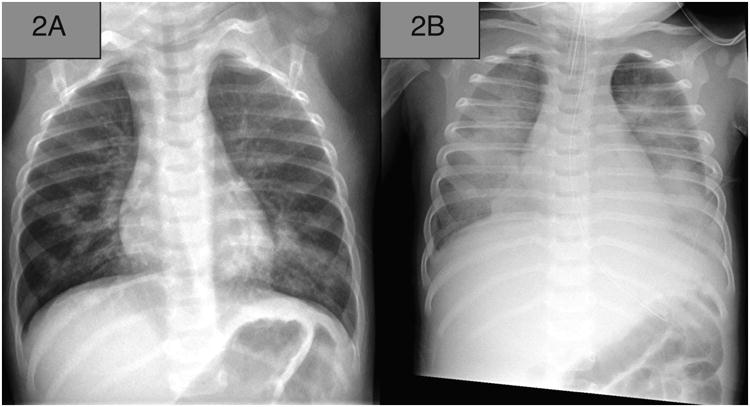
Anterior-posterior chest radiographs of the patient with fatal methicillin-resistant Staphylococcus aureus pneumonia: (A) chest radiograph revealing multifocal pneumonia; (B) chest radiograph revealing development of pleural effusion and imaged after placement of endotracheal tube.
Due to concerns for bacterial pneumonia, the infant received a dose of ampicillin 50 mg/kg in the ED according to national guidelines.6 The patient was admitted to the intensive care unit (ICU) because of increased respiratory distress, and a regimen of ceftriaxone (50 mg/kg) was initiated, of which one dose was administered, to provide broader empiric coverage. The infant deteriorated rapidly, progressing to respiratory failure requiring intubation, during which he suffered cardiac arrest requiring institution of extracorporeal membrane oxygenation. At this time, a new pleural effusion was discovered, requiring a tube thoracotomy (Figure 2B), and the patient was formally diagnosed with sepsis (Figure 1).7 Gentamicin (25 mg/kg) was initiated, then vancomycin (135 mg was dosed intermittently) and piperacillin/tazobactam (100 mg/kg every 6 hours) were initiated to provide broader antibiotic coverage. Despite these efforts, there was no improvement, and care was withdrawn; the infant died 43.2 hours after admission to the ICU. The cause of death on autopsy was severe necrotizing and hemorrhagic pneumonia caused by S. aureus infection, which was determined by postmortem respiratory culture of MRSA.
Patients with Influenza Pneumonia
The CARPE DIEM study enrolls previously healthy children with no chronic illnesses, aged 3 months to 18 years, who present to the ED with signs and symptoms of lower respiratory tract infection (e.g., cough, chest pain, dyspnea/shortness of breath) and are diagnosed with pneumonia confirmed by a focal opacity on chest radiograph. The attending ED physician is asked for the overall clinical impression of their patients, including the suspected probability (0–100%) and likelihood (highly unlikely, unlikely, likely, highly likely) that the patient would progress to severe disease or develop complications of pneumonia. A blood, nasal swab, and urine sample are collected at the time of enrollment. A respiratory viral polymerase chain reaction (PCR) panel and Mycoplasma pneumoniae PCR are performed on the nasal swab sample. The viral panel tests for influenza A, influenza B, parainfluenza 1, parainfluenza 2, parainfluenza 3, respiratory syncytial virus, human metapneumovirus, and human enterovirus/rhinovirus. Clinicians are blinded to the results of these tests since they are performed for research purposes only. Four patients enrolled in the CARPE DIEM study who had positive influenza PCR nasal swab results were included in this analysis as the influenza pneumonia patient cohort.
Healthy Controls
The healthy control cohort was part of a cross-sectional population study entitled the Genomic Control Cohort (GCC), conducted between 2007 and 2010.8 In the GCC study, urine samples were collected from healthy children, aged 3-18 years, living within the greater Cincinnati region. Seven urine samples from the GCC were age range and sex matched to the patients with influenza pneumonia included in the analysis.
Urine Sample Collection
Urine samples were collected in specimen collection cups coated with 10% sodium azide solution to inhibit bacterial growth in the sample.9 They were then centrifuged for 10 minutes and aliquots (1 mL) of supernatant were stored (-80°C) until the time of assay.
Nuclear Magnetic Resonance Data Acquisition and Spectral Analysis
Frozen urine samples were shipped to the University of Michigan's Nuclear Magnetic Resonance (NMR) Metabolomics Laboratory. At the time of the assay, samples were thawed at room temperature. The pH of each sample was adjusted with sodium hydroxide or hydrochloric acid to achieve a pH of 7.0 ± 0.25.10 A volume equivalent to 10% of the total sample volume of 5.4 mM of 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS-d6) in deuterium oxide was added to each sample as an internal standard. A one-dimensional 1H-NMR spectrum of each urine sample was acquired using the first increment of the standard NOESY pulse sequence on a Varian (Varian Inc., Palo Alto, CA) Inova-500 MHz NMR spectrometer at the University of Michigan's Biochemical NMR Laboratory as previously described.10-12 All spectra were recorded at 25°C. Spectral analysis was performed using quantitative metabolomics.13, 14 The raw 1H-NMR spectra were processed using Chenomx software (NMR Suite 7.5; Chenomx Inc., Edmonton, Alberta, Canada).10, 11 The resulting quantified metabolites were standardized to urine creatinine by dividing the concentration of each metabolite by the associated creatinine concentration as determined from the NMR spectrum of the same sample.15, 16 The quantified metabolite data (excluding urea) from the samples from the patient with fatal MRSA pneumonia, patients with influenza pneumonia, and healthy controls were cube root transformed and autoscaled to achieve a normal distribution of the data in order to conduct parametric statistical analysis.10, 17
Statistical Analysis of Metabolite Profiles
The normalized metabolite concentration data of the healthy controls and the patients with influenza pneumonia were compared by using an unpaired Student t test. The resulting p values were corrected for multiple comparisons by calculating the false discovery rate (FDR) using the method of Storey, et al.18 To identify potentially relevant metabolites of fatal MRSA pneumonia, the fold change of each metabolite was calculated by dividing each urine metabolite concentration of the patient with fatal MRSA pneumonia by the median urine concentration values of the same metabolite of the patients with influenza pneumonia and healthy controls. The resulting two lists of metabolites were each ordered by descending fold change. The metabolites with at least a 25-fold change that were common to both lists were considered as potentially relevant.19 The bioinformatics tool, Metscape (http://metscape.ncibi.org/)20, a plugin for the open-source network data integration tool, Cytoscape (http://www.cytoscape.org/), was used to visually assess the metabolic relationship between the found relevant metabolites and to construct a network figure. Statistical analyses and the construction of bar graphs were performed by using PRISM 6.0f (https://www.graphpad.com/), and the radar plot was constructed in Microsoft Excel (Microsoft Corp., Redmond, WA).
Results
Based on the infant's appearance in the ED, the attending physician reported a 5-10% probability of this infant progressing to severe disease or developing pneumonia complications, and felt that it would be unlikely for the patient to progress to severe disease. As part of the research study, procalcitonin and a C-reactive protein levels were measured and found to be elevated (14.1 ng/mL and 4.34 mg/dL, respectively) (Table 1).21 The PCR for Mycoplasma pneumoniae was negative. The viral panel was positive only for respiratory syncytial virus.
The mean (range) age in the healthy control group was 9.0 (7.5–11.0) years and 9.5 (7.5–11.4) years for patients with influenza pneumonia; the fatal MRSA pneumonia patient was 8.4 months of age. In addition, 3 of the 4 patients with influenza pneumonia were given one dose of antibiotics prior to the collection of urine: ampicillin (1 patient), ceftriaxone (1 patient), and azithromycin (1 patient).
A total of 75 metabolites (including creatinine and urea) were identified and quantified in each urine sample. Nine metabolite concentrations were statistically different between healthy controls and patients with influenza pneumonia (Table 2). Seven metabolite concentrations were increased by at least 25-fold in the patient with fatal MRSA pneumonia compared with those of both healthy controls and patients with influenza pneumonia (Table 3 and supplement Figures A–E). The magnitudes of difference in all detected metabolites are highlighted in a radar plot of creatinine-corrected metabolite concentrations (Figure 3A). The seven potentially differentiating metabolites of the patient with fatal MRSA pneumonia from healthy controls and patients with influenza pneumonia are depicted in Figure 3B. Concentrations of both acetylcarnitine and carnitine, which are vital to energy production22 and mitochondrial metabolism23, were substantially higher in the patient with fatal MRSA pneumonia than in either the healthy controls or patients with influenza pneumonia (Figure 3B).
Table 2. Potential Differentiating Metabolite Concentrations of Patients with Influenza Pneumonia Compared with Those of Healthy Controls.
| Metabolite | KEGG ID | p Value | FDR (%) |
|---|---|---|---|
| Creatine | C00300 | 0.004 | 28 |
| Guanidoacetate | C00581 | 0.008 | 31 |
| 1-Methylnicotinamide | C02918 | 0.014 | 34 |
| 2-Hydroxy-3-methylvalerate | NA | 0.018 | 34 |
| Trimethylamine N-oxide | C01104 | 0.022 | 34 |
| Citrate | C00158 | 0.029 | 36 |
| N-methyl hydantoin | C02565 | 0.042 | 38 |
| Methanol | C00132 | 0.050 | 38 |
| Threonine | C00188 | 0.050 | 38 |
KEGG = Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/); FDR = false discovery rate; NA = not applicable.
Table 3. Potential Differentiating Metabolite Concentrations with ≥ 25-Fold Change in the Patient with Fatal MRSA Pneumonia Compared with Those of Patients with Influenza Pneumonia and Healthy Controls.
| Patient with Fatal MRSA Pneumonia vs Healthy Controls: | Patient with Fatal MRSA Pneumonia vs Patients with Influenza Pneumonia: | ||
|---|---|---|---|
| Metabolite | Fold change | Metabolite | Fold change |
|
|
|
||
| 3-Hydroxybutyratea | 933 | 3-Hydroxybutyratea | 521 |
| Acetonea | 717 | Acetonea | 510 |
| Ascorbate | 95 | Acetoacetatea | 116 |
| 3-Aminoisobutyratea | 70 | Cholinea | 58 |
| Isobutyrate | 63 | Fumaratea | 34 |
| Acetoacetatea | 63 | 3-Aminoisobutyratea | 34 |
| Fumaratea | 57 | Adipate | 31 |
| Glucosea | 47 | Glucosea | 30 |
| Cholinea | 41 | Azelate | 27 |
| Lactate | 28 | Succinate | 27 |
| 2-Aminobutyrate | 28 | Trimethylamine N-oxide | 26 |
|
|
|
||
MRSA = methicillin-resistant Staphylococcus aureus.
Metabolites common to both comparisons.
Figure 3.
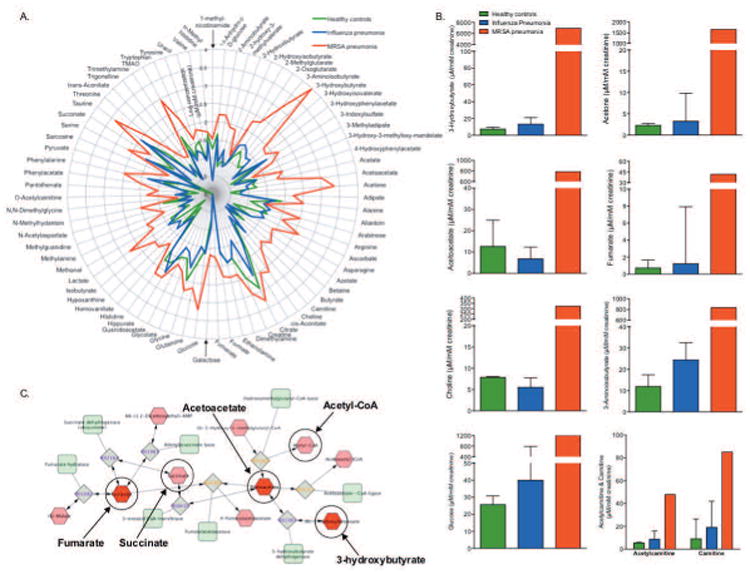
Influenza- and methicillin-resistant Staphylococcus aureus (MRSA)-induced distinct metabolic changes in the urine of children compared with the urine of healthy controls. (A) A radar plot shows the magnitude of the differences in 75 urine metabolites (log μM concentration/mM creatinine) in healthy children (n=7), children with influenza (n=4), and the patient with MRSA pneumonia. (B) Metabolite levels with at least a 25-fold difference between the patient with MRSA pneumonia and both healthy controls and patients with influenza pneumonia are shown in order of descending fold change. Also included are the urine concentrations of acetylcarnitine and carnitine, metabolites important in mitochondrial function, the elevation of which substantiate the magnitude of the perturbation in energy metabolism in the patient with MRSA that is shown by the large difference in the concentrations of urine ketone bodies and glucose. Data are the median (interquartile range). The greatest metabolic change was in ketone bodies (3-hydroxybutyrate, acetoacetate, and acetone), which is further illustrated in the Metscape generated network (C), which shows the relationship between the inputted compounds, acetoacetate, 3-hydroxybutyrate, and fumarate (dark red hexagons); acetone is generated by the nonenzymatic decarboxylation of acetoacetate so it is not shown in the network. The inputted compounds resulted in the generation of a single compound (red hexagons)–reaction (grey squares)–enzyme (green rounded-corner squares) network. This network map exemplifies the range of perturbation in energy metabolism because of the association between ketone bodies and tricarboxylic acid (TCA) metabolites fumarate and succinate, the latter of which was 27-fold higher in the patient with MRSA compared with patients with influenza pneumonia. Of note, acetyl coenzyme (CoA), which initiates the TCA cycle, is produced via glycolysis and carnitine-mediated fatty acid oxidation.
The most changed metabolite concentrations from the patient with fatal MRSA pneumonia were the ketone bodies (3-hydroxybutyrate, acetoacetate, and acetone) and fumarate, a tricarboxylic acid cycle metabolite, which are associated with a single metabolic network (Figure 3C). Notably, the urine urea concentration of the patient with fatal MRSA pneumonia was lower (71,616 μM) than the median (IQR) urea concentrations of the patients with influenza pneumonia (114,871 [97989–165273] μM) and healthy controls (192,506 [167475–217754] μM).
Discussion
This study provides evidence of the potential of metabolomics to identify novel biomarkers of illness severity in advance of clinical presentation and supports the need for further research to identify and validate urinary metabolites as potential biomarkers for early sepsis. This is evidenced by the increase in urine concentrations of several metabolites associated with MRSA infection. This is particularly important since, currently, there are emerging clinical biomarkers that only identify the presence of infection (e.g., elevated white blood cell count or C-reactive protein level) or biomarkers that are moderately predictive of bacterial infection and septic shock in children (e.g., procalcitonin level).24 However, currently available biomarkers neither distinguish among different etiologies (e.g., Streptococcus pneumoniae vs MRSA) nor do they elucidate the severity of infection.25, 26 In this case of fatal MRSA pneumonia, knowledge of the metabolomics profile may have helped with the timely diagnosis of sepsis in the ED, as this diagnosis can be challenging for several reasons, including the presence of common, nonspecific symptoms (e.g., fever, tachycardia) that can occur with more prevalent, less severe infections.27 Had the intensity of this patient's metabolic derangement been known in the ED, more aggressive therapies, including fluid resuscitation, vasopressor support, and broad-spectrum antibiotics, could possibly have been provided earlier in an attempt to avert this fatal outcome.
The findings presented here corroborate and extend the results of previous studies, one of which recognized the utility of metabolomics as a tool for gauging pneumonia severity.28 In that study, children aged 1 month to 11 years old with a diagnosis of sepsis had elevated levels of at least six metabolites (3-hydroxybutyrate, carnitine, creatinine, creatine, glucose, and acetone) compared with those of healthy patients. Additionally, in adults and children, sepsis-induced changes in levels of carnitine and its cometabolite, acetylcarnitine, which are involved in maintaining mitochondrial function, may indicate mitochondrial stress or dysfunction, which is known to occur in sepsis.28-31 The extent of metabolic dysfunction in our patient with fatal MRSA pneumonia was further evidenced by the greater than 25-fold increase in the urinary excretion of ketone bodies, fumarate, and glucose compared with those from the urine samples from patients with influenza pneumonia and healthy controls. These metabolites are different from those identified in a previous study of sepsis in children, most likely due to the host's response to both sepsis and necrotizing pneumonia.32 Importantly, in our patient, the consequence of the high degree of metabolic dysfunction was evident in the urine sample collected prior to the patient's subsequent development of severe septic shock, respiratory failure, and, ultimately, death.
The metabolites choline and 3-aminoisobutyrate were also markedly increased in the patient with fatal MRSA pneumonia when compared with those in the two control groups. In a mouse model, neither of these metabolites were associated with a lung-infected mouse with MRSA.12 However, elevated urine choline has been associated with antibiotic administration,33 which may explain the increase in our patient, as the child's urine sample was collected following administration of a dose of ampicillin. Altered 3-aminoisobutyrate homeostasis is associated with hyper-β-aminoisobutyric aciduria, one of the most common Mendelian metabolic variants in humans.34 In sepsis, increased urine 3-aminoisobutyrate, an end product of nucleic acid metabolism, may be explained by sepsis-induced impairment of skeletal muscle uptake.35
A potential limitation of the study is the overall concentration of the urine samples, which is influenced greatly by the hydration state of the patient. However, the urea concentration in the urine of the patient with fatal MRSA pneumonia was lower, and thus, less concentrated compared with the urea concentrations in healthy controls and patients with influenza pneumonia. Therefore, the less than 2-fold difference in urea concentration (patient with fatal MRSA pneumonia vs patients with influenza pneumonia) does not explain the greater than 100–500-fold increase in ketone body concentrations in the urine of the patient with fatal MRSA pneumonia. In addition, the patient had no clinical signs and symptoms of significant dehydration and did not have prolonged capillary refill time, abnormal skin turgor, sunken eyes, dry mucous membranes, cool extremities, weak pulses, or an absence of tears.36 Furthermore, the patient's laboratory values were inconsistent with dehydration or renal dysfunction, given the normal serum bicarbonate, BUN, and creatinine concentrations at the time of the urine collection.
A second potential limitation is that patient with fatal MRSA pneumonia was 8 months of age compared with the patients with influenza pneumonia and healthy controls who were older, with mean ages of 9.5 and 9 years, respectively (Table 1). Age is known to contribute to the variance of the metabolome; however, in a recent metabolomics study of pediatric sepsis, the magnitudes of age-related metabolic differences were much smaller than those identified in this report.28 Patients with sepsis were categorized into four groups including infants (aged 1 month to 1 year) and school age (aged 6-11 years). Identified metabolites were grouped by these age categories, and carnitine, creatine, and creatinine were the only metabolite concentrations that were elevated in school-age children but not in infants. Based on this finding, we would expect that in our study, in general, these metabolite levels would be much higher in the patients with influenza pneumonia because of their older age, but the metabolite levels from the patient with fatal MRSA pneumonia were always higher. Therefore, we contend that the metabolite level differences among our healthy controls, patients with influenza pneumonia, and the patient with fatal MRSA pneumonia cannot be explained simply based on differences in age.
Conclusion
Quantitative 1H-NMR urine metabolomics revealed a pattern of metabolites in a patient with complicated necrotizing pneumonia caused by MRSA that may serve as a potential tool to assist in early recognition of a severe disease course, namely septic shock. In this study, the profound metabolic change in the patient with fatal MRSA pneumonia was detected in the urine sample prior to many of the standard clinical and laboratory measures suggested severe sepsis. The availability of these data in the ED may have influenced clinical decision making and resulted in the identification of a high-risk patient earlier in the clinical course. However, as the clinical application of metabolomics has yet to be realized, validation of the identified metabolites of patients with fatal MRSA pneumonia is needed as well as further advances in technology to develop point-of-care and/or clinical tests based on the findings from metabolomics studies.
Supplementary Material
Acknowledgments
Sources of support: Dr. Ambroggio receives grant support from the Trustee Award from Cincinnati Children's Hospital Medical Center. Dr. Florin receives grant support from University of Cincinnati (KL2 TR000078-05). Drs. Ambroggio and Florin also receive grant support from the Gerber Research Foundation. Dr. Yeomans' effort is supported in part by a University of Michigan College of Pharmacy Upjohn Award and the Biochemical NMR Core Laboratory. Dr. Stringer's effort is supported in part by a grant from the National Institute of General Medical Sciences (NIGMS; GM111400). The samples from healthy subjects were provided by the Cincinnati Genomic Control Cohort and supported in part by the Cincinnati Children's Research Foundation. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The content is solely the responsibility of the authors and does not necessarily present the official views of the NIGMS or the National Institutes of Health.
References
- 1.Sawicki GS, Lu FL, Valim C, Cleveland RH, Colin AA. Necrotising pneumonia is an increasingly detected complication of pneumonia in children. The European respiratory journal. 2008;6:1285–91. doi: 10.1183/09031936.00099807. [DOI] [PubMed] [Google Scholar]
- 2.Spencer DA, Thomas MF. Necrotising pneumonia in children. Paediatr Respir Rev. 2014;3:240–5. doi: 10.1016/j.prrv.2013.10.001. quiz 45. [DOI] [PubMed] [Google Scholar]
- 3.Krenke K, Sanocki M, Urbankowska E, et al. Necrotizing Pneumonia and Its Complications in Children. Advances in experimental medicine and biology. 2015:9–17. doi: 10.1007/5584_2014_99. [DOI] [PubMed] [Google Scholar]
- 4.Goldstein B, Giroir B, Randolph A International Consensus Conference on Pediatric S. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;1:2–8. doi: 10.1097/01.PCC.0000149131.72248.E6. [DOI] [PubMed] [Google Scholar]
- 5.Scott HF, Deakyne SJ, Woods JM, Bajaj L. The prevalence and diagnostic utility of systemic inflammatory response syndrome vital signs in a pediatric emergency department. Academic emergency medicine : official journal of the Society for Academic Emergency Medicine. 2015;4:381–9. doi: 10.1111/acem.12610. [DOI] [PubMed] [Google Scholar]
- 6.Bradley JS, Byington CL, Shah SS, et al. The management of community-acquired pneumonia in infants and children older than 3 months of age: clinical practice guidelines by the Pediatric Infectious Diseases Society and the Infectious Diseases Society of America. Clin Infect Dis. 2011;7:e25–76. doi: 10.1093/cid/cir531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brierley J, Carcillo JA, Choong K, et al. Clinical practice parameters for hemodynamic support of pediatric and neonatal septic shock: 2007 update from the American College of Critical Care Medicine. Crit Care Med. 2009;2:666–88. doi: 10.1097/CCM.0b013e31819323c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovacic MB, Myers JM, Wang N, et al. Identification of KIF3A as a novel candidate gene for childhood asthma using RNA expression and population allelic frequencies differences. PLoS One. 2011;8:e23714. doi: 10.1371/journal.pone.0023714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernini P, Bertini I, Luchinat C, Nincheri P, Staderini S, Turano P. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. J Biomol NMR. 2011;3-4:231–43. doi: 10.1007/s10858-011-9489-1. [DOI] [PubMed] [Google Scholar]
- 10.Lacy P, McKay RT, Finkel M, et al. Signal intensities derived from different NMR probes and parameters contribute to variations in quantification of metabolites. PloS one. 2014;1:e85732. doi: 10.1371/journal.pone.0085732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slupsky CM, Rankin KN, Wagner J, et al. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal Chem. 2007;18:6995–7004. doi: 10.1021/ac0708588. [DOI] [PubMed] [Google Scholar]
- 12.Slupsky CM, Cheypesh A, Chao DV, et al. Streptococcus pneumoniae and Staphylococcus aureus pneumonia induce distinct metabolic responses. J Proteome Res. 2009;6:3029–36. doi: 10.1021/pr900103y. [DOI] [PubMed] [Google Scholar]
- 13.Stringer KA, Serkova NJ, Karnovsky A, Guire K, Paine R, 3rd, Standiford TJ. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma (1)H-nuclear magnetic resonance quantitative metabolomics and computational analysis. American journal of physiology Lung cellular and molecular physiology. 2011;1:L4–L11. doi: 10.1152/ajplung.00231.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wishart DS. Quantitative metabolomics using NMR. Trends Analyt Chem. 2008;3:228–37. [Google Scholar]
- 15.McClay JL, Adkins DE, Isern NG, et al. (1)H nuclear magnetic resonance metabolomics analysis identifies novel urinary biomarkers for lung function. J Proteome Res. 2010;6:3083–90. doi: 10.1021/pr1000048. [DOI] [PubMed] [Google Scholar]
- 16.Bouatra S, Aziat F, Mandal R, et al. The human urine metabolome. PLoS One. 2013;9:e73076. doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig A, Cloarec O, Holmes E, Nicholson JK, Lindon JC. Scaling and normalization effects in NMR spectroscopic metabonomic data sets. Analytical chemistry. 2006;7:2262–7. doi: 10.1021/ac0519312. [DOI] [PubMed] [Google Scholar]
- 18.Storey J. A direct approach to false discovery rates. Journal of the Royal Statistical Society Series B (Methodological) 2002;Part 3:479–98. [Google Scholar]
- 19.Yang YH, Speed T. Design and Analysis of Comparative Microarray Experiments. Boca Raton, FL: Chapman & Hall/CRC Press; 2003. [Google Scholar]
- 20.Karnovsky A, Weymouth T, Hull T, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;3:373–80. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Florin TA, Ambroggio L. Biomarkers for community-acquired pneumonia in the emergency department. Current infectious disease reports. 2014;12:451. doi: 10.1007/s11908-014-0451-8. [DOI] [PubMed] [Google Scholar]
- 22.Flanagan JL, Simmons PA, Vehige J, Willcox MD, Garrett Q. Role of carnitine in disease. Nutr Metab (Lond) 2010:30. doi: 10.1186/1743-7075-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuter SE, Evans AM. Carnitine and acylcarnitines: pharmacokinetic, pharmacological and clinical aspects. Clin Pharmacokinet. 2012;9:553–72. doi: 10.1007/BF03261931. [DOI] [PubMed] [Google Scholar]
- 24.Lautz AJ, Dziorny AC, Denson AR, et al. Value of Procalcitonin Measurement for Early Evidence of Severe Bacterial Infections in the Pediatric Intensive Care Unit. J Pediatr. 2016;74-81:e2. doi: 10.1016/j.jpeds.2016.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baer G, Baumann P, Buettcher M, et al. Procalcitonin Guidance to Reduce Antibiotic Treatment of Lower Respiratory Tract Infection in Children and Adolescents (ProPAED): A Randomized Controlled Trial. PLoS One. 2013;8:e68419. doi: 10.1371/journal.pone.0068419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esposito S, Tagliabue C, Picciolli I, et al. Procalcitonin measurements for guiding antibiotic treatment in pediatric pneumonia. Respiratory medicine. 2011;12:1939–45. doi: 10.1016/j.rmed.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Hartman ME, Linde-Zwirble WT, Angus DC, Watson RS. Trends in the epidemiology of pediatric severe sepsis*. Pediatr Crit Care Med. 2013;7:686–93. doi: 10.1097/PCC.0b013e3182917fad. [DOI] [PubMed] [Google Scholar]
- 28.Mickiewicz B, Vogel HJ, Wong HR, Winston BW. Metabolomics as a novel approach for early diagnosis of pediatric septic shock and its mortality. American journal of respiratory and critical care medicine. 2013;9:967–76. doi: 10.1164/rccm.201209-1726OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noland RC, Koves TR, Seiler SE, et al. Carnitine insufficiency caused by aging and overnutrition compromises mitochondrial performance and metabolic control. J Biol Chem. 2009;34:22840–52. doi: 10.1074/jbc.M109.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puskarich MA, Finkel MA, Karnovsky A, et al. Pharmacometabolomics of l-carnitine treatment response phenotypes in patients with septic shock. Ann Am Thorac Soc. 2015;1:46–56. doi: 10.1513/AnnalsATS.201409-415OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss SL, Selak MA, Tuluc F, et al. Mitochondrial Dysfunction in Peripheral Blood Mononuclear Cells in Pediatric Septic Shock. Pediatr Crit Care Med. 2014 doi: 10.1097/PCC.0000000000000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langley RJ, Tsalik EL, van Velkinburgh JC, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med. 2013;195:195ra95. doi: 10.1126/scitranslmed.3005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swann JR, Tuohy KM, Lindfors P, et al. Variation in antibiotic-induced microbial recolonization impacts on the host metabolic phenotypes of rats. J Proteome Res. 2011;8:3590–603. doi: 10.1021/pr200243t. [DOI] [PubMed] [Google Scholar]
- 34.Suhre K, Wallaschofski H, Raffler J, et al. A genome-wide association study of metabolic traits in human urine. Nature genetics. 2011;6:565–9. doi: 10.1038/ng.837. [DOI] [PubMed] [Google Scholar]
- 35.Warner BW, James JH, Hasselgren PO, Hummel RP, 3rd, Fischer JE. Effect of sepsis and starvation on amino acid uptake in skeletal muscle. The Journal of surgical research. 1987;4:377–82. doi: 10.1016/0022-4804(87)90172-7. [DOI] [PubMed] [Google Scholar]
- 36.Emond S. Evidence-based emergency medicine/rational clinical examination abstract Dehydration in infants and young children. Ann Emerg Med. 2009;3:395–7. doi: 10.1016/j.annemergmed.2008.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


