Abstract
X-ray free electron lasers (XFELs) have potential to revolutionize macromolecular structural biology due to the unique combination of spatial coherence, extreme peak brilliance and short duration of X-ray pulses. A recently emerged serial femtosecond crystallography (SFX) approach using XFEL radiation overcomes some of the biggest hurdles of traditional crystallography related to radiation damage through the diffraction-before-destruction principle. Intense femtosecond XFEL pulses enable high-resolution room temperature structure determination of difficult to crystallize biological macromolecules, while simultaneously opening up a new era of time-resolved structural studies. Here, we review the latest developments in instrumentation, sample delivery, data analysis, crystallization methods and applications of SFX to important biological questions, and conclude with brief insights into the bright future of structural biology using XFELs.
Keywords: structural biology, lipidic cubic phase, X-ray free electron laser, membrane proteins, protein complexes, molecular movies
XFELs opened up new era in crystallography
Elucidating the three-dimensional structures of proteins is essential for understanding their physiological function. In recent years, the number of available protein structures has increased rapidly, most notably in the field of membrane proteins. While several methods for obtaining macromolecular structures exist, X-ray crystallography prevails as a dominant technique. Since the first protein crystal structures were solved over 50 years ago a number of technical advances have accelerated the rate of structural studies. Major milestones have since been achieved including the development of cryocrystallography (see Glossary), which substantially reduces radiation damage [1], as well as microcrystallography, which enables high-resolution data collection from very small crystals (10–50 µm) [2]. Nevertheless, radiation damage remains a major obstacle for structure determination of many challenging macromolecules and their complexes at synchrotron sources. Radiation damage along with limitations in the intensity and the minimal pulse duration at third generation synchrotron sources also restricts applications of time-resolved crystallography to mainly reversible conformational changes in well-behaved proteins for which relatively large crystals are available.
While cryo-cooling can substantially slow down propagation of the secondary damage effects caused by the ionizing radiation, it was proposed that radiation damage can be essentially eliminated by using sufficiently short X-ray pulses, such as a few femtoseconds in duration [3]. Such an experiment was later simulated, and the results suggested that usable diffraction data can be collected from small protein crystals before they disintegrate by using extremely bright and short X-ray pulses, thereby introducing the “diffraction-before-destruction” principle [4] and inspiring construction of high-energy, hard X-ray free electron lasers (XFELs).
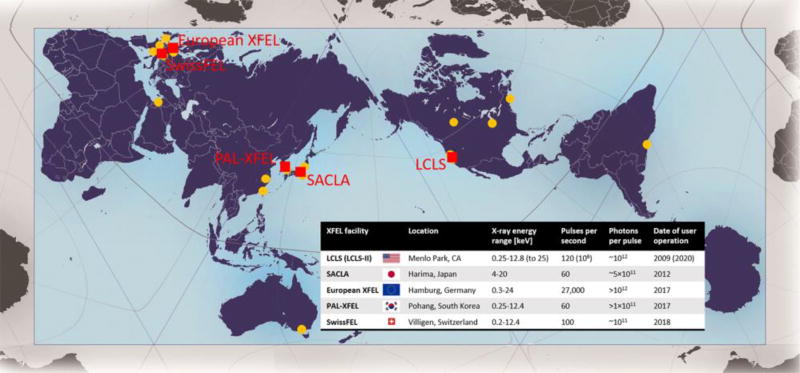
The first hard XFEL, the Linac Coherent Light Source (LCLS) in Menlo Park, USA, was commissioned in 2009 [5], followed by the Spring-8 Angstrom Coherent Laser (SACLA) in Harima, Japan, in 2011, PAL-XFEL in Pohang, South Korea and the European XFEL in Hamburg, Germany in 2017; with SwissFEL in Villigen, Switzerland planned in 2018 (Figure 1). These sources are capable of producing coherent X-rays with energies up to ~13 keV (25 keV for some sources) and a peak brilliance that is about nine to ten orders of magnitude stronger than that of a third generation synchrotron. They deliver extremely short pulses, in the order of femtoseconds (fs) in duration, allowing data collection before the onset of radiation damage. An early application of XFEL radiation to structural biology was the development of serial femtosecond crystallography (SFX), enabling data collection from hundreds of thousands of small, fully hydrated randomly oriented protein crystals streamed across the beam using a “one crystal, one shot” approach [6]. Since XFELs typically operate at high pulse repetition rates, SFX experiments generate millions of detector images within a short period of time. Novel protocols and data management strategies had to be developed to handle such large amounts of data. Additionally, since crystals remain essentially static on the time scale of an XFEL pulse, only partial reflections are recorded, which called for novel developments of indexing, integrating and merging such data.
Figure 1. Locations and parameters of XFEL facilities across the world (red squares).
For reference, locations of the most widely used third generation synchrotron sources are indicated by orange dots (including all 16 facilities with an average of 100 or more deposited structures per year between 2013 and 2015, as well as new facilities in Jordan and Brazil that are not yet commissioned). Locations are approximate. AuthaGraph map projection used with kind permission of Hajime Narukawa.
While data collection from small crystals (10–50 µm) is feasible at microfocus beamlines on third generation synchrotron sources, onset of radiation damage often limits the amount of information from such crystals even at cryocooled conditions. By using XFELs, these issues can be successfully circumvented since XFELs allow for damage-free room temperature data collection from micrometer and sub-micrometer sized crystals. Room temperature structures enabled by XFELs correctly describe conformational heterogeneity of a protein, which may be perturbed by cryocooling [7, 8], and give access to protein structure and dynamics via pump-probe time-resolved experiments [9].
Initial experiments at LCLS were mainly focused on low-resolution proof-of-principle experiments of various targets including photosystem I [6] and the photosynthetic reaction center [10]. Since then, technical upgrades allowed for high-resolution X-ray data to be recorded [11], including the high-resolution structure of cathepsin B bound to its native inhibitor [12], a potential drug target against sleeping sickness. Additionally, structures of several biomedically important G protein-coupled receptors (GPCRs) have been successfully solved with XFELs [13–19], including the rhodopsin-arrestin complex [20]. Furthermore, recent demonstrations of the possibility of de novo phasing [21–26] as well as ultra-fast time-resolved studies of proteins [27–30] and nucleic acids [31] further unveil the potential of XFELs for structural biology.
In this review we describe and discuss the latest advances in the SFX field, including hardware, software and conceptual developments. We also provide an outlook on what the exciting future of SFX holds in terms of structural biology, and the field of membrane protein crystallography and time-resolved studies in particular.
Experimental setup
Much of the recent success for SFX can be attributed to use of the CXI (Coherent X-ray Imaging) beamline [32] at LCLS (Figure 2). This beamline operates under vacuum to reduce background scattering and is equipped with a Cornell-SLAC Pixel Array Detector (CSPAD) [33], which can acquire data at the maximum repetition rate of LCLS (120 Hz, i.e. 120 frames per second). The beam is focused to a size of 0.1 or 1 µm by a pair of Kirkpatrick-Baez mirrors. The samples are typically introduced into the XFEL beam by a sample delivery system [34]. Furthermore this beamline has a tunable wavelength with energies ranging from 5 to 12 keV and is equipped with tunable pump lasers, enabling a range of different experiments, including de novo phasing and time-resolved pump-probe experiments as well as concurrent spectroscopy applications [35], and solution X-ray scattering experiments (SAXS/WAXS) [36] that are especially useful to investigate characteristic timescales for time-resolved experiments.
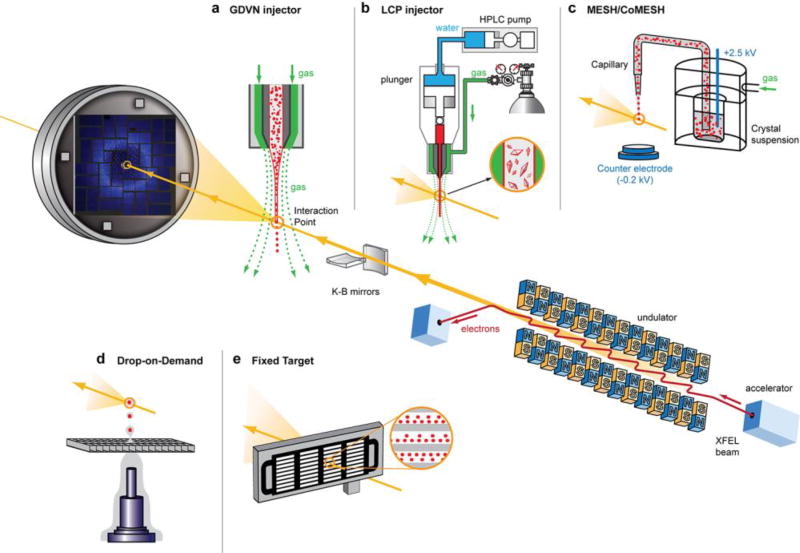
Figure 2. X-ray generation by FELs and an SFX beamline setup.
An electron bunch is accelerated to a relativistic speed and passed through an over 100 m long undulator, generating an extremely bright and short pulse of coherent X-rays, which is further focused by a pair of Kirkpatrick-Baez (K-B) mirrors or other X-ray optics to micron or submicron-sized spot inside a sample delivery chamber. Inserts demonstrate different methods of sample delivery into the focal point of the XFEL beam. Diffracted X-rays are recorded by a fast-reading detector while the direct beam passes through a hole in the detector and potentially can be re-focused and used for another experiment downstream. Different parts in this figure are not drawn to scale.
XFELs, with their unique spatial and temporal capabilities, enable breakthrough experiments across a wide range of disciplines. However, the impact and success of SFX in structural biology has motivated the commissioning of additional instruments, such as the XPP (X-ray Pump-Probe) and MFX (Macromolecular Femtosecond Crystallography) at LCLS [37], BL3/BL2 at SACLA [38], FXS (Femtosecond X-ray Scattering) and CXI (Coherent X-ray Imaging) at PAL-XFEL [39], and SPB/SFX (Single Particles, Clusters and Biomolecules & Serial Femtosecond Crystallography) at the European XFEL [40]. Some of these instruments operate using an ambient pressure sample chamber typically filled with helium, which alleviates potential issues with sample freezing upon injection into vacuum as well as bypass other technical challenges associated with sample delivery or exchange in the vacuum environment.
Detectors for XFELs
XFELs present specific challenges for detector development compared to synchrotron sources due to their extreme brightness, short pulse duration and fast repetition rate. The current setup at the CXI beamline at LCLS employs a 2.3 million pixel CSPAD detector consisting of 64 tiles (each 194 pixels by 185 pixels) with a pixel size of 110×110 µm2 [33]. The 64 tiles are arranged in quadrants, positioned in such a way that the strong direct beam can pass through the hole in the center. The unused beam can then be refocused downstream, allowing for several experiments to be performed simultaneously [41].
Currently, second-generation detectors for XFELs are being developed such as ePix10k [42], JUNGFRAU [43] and AGIPD [44]. These detectors are designed to fulfill the potential of higher repetition rates and are therefore capable of faster readout (up to 4.5 MHz). In addition, these novel detectors possess a higher dynamic range, enabling faster and more precise SFX data collection and are particularly useful for time-resolved experiments.
Sample delivery technology
Due to its extreme brightness, focused XFEL beam destroys any material with which it interacts. Therefore, a reliable way of replenishing microcrystals at the intersection point with the XFEL beam is required. This can be done in two principal ways: 1) the sample can be streamed across the beam using a special injector device, or 2) the sample can be fixed on a solid support, which is then rastered by the beam (fixed target).
The Gas Dynamic Virtual Nozzle (GDVN) injector [45] was one of the first and remains one of the most widely used injectors for SFX experiments to date (Figure 2a). Crystals in their mother liquor are jetted through a 50 µm capillary, and the stream is focused down to a few µm in diameter by a sheath of gas. The main drawback of the GDVN injector is the high rate of sample consumption. With a typical flow rate of 10–40 µl/minute [10, 46], a full dataset requires up to a few hundred milligrams of crystallized protein, which for many targets, in particular for membrane proteins, is not feasible. Recently, modifications of GDVN have being developed reducing sample consumption up to eight fold [46].
In order to circumvent this problem and reduce sample consumption, a special viscous media injecting device, also known as lipidic cubic phase (LCP) injector (Figure 2b), has been developed [14]. LCP is a liquid crystalline gel-like mesophase that mimics the native membrane environment and supports crystallization of membrane proteins. The LCP crystallization method has proven highly successful for a variety of membrane proteins, including G protein-coupled receptors (GPCRs), microbial rhodopsins, ion channels, transporters, enzymes, photosynthetic complexes, and β-barrel outer membrane proteins [47]. Crystals of soluble proteins can also be grown in LCP or mixed with LCP post-growth for a more efficient sample delivery [48]. The LCP injector consists of a reservoir and a hydraulic plunger which is driven by an HPLC system to extrude the LCP through a narrow capillary. The resulting LCP stream is stabilized by co-flowing gas. The gel-like consistency of LCP allows for better control over the crystal flow rate, which can then be matched to the XFEL pulse repetition rate to enable efficient use of crystals. The crystal flow rate is typically kept to 10–300 nl per minute for nozzles with an internal diameter of 10–50 µm, lowering sample consumption to under 0.3 mg of crystallized protein per dataset [49]. The versatility of the LCP injector becomes further evident by its capability to inject crystals in other viscous media such as agarose, which can support delivery of both soluble and membrane protein crystals [50]. Other viscous media used for XFEL sample delivery include a grease matrix [51] and Vaseline [52], as well as various viscous hydrophilic injection matrices, such as hyaluronic acid [53], hydroxyethyl cellulose [54], high molecular weight polyethylene oxide [55], sodium carboxymethyl cellulose and Pluronic F-127 [56]. Alternative injectors have been developed with the goal of reducing crystal consumption include the voltage driven electrokinetic injector (Figure 2c), also known as MESH (Microfluidic Electric Sample Holder) [57] and CoMESH (Concentric Microfluidic Electric Sample Holder) [58], as well as different variations of acoustic injectors (Figure 2d) for drop-on-demand data collection [59, 60]. The CoMESH injector was specifically designed to prevent freezing of crystals in their native mother liquor, by supplying an outer sheath of a cryo-protecting solution in a mixer just before interaction with the XFEL beam. A recent implementation of drop-on-demand delivery approach couples acoustic droplet ejection with a conveyer belt drive to optimize simultaneous time-resolved SFX and spectroscopic measurements over a wide range of time scales [61].
Time-resolved SFX studies of reactions initiated by binding of diffusible ligands impose their own requirements on the sample delivery systems. The fast turnover rate of enzymatic reactions demands a fast protein sample and reactant mixing and an adjustable delay time between the reaction initiation and data collection to be able to grasp different time point of the reaction process. Several liquid sample mixing injectors have been developed to satisfy those requirements, such as hydrodynamic mixers [62, 63] and T-junctions [31, 64].
Conceptually different from liquid jet injection is sample delivery using a fixed target approach (Figure 2e) [65, 66]. Typically, crystals, grown in or mounted on a chip, or harvested in a loop, are rapidly rastered with the XFEL beam. This approach minimizes sample waste and allows for better control over the time delay in time-resolved pump-probe experiments. Utilizing X-ray transparent materials, crystals can be grown directly inside a fixed target chip and data collected in situ, avoiding the need to harvest crystals [67]. Additionally, goniometer-based setups similar to those used at synchrotron sources allow for data collection under cryogenic conditions, mimicking rotation data collection at synchrotrons [68]. For large crystals, radiation damage-free diffraction data can be collected from single crystals if the distance between subsequent irradiation points is sufficiently large [69].
Crystallization techniques
The unique character of SFX data collection call for the development of new crystallization methods, or adaptation of existing methods to suit a particular sample delivery approach. In case of injector-based sample delivery systems the goal is to produce a homogeneous sample with a sufficient volume as well as optimal crystal size and density to ensure reliable crystal delivery with high crystal hit rates [70]. This is an important distinction to traditional crystallography, where single large crystals are desired since a whole dataset is typically collected from a single (or a few) crystals.
Crystallization in solution
Crystals grown in solution are most often initially obtained by high-throughput screening in plates and then optimized for batch setups (either in plates or in vials) which are then injected into the XFEL beam in their native mother liquor [70–72]. Crystals grown in solution can easily be concentrated by centrifugation and then filtered prior to injection thereby removing large crystals and impurities to avoid clogging the injector. Microseeding has been shown to improve crystal monodispersity and quality [73]. Larger crystals have been crushed into smaller ones by vortexing with beads without impacting their diffraction quality [74]. Alternatively, the free-interface diffusion (FID) method has been used to obtain a high concentration of very small, highly stable crystals [70].
For crystals grown in solution, an anti-settling device [75] is often employed to prevent crystals from settling at the bottom and on the sides of the reservoir. The size distribution, density and quality of microcrystals prepared for SFX data collection are typically characterized using the following techniques: SONICC (Second-Order Nonlinear Imaging of Chiral Crystals), DLS (Dynamic Light Scattering) and TEM (Transmission Electron Microscopy) [31, 76, 77].
Crystallization in LCP
Membrane proteins, which comprise about a third of the human proteome and represent targets for almost two thirds of marketed drugs [78, 79], are notoriously difficult to crystallize due in part to their amphiphilic nature and their inherent flexibility. A major breakthrough in structural biology of membrane proteins, in particular GPCRs, was the development of crystallization in a stabilizing, native-like lipid membrane environment of LCP [80]. The most commonly used host lipid for crystallization purposes is monoolein, which spontaneously forms the lipidic cubic phase (LCP) when combined with protein solution. The resulting lipid-protein mixture is then overlaid with a precipitant solution to induce crystallization. Initial crystallization conditions are identified using high-throughput nanovolume crystallization in 96-well glass sandwich plates and later transferred and optimized in Hamilton gas-tight syringes to yield a high density of microcrystals homogenous in size [81, 82]. Due to the gel-like consistency of LCP, the crystals are practically impossible to concentrate and filter post-growth, therefore, special precautions should be employed to avoid trapping, in LCP, large dust particles and fibers that can clog the injector.
In vivo crystallization
In special cases, tedious protein purification and sample preparation can be circumvented altogether, making use of the propensity of certain proteins to spontaneously form crystals inside cells, or cellular compartments in vivo. For example, overexpressing procathepsin B in Sf9 insect cells leads to spontaneous crystallization and, after cell lysis, allowed for data collection and structure solution using an SFX approach at LCLS [12]. In cellulo SFX of yeast alcohol oxidase has been attempted using XFEL diffraction of whole cells, or the isolated subcellular peroxisome fraction, where crystals were found to form, although data could not be indexed due to low resolution [83]. Finally, Cry3A crystals grown in bacterial cells could be exposed to X-rays by streaming whole cells into the XFEL beam, yielding a structure at 2.9 Å resolution [84].
Data processing developments
Due to the nature of the sample delivery process in an SFX experiment, not every X-ray pulse hits a crystal and many blank images are therefore generated. In addition, tens of thousands of images are collected within a short time frame, therefore the first step in data processing is to identify useful patterns containing Bragg reflections, referred to as crystal hits. The program Cheetah [85] evaluates each image on metrics such as the number of Bragg peaks, maximum resolution and detector oversaturation levels, while simultaneously performing pre-processing steps necessary for downstream analysis. Additionally some preliminary data quality metrics can be obtained through the generation of virtual powder diffraction patterns and statistics on hit rate and resolution.
After hits have been identified, the data are further processed using one of the available programs specifically designed or adapted for SFX, such as cctbx.xfel [86], CrystFEL [87] or nXDS [88]. CrystFEL performs indexing by calling DirAx [89], XDS [90] or Mosflm [91], after which diffraction spots are modeled as circular regions while masking surrounding background pixels. Partial reflections are typically merged using a Monte Carlo approach [92], which relies on high redundancy of individual observations. Various parameters affecting observed intensities have been actively researched in recent years and several enhanced algorithms that improved the accuracy of obtained structure factors emerged. Those include refinement of the detector geometry [93], inclusion of linear and Debye-Waller scaling terms [94, 95], refinement of crystal parameters before integration [96, 97] and estimation of reflection partialities and post-refinement [94, 98].
Another important improvement to the initial data processing algorithms is the possibility to resolve indexing ambiguities, which arise in certain crystal symmetries that allow multiple choices of lattice indexing. Several similar algorithms for resolving the indexing ambiguity have been recently implemented [87, 88, 99], based on the comparison of correlation coefficients between the intensities of each crystal and the intensities of all other crystals with the same or opposite indexing assignment. The final choice of the indexing assignment for a given pattern is based on a stronger correlation with the corresponding group.
Structure determination and de novo phasing
Most structures to date that employed XFEL radiation have been solved using the molecular replacement (MR) method, where a structural model of a homologous protein is used for the initial phase estimates. However, often no template structures of sufficient quality for MR are available for the studied target, thus requiring de novo phasing. While being relatively routine at synchrotron sources, de novo phasing has proven difficult at XFELs, because of the inherent inaccuracy in the amplitudes of the structure factors derived from SFX data due to variations in the XFEL pulse energy and intensity, crystal size and mosaicity, partiality of recorded Bragg reflections and other factors. Therefore, high data multiplicity combined with advanced merging techniques are crucial for the success of experimental phasing. The first demonstration of successful phasing relied on the strong anomalous signal of a gadolinium derivative of lysozyme using the single wavelength anomalous diffraction (SAD) method [21]. This experiment required as many as 60,000 indexed patterns to succeed. However, after improvements in data processing, which included employment of scaling procedures and optimization of the detector geometry, the number of patterns necessary for successful experimental phasing could be reduced to 7,000 [26]. Another study reported structure determination of luciferin-regenerating enzyme by single isomorphous replacement with anomalous scattering (SIRAS) using a mercury derivative [23] and requiring 10,000 patterns for each of the native and derivative datasets. Most recently, a novel structure of a mosquito larvicide BinAB has been solved by multiple isomorphous replacement with anomalous scattering (MIRAS) using three different derivatives: mercury, gadolinium and iodine [100]. Another approach successfully used for experimental phasing was based on the use of iodine-containing detergent to purify and crystallize membrane proteins, and serve as a source of anomalous signal for SAD, SIR or SIRAS phase retrieval methods [25].
While these studies provided important insights into the parameters crucial for a successful phasing experiment, they relied on the availability of well diffracting derivative crystals with a strong anomalous signal. Utilization of much weaker anomalous signal from endogenous sulfur atoms can alleviate many problems related to the use of heavy-atom compounds and production of derivatized crystals. Initial studies employing sulfur for SAD phasing at XFELs have been applied to soluble test proteins: lysozyme [22] and thaumatin [26]. More recently, de novo phasing by sulfur-SAD was accomplished with the first membrane protein, human A2A adenosine receptor, resulting in a room temperature 1.9 Å resolution structure [24]. Data analysis from this experiment suggested that ~600,000 indexed patterns collected at 6 keV X-ray energy would be sufficient for phasing a protein containing only 12 sulfur atoms per 447 residues (2.7%), which is lower than in about 90% of human proteins, indicating that most proteins should be amenable for de novo phasing using XFELs. Thus, de novo phasing is likely going to be a routine procedure at XFELs in the near future, only currently limited by the scarcity of available XFEL beam time.
SFX applications
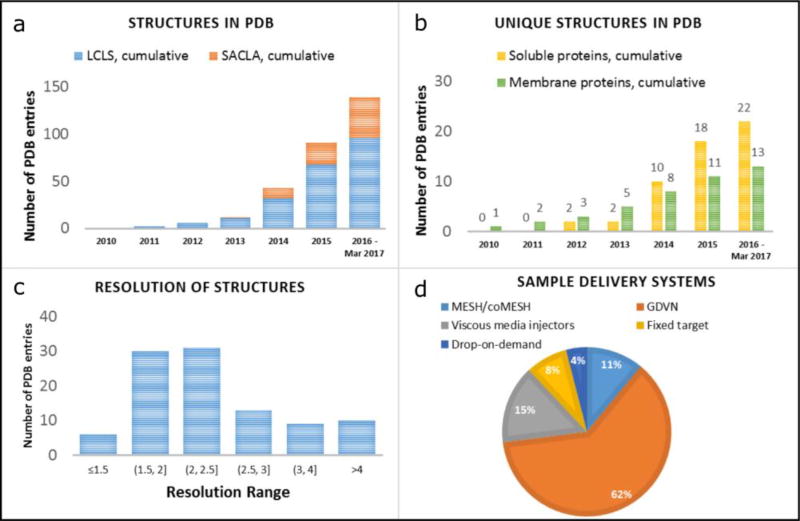
Between the first proof-of-principle structure of Photosystem I (published in February of 2011 [6]) and March of 2017, 139 entries obtained at XFELs (96 at LCLS and 43 at SACLA), corresponding to 35 unique proteins (22 soluble and 13 membrane), have been deposited to PDB (Figure 3a,b). The majority of the structures (75%) are solved at a resolution within the 1.5 – 3 Å range (Figure 3c), with the highest resolution structure, that of proteinase K solved to 1.20 Å [54], pushing the atomic resolution level. Such resolution is not routinely accessible for room temperature structure determination at synchrotron sources due to radiation damage. The smallest protein crystals that yielded high-resolution structure by SFX had an average volume of less than 0.016 µm3 and contained 9,000 unit cells [101].
Figure 3. Statistics on the macromolecular structures solved using XFELs to date.
(a) Growth in the number of macromolecular structures deposited to PDB using the data obtained at LCLS (blue bars) and SACLA (orange bars) (b) Growth in the number of “unique” (each protein is counted only once at the earliest deposition date) macromolecular structures deposited to PDB that were solved using XFELs. Structures of soluble proteins are shown as yellow bars and structures of membrane proteins are shown as green bars. (c) Number of structures falling into the respective resolution ranges. (d) Number of structures solved using different sample delivery methods.
One of the most successful applications of SFX has been the structural study of membrane proteins using LCP as the crystallization and crystal delivery medium, which led to structure determination of several GPCRs [13–19] including the signaling complex between GPCR and arrestin [20], the membrane enzyme diacylglycerol kinase [102], and the light-activated proton pump bacteriorhodopsin [103].
GPCRs constitute the most abundant protein superfamily in the human genome with over 800 members. They mediate cellular signaling, regulate the majority of physiological processes and serve as targets for about a third of marketed drugs [79]. Initial development and implementation of SFX data collection on GPCR microcrystals grown and delivered in LCP was done using the human serotonin receptor 5-HT2B [13]. Compared with the structure solved by using traditional microcrystallography from cryo-cooled crystals the room temperature XFEL structure displayed a distinct distribution of thermal motions and conformations of residues that likely more accurately represent the receptor structure and dynamics in native cellular environments. For the smoothened receptor, a critical tumor target, SFX enabled the determination of its structure bound to the teratogen cyclopamine, where high mosaicity had prevented synchrotron data collection on large crystals, but small crystals were found to diffract to 3.2 Å at LCLS [14]. Similarly, whereas the synchrotron-generated structure for the δ-opioid receptor (with a resolution of 3.4 Å) could not unambiguously identify the binding mode of a bifunctional peptide pain-killer with reduced tolerance and dependency, the 2.7 Å structure obtained at LCLS revealed the correct conformation of the bound peptide [15]. Furthermore, the angiotensin receptor AT1R, which is antagonized by many antihypertensive drugs, represents the first novel GPCR structure solved by SFX [16].
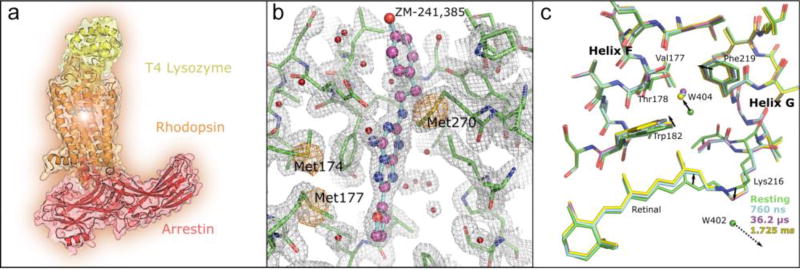
More recently, SFX was instrumental for the structure determination of a GPCR-arrestin signaling complex (Figure 4a). Arrestins play a dual role in cell signaling. First, they bind to activated and phosphorylated receptors abolishing signaling through G proteins and leading to receptor desensitization and internalization [104]. In addition to terminating G protein signaling, arrestins initiate G protein-independent signaling pathways leading to distinct cellular responses. After extensive efforts in stabilizing the rhodopsin-arrestin complex, small (10–15 µm in size) crystals were obtained in LCP, which diffracted only to ~8 Å at a synchrotron source. The same crystals diffracted to ~3.3 Å (anisotropic) resolution at LCLS leading to the first high-resolution structure of a GPCR-arrestin complex [20]. This pioneering study is a great example of the potential of XFELs applied to structural biology, where systems not tractable by traditional synchrotron-based structure determination can be successfully studied by using the high intensity XFEL beams.
Figure 4. Representative protein structures solved using XFELs.
(a) The structure of a major signaling complex between a GPCR and arrestin. (b) Native phasing of XFEL data: the structure of a GPCR, adenosine A2A receptor, was solved using sulfur SAD. The A2A receptor is shown as green sticks, the ligand ZM-241,385 is shown in ball-and-stick representation, the 2mFo-DFc electron density map is shown as a grey mesh and contoured at 1.5 σ, and the anomalous difference map is shown as an orange mesh and contoured at 5 σ. (c) Time resolved atomic structure of bacteriorhodopsin. The structures at different time points (760 ns – cyan. 36.2 µs – magenta, 1.725 ms – yellow) are superimposed with the ground structure (green). Solid arrows indicate the direction of changes in the structure with time, the dashed arrow indicates water molecule W402 becoming unstructured after the retinal isomerization. Adapted from Ref. [30].
Another major application of XFELs in structural biology is to utilize the inherent temporal resolution capacity of XFELs to study reaction intermediates or other structural changes. Currently the best time resolution achievable at third generation synchrotrons is ~100 ps, however many key structural changes occur at time scales much faster than that, such as sub-picosecond collective motions in CO myoglobin [29]. Hence, utilizing the temporal resolution of an XFEL can significantly improve the accessibility of ultra-fast reactions and internal motions. Furthermore, the use of XFELs can overcome the problem of radiation damage [4, 105] and simultaneously evade the problem of studying irreversible reactions since each crystal is exposed to the XFEL beam only once. An additional advantage of using small crystals is that the induced conformational changes are more uniform in time and space throughout the crystal. A number of recent studies have shown the potential of using XFELs for time-resolved crystallography, following a pump-probe approach that uses an optical laser as a reaction trigger, including photosystem II (PSII) [27, 35, 106], photoactive yellow protein PYP [28, 107] and light-driven proton pump bacteriorhodopsin [30]. PSII, a large multi-subunit membrane protein complex, is part of the photosynthetic machinery which catalyzes the light-driven splitting of water to oxygen, a process which is essential to maintain the Earth’s atmosphere. XFEL time-resolved experiments allowed for probing several intermediates of the PSII photocycle providing mechanistic insights into the oxygen evolution reaction.
In a recent study, high quality electron density maps of PYP at 1.6 Å were obtained for time points between 100 fs and 3 ps, enabling the visualization of one of the fastest reactions in biomolecules, the cis/trans isomerization of a chromophore [107]. For bacteriorhodopsin, a molecular movie consisting of 13 snapshots of conformational changes evenly spaced on a logarithmic scale between 16 ns and 1.725 ms after photoactivation were reported [30] (Figure 4c).
Besides fast, light-triggered reactions, slower processes, allowing the investigation of more general, substrate-triggered biological reactions can be studied by mix-and-inject serial crystallography [108]. With diffusion rates strongly depending on the crystal size, the small crystal size amenable to structural studies with XFELs facilitates a millisecond time resolution, which is currently not possible with larger crystals required for time-resolved synchrotron studies [108]. Using mix-and-inject approach, conformational changes in an RNA riboswitch have been probed at 10 s and 10 min after binding of an adenine ligand [31]. This study captured a ligand-bound intermediate state of the riboswitch with implications for understanding the molecular mechanisms of regulation of gene expression [31]. More recently, binding of an antibiotic to β-lactamase from M. tuberculosis has been demonstrated [64]. Taken together, these groundbreaking experiments have enabled time-resolved structural studies of enzymatic and other ligand-driven biological processes [109].
What will the future bring?
While the SFX field is still relatively young, it has already led to multiple breakthroughs in structural biology enabling damage-free room temperature structure determination of important biomolecules and complexes, as well as the recording of molecular movies with sub-picosecond temporal resolution. With the advent of new XFEL facilities being built around the world and upgrades of current XFELs (LCLS-II) (Figure 1), the speed of technological development and subsequent scientific discoveries will only accelerate.
One of the most important features of the new XFELs that use superconducting accelerators is a faster X-ray pulse repetition rate, which can significantly shorten the data collection time, allowing for multiple sets of data collection within a single beamtime shift. Additionally, multiplexing experiments by switching beams between experimental stations or refocusing ‘unused’ beam will also increase the available beamtime. Better detectors with higher dynamic range and faster acquisition rates will not only lead to improved data but will also increase data collection efficiency.
With development of new sample delivery methods such as fixed targets or drop-on demand injectors, sample consumption can be further reduced, while simultaneously increasing the efficiency of data collection by synchronization to the XFEL pulses. Furthermore, standardization of sample injection protocols should allow for automated data collection, with the possibility of shipping samples and collecting data remotely, similar to what currently is implemented at synchrotrons.
XFELs begin to blur the boundaries between crystallographic studies and single-molecule structure determination. While recent technological breakthroughs in cryo-electron microscopy enabled the structure determination from single molecules with better than 3 Å resolution [110], thus bypassing the major bottleneck of crystallization, analysis of recent SFX data indicated that current XFELs can be used for high-resolution structure determination from protein crystals with as low as 100 unit cells, suggesting that room temperature single-molecule imaging of biological macromolecules soon could be within reach [101].
In conclusion, the rapid development of tools, protocols, instrumentation and data processing over the past few years is a tremendous achievement and the future indeed looks bright for overcoming new frontiers in structural biology using XFEL radiation.
Trends box.
X-ray free electron lasers (XFELs) allow radiation damage to be overcome, enabling high-resolution room temperature structures of difficult-to-crystallize proteins, for which only small crystals are available, and extremely radiation sensitive macromolecules, such as metalloenzymes.
De novo phasing at XFELs has been established on several different targets and by different methods thereby enabling structure determination of novel macromolecules of biological interest.
The extremely short femtosecond duration XFEL pulses provide access to dynamic information about unstable intermediate states and irreversible reactions.
The development of new injectors, crystal delivery media and fixed target devices has dramatically reduced the amount of required crystallized protein. Mixing injectors provide an important avenue to study biological interactions in four-dimensional space.
Developments in data processing software reduce the required amount of data and improve data quality.
Outstanding questions (box).
-
-
What is the minimal number of diffraction images required for structure determination by SFX?
-
-
How quickly can a complete data set can be routinely collected by SFX? Can SFX data collection surpass the throughput of traditional crystallography?
-
-
What is the effect of primary radiation damage from an XFEL pulse, and how does it depend on the pulse duration and intensity, X-ray energy, and the distribution of electron-rich clusters in the protein structure, such as those in metalloenzymes?
-
-
What is the limit in the number of unit cells in the crystal sufficient for high-resolution data collection at XFELs? Will it be possible to collect atomic resolution data from single protein molecules, and when?
-
-
Which new technologies are needed to increase access to XFEL beamtime and improve the efficiency of the XFEL beamtime usage?
Acknowledgments
The authors would like to thank Drs. S. Boutet, A. Mancuso, K. Tono, and Prof. I.S. Ko for the helpful discussion of current and future XFEL instrumentation, and Prof. H. Narukawa for the permission to use the AuthaGraph map projection. Research work in the authors’ laboratory related to this review is funded by the National Institutes of Health grant R01 GM108635 and the National Science Foundation award 1231306. L.C.J would like to acknowledge fellowship funds from the Human Frontier Science Program Organization (HFSPO).
Glossary
- Crystal hit rate
the ratio between the number of detector images with identified diffraction spots and the total number of collected images.
- Cryocrystallography
cryo-cooling crystals during crystallographic data collection in order to reduce radiation damage effects.
- De novo phasing
experimental approaches for deriving phases of structure factors from measured intensities using such methods as SIR/MIR (Single/Multiple Isomorphous Replacement, SAD/MAD (Single/Multiple-wavelength Anomalous Dispersion) or SIRAS/MIRAS (Single/Multiple Isomorphous Replacement combined with Anomalous Scattering).
- Fixed target
a concept for sample delivery that relies on immobilizing the sample on a solid support for serial data collection by rastering with the XFEL beam.
- Goniometer
a device used to position the crystal in selected orientations for optimization of data collection.
- Lipidic Cubic Phase (LCP)
a gel-like membrane mimetic matrix that supports membrane protein crystallization from a native-like environment and can be used for efficient sample delivery.
- Microfocus beamline
a synchrotron beamline equipped with beam-focusing optics and precise goniometers to enable data collection from micrometer-sized crystals.
- Molecular replacement
a method for solving the phase problem (see below) by using initial phases from a similar (homologous) protein.
- Phase problem
reconstitution of electron density requires the knowledge of structure factor phases, however, in a crystallographic experiment only structure factor amplitudes, but not their phases, are measured.
- Pump-probe
a time-resolved experiment in which an optical pulse (pump) triggers a reaction followed by an XFEL pulse (probe) at a variable time delay.
- Radiation damage (primary, secondary)
Primary radiation damage implies immediate ionization effects caused by absorbed photons and ejected photoelectrons while secondary radiation damage occurs through (mainly) the formation of free radicals (caused by primary ionization events).
- Resolution
a measure of the visibility of details in a density map. In crystallography, resolution is traditionally measured in Ångströms (1 Å=0.1 nm). The lower the value, the more details can be seen. At approximately 3.5 Å, individual amino acid side chains are visible and at 2.5 Å, waters are discernible, at better than 2 Å resolution multiple amino acid conformations could be modeled.
- Self-Amplified Spontaneous Emission (SASE)
a process by which a laser beam is generated from a high-energy electron beam. An electron bunch is accelerated to a relativistic speed and passed through a long undulator, which supplies a transverse magnetic field that is periodically changing in space, encouraging electrons to follow a sinusoidal path emitting X-ray photons. Interaction between spontaneously emitted photons and electrons induces alignment of electrons (“microbunching”), thereby causing them to emit X-ray radiation coherently.
- Serial Femtosecond Crystallography (SFX)
a data collection paradigm where diffraction data are collected from a large number of microcrystals in a “one crystal, one shot” approach using an XFEL.
- Third generation synchrotron source
a synchrotron source equipped with straight sections containing undulator or wiggler magnets leading to increased brilliance as well as spatial and temporal coherence of X-ray beams compared to earlier generation synchrotron sources.
- Time-resolved crystallography
a method used to visualize conformational transitions occurring within a protein as it performs its function with a high spatial and temporal resolution. The time-component is achieved by collecting X-ray data at several time-points after initiation of the reaction.
- X-ray free electron laser (XFEL)
a latest generation light source producing ultra-bright pulses of coherent X-rays with ultra-short duration employing the SASE principle on freely moving electrons.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Garman E. Cool data: quantity and quality. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1641–1653. doi: 10.1107/s0907444999008653. [DOI] [PubMed] [Google Scholar]
- 2.Smith JL, et al. Micro-crystallography comes of age. Curr. Opin. Struct. Biol. 2012;22:602–12. doi: 10.1016/j.sbi.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solem JC. Imaging biological specimens with high-intensity soft x rays. J. Opt. Soc. Am. B. 1986;3:1551–1565. [Google Scholar]
- 4.Neutze R, et al. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature. 2000;406:752–7. doi: 10.1038/35021099. [DOI] [PubMed] [Google Scholar]
- 5.Emma P, et al. First lasing and operation of an ångstrom-wavelength free-electron laser. Nat. Photon. 2010;4:641–647. [Google Scholar]
- 6.Chapman HN, et al. Femtosecond X-ray protein nanocrystallography. Nature. 2011;470:73–7. doi: 10.1038/nature09750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser JS, et al. Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16247–16252. doi: 10.1073/pnas.1111325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keedy DA, et al. Crystal cryocooling distorts conformational heterogeneity in a model Michaelis complex of DHFR. Structure. 2014;22:899–910. doi: 10.1016/j.str.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spence J. XFELs for structure and dynamics in biology. IUCrJ. 2017;4 doi: 10.1107/S2052252517005760. Published online May 10, 2017. http://dx.doi.org/10.1107/S2052252517005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johansson LC, et al. Lipidic phase membrane protein serial femtosecond crystallography. Nat. Methods. 2012;9:263–5. doi: 10.1038/nmeth.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boutet S, et al. High-Resolution Protein Structure Determination by Serial Femtosecond Crystallography. Science. 2012;337:362–364. doi: 10.1126/science.1217737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Redecke L, et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science. 2013;339(6116):227–30. doi: 10.1126/science.1229663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W, et al. Serial femtosecond crystallography of G protein-coupled receptors. Science. 2013;342:1521–4. doi: 10.1126/science.1244142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weierstall U, et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 2014;5:3309. doi: 10.1038/ncomms4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenalti G, et al. Structural basis for bifunctional peptide recognition at human delta-opioid receptor. Nat. Struct. Mol. Biol. 2015;22:265–8. doi: 10.1038/nsmb.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, et al. Structure of the Angiotensin receptor revealed by serial femtosecond crystallography. Cell. 2015;161:833–44. doi: 10.1016/j.cell.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, et al. Structural basis for selectivity and diversity in angiotensin II receptors. Nature. 2017;544:327–332. doi: 10.1038/nature22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, et al. Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand. Nat. Commun. 2017;8:15383. doi: 10.1038/ncomms15383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, et al. Structure of the full-length glucagon class B G-protein-coupled receptor. Nature. 2017;546:259–264. doi: 10.1038/nature22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang Y, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–7. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barends TR, et al. De novo protein crystal structure determination from X-ray free-electron laser data. Nature. 2014;505:244–7. doi: 10.1038/nature12773. [DOI] [PubMed] [Google Scholar]
- 22.Nakane T, et al. Native sulfur/chlorine SAD phasing for serial femtosecond crystallography. Acta Crystallogr. D Biol. Crystallogr. 2015;71:2519–25. doi: 10.1107/S139900471501857X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamashita K, et al. An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography. Sci. Rep. 2015;5:14017. doi: 10.1038/srep14017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batyuk A, et al. Native phasing of x-ray free-electron laser data for a G protein-coupled receptor. Sci. Adv. 2016;2:e1600292. doi: 10.1126/sciadv.1600292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakane T, et al. Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent. Proc. Natl. Acad. Sci. U. S. A. 2016;113:13039–13044. doi: 10.1073/pnas.1602531113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nass K, et al. Protein structure determination by single-wavelength anomalous diffraction phasing of X-ray free-electron laser data. IUCrJ. 2016;3:180–91. doi: 10.1107/S2052252516002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kupitz C, et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature. 2014;513:261–5. doi: 10.1038/nature13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenboer J, et al. Time-resolved serial crystallography captures high-resolution intermediates of photoactive yellow protein. Science. 2014;346:1242–6. doi: 10.1126/science.1259357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barends TR, et al. Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science. 2015;350:445–50. doi: 10.1126/science.aac5492. [DOI] [PubMed] [Google Scholar]
- 30.Nango E, et al. A three-dimensional movie of structural changes in bacteriorhodopsin. Science. 2016;354:1552–1557. doi: 10.1126/science.aah3497. [DOI] [PubMed] [Google Scholar]
- 31.Stagno JR, et al. Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography. Nature. 2017;541:242–246. doi: 10.1038/nature20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boutet S, J Williams G. The Coherent X-ray Imaging (CXI) instrument at the Linac Coherent Light Source (LCLS) New J. Phys. 2010;12:035024. [Google Scholar]
- 33.Hart P, et al. The Cornell-SLAC Pixel Array Detector at LCLS. SLAC-PUB-15284 2012 [Google Scholar]
- 34.Weierstall U. Liquid sample delivery techniques for serial femtosecond crystallography. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130337. doi: 10.1098/rstb.2013.0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kern J, et al. Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy. Nat. Commun. 2014;5:4371. doi: 10.1038/ncomms5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnlund D, et al. Visualizing a protein quake with time-resolved X-ray scattering at a free-electron laser. Nat. Meth. 2014;11:923–926. doi: 10.1038/nmeth.3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boutet S, et al. The New Macromolecular Femtosecond Crystallography (MFX) Instrument at LCLS. Synchrotron Radiation News. 2016;29:23–28. doi: 10.1080/08940886.2016.1124681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yabashi M, et al. Overview of the SACLA facility. J. Synchrotron Radiat. 2015;22:477–484. doi: 10.1107/S1600577515004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park J, et al. Current status of the CXI beamline at the PAL-XFEL. J. Korean Phys. Soc. 2016;69:1089–1093. [Google Scholar]
- 40.Altarelli M, Mancuso AP. Structural biology at the European X-ray free-electron laser facility. Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130311. doi: 10.1098/rstb.2013.0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boutet S, et al. Characterization and use of the spent beam for serial operation of LCLS. J. Synchrotron Radiat. 2015;22:634–643. doi: 10.1107/S1600577515004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blaj G, et al. X-ray detectors at the Linac Coherent Light Source. J. Synchrotron Radiat. 2015;22:577–83. doi: 10.1107/S1600577515005317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mozzanica A, et al. Characterization results of the JUNGFRAU full scale readout ASIC. J. Instrum. 2016;11:C02047. [Google Scholar]
- 44.Henrich B, et al. The adaptive gain integrating pixel detector AGIPD a detector for the European XFEL. Nucl. Instrum. Meth. Phys. Res. A. 2011;633:S11–S14. [Google Scholar]
- 45.DePonte DP, et al. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D. 2008;41:195505. [Google Scholar]
- 46.Oberthuer D, et al. Double-flow focused liquid injector for efficient serial femtosecond crystallography. Sci. Rep. 2017;7:44628. doi: 10.1038/srep44628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caffrey M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr. F. 2015;71:3–18. doi: 10.1107/S2053230X14026843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fromme R, et al. Serial femtosecond crystallography of soluble proteins in lipidic cubic phase. IUCrJ. 2015;2:545–51. doi: 10.1107/S2052252515013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu W, et al. Femtosecond crystallography of membrane proteins in the lipidic cubic phase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369(1647):20130314. doi: 10.1098/rstb.2013.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conrad CE, et al. A novel inert crystal delivery medium for serial femtosecond crystallography. IUCrJ. 2015;2:421–30. doi: 10.1107/S2052252515009811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugahara M, et al. Grease matrix as a versatile carrier of proteins for serial crystallography. Nat. Methods. 2015;12:61–3. doi: 10.1038/nmeth.3172. [DOI] [PubMed] [Google Scholar]
- 52.Botha S, et al. Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams. Acta Crystallogr. D Biol. Crystallogr. 2015;71:387– 97. doi: 10.1107/S1399004714026327. [DOI] [PubMed] [Google Scholar]
- 53.Sugahara M, et al. Oil-free hyaluronic acid matrix for serial femtosecond crystallography. Sci. Rep. 2016;6:24484. doi: 10.1038/srep24484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Masuda T, et al. Atomic resolution structure of serine protease proteinase K at ambient temperature. Sci. Rep. 2017;7:45604. doi: 10.1038/srep45604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin-Garcia JM, et al. Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation. IUCrJ. 2017;4 doi: 10.1107/S205225251700570X. Published online May 24, 2017. http://doi.org/10.1107/S205225251700570X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kovacsova G, et al. Viscous hydrophilic injection matrices for serial crystallography. IUCrJ. 2017;4 doi: 10.1107/S2052252517005140. Published online May 5, 2017. http://doi.org/10.1107/S2052252517005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sierra RG, et al. Nanoflow electrospinning serial femtosecond crystallography. Acta Crystallogr. D Biol. Crystallogr. 2012;68:1584–1587. doi: 10.1107/S0907444912038152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sierra RG, et al. Concentric-flow electrokinetic injector enables serial crystallography of ribosome and photosystem II. Nat Methods. 2016;13:59–62. doi: 10.1038/nmeth.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mafune F, et al. Microcrystal delivery by pulsed liquid droplet for serial femtosecond crystallography. Acta Crystallogr. D Struct. Biol. 2016;72:520–3. doi: 10.1107/S2059798316001480. [DOI] [PubMed] [Google Scholar]
- 60.Roessler CG, et al. Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography. Structure. 2016;24:631–40. doi: 10.1016/j.str.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fuller FD, et al. Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers. Nat. Methods. 2017;14:443–449. doi: 10.1038/nmeth.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang D, et al. Double-focusing mixing jet for XFEL study of chemical kinetics. J. Synchrotron Radiat. 2014;21:1364–6. doi: 10.1107/S160057751401858X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Calvey GD, et al. Mixing injector enables time-resolved crystallography with high hit rate at X-ray free electron lasers. Struct. Dyn. 2016;3:054301. doi: 10.1063/1.4961971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kupitz C, et al. Structural enzymology using X-ray free electron lasers. Struct. Dyn. 2017;4:044003. doi: 10.1063/1.4972069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunter MS, et al. Fixed-target protein serial microcrystallography with an x-ray free electron laser. Sci. Rep. 2014;4:6026. doi: 10.1038/srep06026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mueller C, et al. Fixed target matrix for femtosecond time-resolved and in situ serial micro-crystallography. Struct. Dyn. 2015;2:054302. doi: 10.1063/1.4928706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baxter EL, et al. High-density grids for efficient data collection from multiple crystals. Acta Crystallogr. D Struct. Biol. 2016;72:2–11. doi: 10.1107/S2059798315020847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cohen AE, et al. Goniometer-based femtosecond crystallography with X-ray free electron lasers. Proc. Natl. Acad. Sci. U. S. A. 2014;111:17122–7. doi: 10.1073/pnas.1418733111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hirata K, et al. Determination of damage-free crystal structure of an X-ray-sensitive protein using an XFEL. Nat. Methods. 2014;11:734–6. doi: 10.1038/nmeth.2962. [DOI] [PubMed] [Google Scholar]
- 70.Kupitz C, et al. Microcrystallization techniques for serial femtosecond crystallography using photosystem II from Thermosynechococcus elongatus as a model system. Philos. Trans. R. Soc. B Biol. Sci. 2014;369:20130316. doi: 10.1098/rstb.2013.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coe J, et al. Crystallization of Photosystem II for Time-Resolved Structural Studies Using an X-ray Free Electron Laser. Methods Enzymol. 2015;557:459–82. doi: 10.1016/bs.mie.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mahon BP, et al. Microbatch Mixing: “Shaken not Stirred”, a Method for Macromolecular Microcrystal Production for Serial Crystallography. Cryst. Growth Des. 2016;16:6214–6221. [Google Scholar]
- 73.Ibrahim M, et al. Improvements in serial femtosecond crystallography of photosystem II by optimizing crystal uniformity using microseeding procedures. Struct. Dyn. 2015;2:041705. doi: 10.1063/1.4919741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edlund P, et al. The room temperature crystal structure of a bacterial phytochrome determined by serial femtosecond crystallography. Sci. Rep. 2016;6:35279. doi: 10.1038/srep35279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lomb L, et al. An anti-settling sample delivery instrument for serial femtosecond crystallography. J. Appl. Crystallogr. 2012;45:674–678. [Google Scholar]
- 76.Barnes CO, et al. Assessment of microcrystal quality by transmission electron microscopy for efficient serial femtosecond crystallography. Arch. Biochem. Biophys. 2016;602:61–68. doi: 10.1016/j.abb.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Darmanin C, et al. Protein crystal screening and characterization for serial femtosecond nanocrystallography. Sci. Rep. 2016;6:25345. doi: 10.1038/srep25345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Overington JP, et al. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 79.Santos R, et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cherezov V. Lipidic cubic phase technologies for membrane protein structural studies. Curr. Opin. Struct. Biol. 2011;21:559–566. doi: 10.1016/j.sbi.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu W, et al. Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nat. Protoc. 2014;9:2123–34. doi: 10.1038/nprot.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ishchenko A, et al. Preparation and Delivery of Microcrystals in Lipidic Cubic Phase for Serial Femtosecond Crystallography. J. Vis. Exp. 2016;115:e54463. doi: 10.3791/54463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jakobi AJ, et al. In cellulo serial crystallography of alcohol oxidase crystals inside yeast cells. IUCrJ. 2016;3:88–95. doi: 10.1107/S2052252515022927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sawaya MR, et al. Protein crystal structure obtained at 2.9 A resolution from injecting bacterial cells into an X-ray free-electron laser beam. Proc. Natl. Acad. Sci. U. S. A. 2014;111:12769–74. doi: 10.1073/pnas.1413456111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barty A, K R, Maia FR, Hantke M, Yoon CH, White TA, Chapman H. Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 2014;47:1118–1131. doi: 10.1107/S1600576714007626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hattne J, et al. Accurate macromolecular structures using minimal measurements from X-ray free-electron lasers. Nat. Methods. 2014;11(5):545–8. doi: 10.1038/nmeth.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.White TA, et al. Recent developments in CrystFEL. J. Appl. Crystallogr. 2016;49:680–689. doi: 10.1107/S1600576716004751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kabsch W. Processing of X-ray snapshots from crystals in random orientations. Acta Crystallogr. D Biol. Crystallogr. 2014;70:2204–16. doi: 10.1107/S1399004714013534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Duisenberg A. Indexing in single-crystal diffractometry with an obstinate list of reflections. J. Appl. Crystallogr. 1992;25:92–96. [Google Scholar]
- 90.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Battye TGG, et al. IMosflm: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–81. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kirian RA, et al. Femtosecond protein nanocrystallography-data analysis methods. Opt. Express. 2010;18:5713–23. doi: 10.1364/OE.18.005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yefanov O, et al. Accurate determination of segmented X-ray detector geometry. Opt. Express. 2015;23:28459–70. doi: 10.1364/OE.23.028459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sauter N. XFEL diffraction: developing processing methods to optimize data quality. J. Synchrotron Radiat. 2015;22:239–248. doi: 10.1107/S1600577514028203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Uervirojnangkoorn M, et al. Enabling X-ray free electron laser crystallography for challenging biological systems from a limited number of crystals. Elife. 2015;4:e05421. doi: 10.7554/eLife.05421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ginn HM, et al. A revised partiality model and post-refinement algorithm for X-ray free-electron laser data. Acta Crystallogr. D Biol. Crystallogr. 2015;71:1400–10. doi: 10.1107/S1399004715006902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sauter NK, et al. Improved crystal orientation and physical properties from single-shot XFEL stills. Acta Crystallogr. D Biol. Crystallogr. 2014;70:3299–3309. doi: 10.1107/S1399004714024134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.White TA. Post-refinement method for snapshot serial crystallography. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369:20130330. doi: 10.1098/rstb.2013.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brehm W, Diederichs K. Breaking the indexing ambiguity in serial crystallography. Acta Crystallogr. D Biol. Crystallogr. 2014;70:101–9. doi: 10.1107/S1399004713025431. [DOI] [PubMed] [Google Scholar]
- 100.Colletier JP, et al. De novo phasing with X-ray laser reveals mosquito larvicide BinAB structure. Nature. 2016;539:43–47. doi: 10.1038/nature19825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gati C, et al. Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2247–2252. doi: 10.1073/pnas.1609243114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li D, et al. Ternary structure reveals mechanism of a membrane diacylglycerol kinase. Nat. Commun. 2015;6:10140. doi: 10.1038/ncomms10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nogly P, et al. Lipidic cubic phase injector is a viable crystal delivery system for time-resolved serial crystallography. Nat. Commun. 2016;7:12314. doi: 10.1038/ncomms12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Luttrell LM, Lefkowitz RJ. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J. Cell. Sci. 2002;115:455–65. doi: 10.1242/jcs.115.3.455. [DOI] [PubMed] [Google Scholar]
- 105.Johansson LC, et al. Structure of a photosynthetic reaction centre determined by serial femtosecond crystallography. Nat. Commun. 2013;4:2911. doi: 10.1038/ncomms3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kern J, et al. Room temperature femtosecond X-ray diffraction of photosystem II microcrystals. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9721–6. doi: 10.1073/pnas.1204598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pande K, et al. Femtosecond structural dynamics drives the trans/cis isomerization in photoactive yellow protein. Science. 2016;352:725–9. doi: 10.1126/science.aad5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schmidt M. Mix and Inject: Reaction Initiation by Diffusion for Time-Resolved Macromolecular Crystallography. Adv. Condens. Matter Phys. 2013;2013:167276. [Google Scholar]
- 109.Spence J, Lattman E. Imaging enzyme kinetics at atomic resolution. IUCrJ. 2016;3:228–229. doi: 10.1107/S2052252516010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bai X-C, et al. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]