Abstract
Objective
The objective of this review is to explain the science of metabolomics—a science of systems biology that measures and studies endogenous small molecules (metabolites) that are present in a single biological sample—and its application to the diagnosis and treatment of sepsis. In addition, we discuss how discovery through metabolomics can contribute to the development of precision medicine targets for this complex disease state and the potential avenues for those new discoveries to be applied in the clinical environment.
Methods
A nonsystematic literature review was performed focusing on metabolomics, pharmacometabolomics, and sepsis. Human (adult and pediatric) and animal studies were included.
Main Results
Metabolomics has been investigated in the diagnosis, prognosis, and risk stratification of sepsis, as well as for the identification of drug target opportunities. Metabolomics elucidates a new level of detail, when compared with other systems biology sciences, with regard to the metabolites that are most relevant in the pathophysiology of sepsis, as well as highlighting specific biochemical pathways at work in sepsis. Metabolomics also highlights biochemical differences between sepsis survivors and nonsurvivors at a level of detail greater than that demonstrated by genomics, transcriptomics, or proteomics, potentially leading to actionable targets for new therapies. The application of pharmacometabolomics and its integration with other systems pharmacology to sepsis therapeutics could be particularly helpful in differentiating drug responders and nonresponders and furthering knowledge of mechanisms of drug action and response.
Conclusion
The accumulated literature on metabolomics suggest that it is a viable tool for continued discovery around the pathophysiology, diagnosis and prognosis, and treatment of sepsis in both adults and children, and provides a greater level of biochemical detail and insight than other systems biology approaches. However, the clinical application of metabolomics in sepsis has not yet been fully realized. For this to be achieved, prospective validation studies are needed to translate metabolites from the discovery phase into the clinical utility phase.
Keywords: pediatrics, systems biology, systems pharmacology, pharmacometabolomics, pharmacotherapy, critical care
Introduction
Sepsis is one of the leading causes of mortality in the world.1 Health care costs are significant, as septic patients often require care in the intensive care unit (ICU). Sepsis makes a significant contribution to the overall expense of intensive care medicine, which collectively represents close to 1% of the United States' gross domestic product.2 Furthermore, the incidence of sepsis is on the rise, which is consistent with an aging population and with an increasing number of immunosuppressed individuals. The most vulnerable populations for sepsis include the elderly and neonates, with a wide variation in incidence between these groups. Sepsis is diagnosed in 0.56 per 1000 children per year in the United States, with a 10.3% mortality rate3 but accounts for 40% of deaths in children younger than 5 years old worldwide.4 In adults, the incidence of sepsis is 18.6 per 1000 hospitalizations, with a mortality rate of up to 50% in fulminant septic shock.5
Etiology and Definition of Sepsis
The etiology of sepsis varies by age and can include bacterial, viral, and fungal pathogens. In children, the most common pathogens are Staphylococcus species, followed by fungal infections, the latter of which are more common in children with cancer.6 Viral pathogens are common in younger children but are frequently accompanied by bacterial co-infection.3 In adults, about half of the infections are caused by gram-positive bacteria, about 40% from gram-negative bacteria, and less than 6% from anaerobes and fungi.7
Although the pathogens that cause sepsis have not significantly changed, the definition of sepsis was recently updated after more than 20 years. This was prompted by a call to reassess the definition due to an emergence of new knowledge in the field.8-11 However, these revised Third International Consensus (Sepsis-3) definitions only apply to adults (Table 1),11 as in children, hypotension presents much later and may indicate nonreversible cardiac failure.12 In addition, the classification of severe sepsis was dropped in the most recent adult definitions but remains in the pediatric definition.
Table 1. Consensus Definitions of Sepsis.
| Sepsis-Related Term | Adult11 | Pediatric15 |
|---|---|---|
| Systemic inflammatory response syndrome (SIRS) | Two or more of the following:
|
Two or more of the following: |
| Sepsis | Life-threatening organ dysfunction due to a dysregulated host response to infection | SIRS that results from or occurs in the presence of suspected or proven infection |
| Severe Sepsis | Definition no longer used | Sepsis plus one of the following:
|
| Septic Shock | A subset of sepsis that includes persistent hypotension that requires vasopressors to maintain a MAP ≥ 65 mm Hg and a serum lactate level of > 2 mmol/L despite adequate fluid resuscitation | Sepsis and, despite administration of isotonic intravenous fluid bolus >40 mL/kg in 1 hour, one of the following:
Or two of the following:
|
PaC02 = partial pressure of arterial carbon dioxide; MAP = mean arterial pressure; SBP = systolic blood pressure.
Age-specific vital signs and laboratory variables are provided in reference 15.
Organ dysfunction criteria are provided in reference 15.
The diagnosis, risk stratification, and treatment of sepsis in both children and adults is challenging due to its inherent heterogeneity and the absence of a gold standard for diagnosis. This has traditionally led to poor clinical outcomes and has contributed to a plethora of failed pharmacotherapy clinical trials.13, 14 Diagnostic and prognostic tools are sparse and are largely based on clinical signs and symptoms. These parameters can vary in children, depending on age.15 For example, tachycardia in an 8-month-old infant is defined as > 180 beats per minute whereas it is > 140 beats per minute in a 3-year-old. In addition, although the Sequential Organ Failure Assessment (SOFA) score is predictive of outcome in adult patients treated in the intensive care unit, it is not sufficient for pediatric patients with sepsis. The Pediatric Logistic Organ Dysfunction Score is the only pediatric scoring system that has been validated in clinical trials.16 Collectively, use of scoring systems are cumbersome and typically are not routinely employed as part of patient care. As such, the Sepsis-3 task force recommended that a SOFA score ≥ 2 be used as a criterion for sepsis because it can be calculated from clinical data that are more readily available.9 Nevertheless, these clinical tools have a low level of accuracy, which creates a particular challenge for achieving precision medicine in sepsis and has hindered the identification of pharmacotherapies targeted at specific subpopulations with this disease. For instance, patients with sepsis can have immune responses ranging from an overactive inflammatory cascade to a highly immunosuppressed phenotype.17 These phenotypes may not initially be easily differentiated at the bedside or correlate with observable clinical parameters. These points are illustrated in the accompanying case report of pediatric sepsis.18
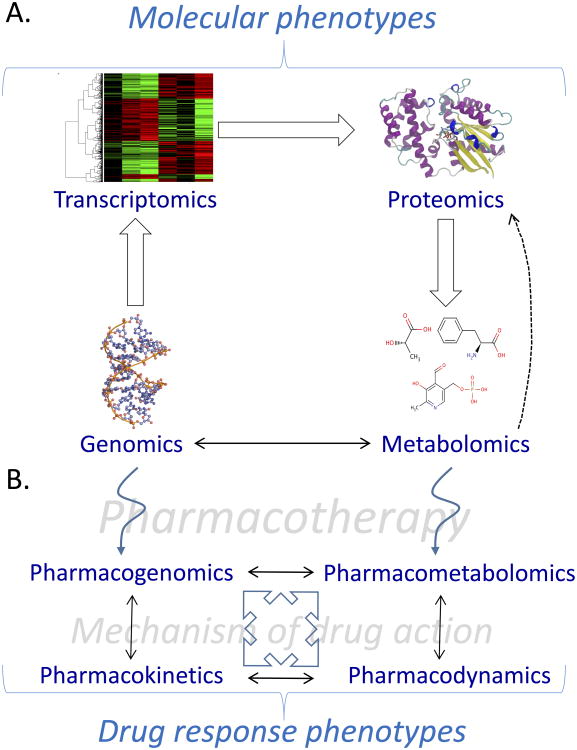
Accurate and early identification of sepsis could influence clinical decision making and direct more precise therapeutic intervention. Therefore, there is an enormous need for new approaches to more accurately phenotype sepsis. The systems biology and pharmacology sciences (Figures 1A and 1B), including metabolomics and pharmacometabolomics, have great potential to aid in defining specific sepsis phenotypes and find much needed predictive and prognostic biomarkers that can lead to more personalized management and therapeutics.19 In this article, we present an overview of the current knowledge of sepsis and the role that metabolomics and pharmacometabolomics could play in advancing a precision medicine initiative for sepsis. In addition, we depict the utility of metabolomics for the timely identification of pediatric sepsis in the accompanying case report.18
Figure 1.
Overview of the interactions between systems biology and pharmacology sciences. (A) Systems biology consists of genomics, transcriptomics, proteomics, and metabolomics. Although the transition of these sciences is often viewed as linear, it is likely that there are bidirectional interactions among them. For example, metabolites serve as signaling molecules for gene and protein regulation. (B) Systems pharmacology includes pharmacogenomics, pharmacometabolomics, pharmacokinetics, and pharmacodynamics. These sciences interact in such a way that they can inform each other so that more detail about mechanisms of drug action and drug response phenotypes can be learned.
Overview of Metabolomics
Metabolomics is a science of systems biology (Figure 1A) that measures and studies endogenous small molecules that are present in a single biological sample.20, 21 The metabolome refers to the complete set of metabolites in any given biofluid, cell, tissue, or organism.22 These small molecules, or metabolites, are typically less than 1500 daltons in size and are produced from metabolic processes (e.g., the tricarboxylic acid cycle) and complex biological interactions within an organism as well as interactions between the host and microbes. Unlike genomics, in which a genetic mutation may have little or no impact on the function of a protein or proteomics, which may not identify a functional change in a protein, clinical metabolomics detects the direct result of a biochemical response to internal and external factors. In addition, virtually any type of biological sample can be assayed, including blood, urine, and tissue.
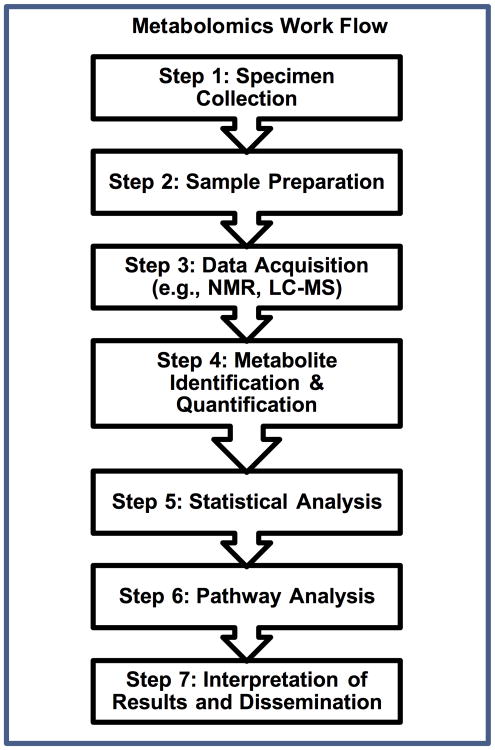
There are multiple steps that should be followed to conduct a metabolomics study (Figure 2). The generation of reliable metabolomics starts with sample collection (step 1). Use of a standard operating procedure is essential and is particularly important for multicenter studies, as is expeditious and consistent sample handling and storage (e.g., −80°C).23 For specific analytical platforms and, most often, with the exception of urine, macromolecules need to be removed from the sample (Figure 2, step 2) before assay. There are a number of options for this including sample ultrafiltration or methanol precipitation. The choice depends on the type of sample (e.g., tissue samples require more processing) and the objective of the metabolomics study. For example, if serum samples are filtered, the lipid metabolites will be retained and forfeited in the filter, whereas use of a methanol-chloroform extraction will yield both an aqueous and nonaqueous portion of the sample; this permits dual-platform assays for aqueous and lipid metabolites from a single sample.24
Figure 2.
Representative steps in a typical metabolomics work flow. If prospective sampling is planned, samples should be collected (step 1) using a standard operating procedure to ensure consistent procedures and sample processing. Sample preparation (step 2) will vary depending on the sample type. The most common analytical platforms for metabolomics data acquisition include nuclear magnetic resonance (NMR) and liquid (or gas) chromatography followed by mass spectrometry (LC-MS). A number of different publically and commercially available platforms exist for spectral processing and metabolite identification and quantification (step 4). These include Chenomx (chenomx.com), XCMS (https://xcmsonline.scripps.edu/) and MS-Dial (http://prime.psc.riken.jp/Metabolomics_Software/MS-DIAL/index.html). Statistical analysis (step 5) can be performed using quantified or nonquantified data using a number of different approaches (see text). Pathway analysis or the mapping of metabolites to metabolic networks can be achieved using a number of different tools including Metscape (http://metscape.ncibi.org/) or MetaboAnalyst (http://www.metaboanalyst.ca/).
Metabolites can be measured by a number of different analytical platforms, but the two most common are one-dimensional (1D) proton (1H) nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) (Figure 2, step 3). The advantages of NMR include that it is routinely quantitative and is nondestructive to the sample. It is especially useful for the detection of medium to high abundant metabolites, polar compounds, and metabolites with a molecular weight less than ∼100 daltons, which liquid chromatography (LC)-MS may miss. However, even though nearly every compound has a distinct NMR spectrum, it can be challenging to identify and quantify metabolites that have overlapping spectral peaks that can occur in complex mixtures.25 Use of commercial spectral processing and analysis software such as Chenomx (www.chenomx.com; NMR Suite 8.0; Chenomx Inc., Edmonton, Alberta, Canada) can be used to optimize metabolite identification and quantification from raw NMR spectra.
MS platforms such as gas chromatography (GC)-MS and LC-MS are more sensitive than NMR and are therefore better suited for the measurement of low-abundant metabolites (< 5 μM) and for the detection of lipid and volatile compounds. Presently, LC-MS is commonly used for what is referred to as “untargeted” metabolomics because it effectively detects a broad range of different types of metabolites. Analytical and spectral processing technology is rapidly evolving such that aqueous and lipid compounds can be simultaneously detected, but LC-MS is highly variable, which makes cross-center studies challenging. In addition, there is no universal LC-MS metabolite library that can be applied to different instruments. This makes careful metabolite identification and quantification particularly important. For more detail, the reader is referred to recent reviews of analytical metabolomics.26, 27 1H-NMR spectral acquisition is relatively straightforward, but there are specifications that need to be followed (e.g., the pulse sequence).28 This is critical for quantitative NMR metabolomics for which a reliable internal standard peak is necessary. If done correctly and the same across clinical centers, the data are highly reproducible. Spectral analysis for LC-MS is more automated than it is for NMR. Most manufacturers of LC-MS instrumentation have special software that finds and names peaks. Confirmation of metabolite identification and quantification is then done using known standards (Figure 2, step 4).
Following the generation of a metabolomics data set, the statistical analysis is the next step (Figure 2, step 5). This can be quite complicated and typically begins with unsupervised and supervised learning methods29 such as principal component analysis (PCA) and partial least squares–discriminant analysis (PLS-DA), respectively. Unsupervised methods aid in describing the data based on data-driven group classification and trends that exist within the data. In addition to PCA, other common unsupervised methods include K-means clustering and hierarchical clustering. Supervised learning methods are used to determine the predictive power of the identified metabolites and the outcome (e.g., sepsis vs no sepsis, or severe sepsis vs sepsis). These methods include PLS-DA and support vector machine and are often employed for discovering biomarker candidates.29 These analyses are often performed using qualitative metabolomics data and are referred to as chemometric analyses. However, nonparametric and parametric statistics can be employed for metabolomics data analysis, particularly for the analysis of quantified data to identify differentiating metabolites. This is preceded by data transformation (e.g., log normalization and range scaling) to achieve a normal distribution. Following the generation of p values, a scheme for the correction for multiple comparisons, such as the calculation of false discovery rate30, 31, should be employed. Metabolomics data analysis can be accomplished using a number of publically available platforms including MetaboAnalyst (http://www.metaboanalyst.ca/).32 Importantly, a single study is not sufficient to conduct a robust validation analysis, so it is important that findings are validated using samples generated from separate, independent studies.33 This level of rigor is critical to test reproducibility and the testing of predictive models that is needed to build biomarker credentials. Following statistical analysis, pathway mapping using tools like Metscape (http://metscape.ncibi.org/), which is a plugin for Cytoscape (http://cytoscape.org/), can be employed to visualize metabolomics data and their associated metabolic pathways.34
Although the overarching principles of the above approach remain constant and are independent of the disease process being studied, there are specific considerations when studying the metabolomics of sepsis. The location of the patient within the hospital and at the time of sample collection are important factors to consider. Unlike some other disease states that have been studied using metabolomics (diabetes mellitus, cardiovascular disease), time of sample collection is key in the study of sepsis because the rapid progression of the illness can impact key metabolic pathways. While point-in-time estimates certainly offer information, sequential sample collection from the same subject is an ideal way to study these changes. In addition, a newly identified patient with sepsis who has just arrived in the emergency department may have a markedly different metabolomic profile compared with a patient receiving multiple interventions for several days in the ICU.
Pharmacometabolomics is the application of metabolomics to the prediction of drug response (Figure 1B).35 The previously discussed principles of metabolomics with regard to study design, work flow, and assays all apply to pharmacometabolomics. Although the strict definition of pharmacometabolomics limits it to prediction, it is important to note that the extent of found associations between pretreatment levels of metabolites and the divergence of drug-induced metabolic changes and phenotypes needs to be prospectively tested in order to accurately and reliably forecast drug response. In addition to the prediction of drug response, pharmacometabolomics could be a particularly powerful tool for furthering knowledge of the mechanistic underpinnings of diverse drug responses, mechanisms of drug action, and adverse drug reactions either alone or integrated with other systems pharmacology sciences.36
Metabolic Diagnosis of Sepsis
The inflammatory response, evidenced by vital sign aberrations, can result from sterile inflammation (surgery, trauma) or as sequelae of infection (sepsis) (Table 1). Therefore, identifying patients who have underlying infection and potential for progression through the sepsis continuum can be clinically difficult for both adults and children. The absence of a validated diagnostic test and the low incidence of detectable bacteria in the blood make sepsis diagnosis particularly challenging. Metabolomics has the potential to provide information to guide this key clinical decision. Metabolite profiles have consistently demonstrated the ability to discriminate sterile inflammation from sepsis in both human and animal studies.37-39 Among adults admitted to an ICU following traumatic injury, metabolite profiles on admission successfully identified those who proceeded to develop sepsis from those who did not.40 Similar differences are seen in children, where metabolite profiles clearly differentiate sepsis from sterile inflammation and survivors from nonsurvivors.41 Among neonates, metabolite profiles discriminate healthy controls from those with sepsis and display distinct patterns between early- and late-onset sepsis.42 In early sepsis in both adults and children, metabolites involved in energy metabolism show consistent directional changes. Ketone bodies are increased, and levels of citrate, pentose phosphate pathway compounds, ribitol, and ribonic acid are decreased.41, 42 Furthermore, several studies have identified glucose, lactate, acetate, and citrate as metabolites that differentiate sepsis and systemic inflammatory response syndrome (SIRS) (Tables 1 and 2).39, 41-44 These findings are presented in a case of a pediatric sepsis where 1H-NMR urine metabolomics detected a large shift in energy metabolites, including increased levels of ketone bodies, which theoretically could have aided in the early diagnosis of sepsis in this patient.18
Table 2. Metabolic Pathways and Associated Metabolites That Are Altered in Sepsis.
| Metabolic pathway | Sepsis response | Representative metabolites and direction of change |
|---|---|---|
| Amino acid metabolism | Increased catabolism of body tissues for energy production | ↓ kynurenine, a by-product of tryptophan metabolism found in sepsis survivors, suggests efficient transition to noncatabolic pathways37 ↓ decreased amino acids correlate with bacteremic sepsis40, 53, 62 ↑ amino acids in response to effective treatment57 |
| Mitochondrial energy metabolism | Increased to meet energy requirement/demand | ↑ 3-hydroxybutyrate and acetoacetate (ketone bodies) increase in nonsurvivors suggests compensatory response to decreased ATP51 ↑ lysophosphatidylcholines37 ↑ acylcarnitines (transport long-chain fatty acids across mitrochondrial membrane) in nonsurvivors suggests defect in free fatty acid metabolism42, 44, 48 ↑ linoleic acid in response to effective treatment57 |
| Tricarboxylic acid cycle | Utilized substrates for aerobic catabolism | ↑ amounts of substrate (citrate, malate, pyruvate, acetate, lactate) in nonsurvivors due to inability to metabolize42, 48 |
| Pentose phosphate pathway | Utilized as alternate pathway for glucose metabolism | ↓ THBA, ribitol, and ribonic acid suggest compensatory response to decreased ATP42 |
ATP = adenosine triphosphate; THBA = 2,3,4-trihydroxybutyric acid.
Metabolomics for Prognosis and Monitoring Sepsis
Ultimately, the short-term outcomes of interest for clinicians are mortality and end-organ damage. These endpoints can occur regardless of the specific pathogen and are likely due to a complex interplay between pathogen, patient, and environment. It is beneficial for clinicians to be able to identify at-risk patients early. Patients with evolving sepsis display metabolite profiles that are consistent regardless of pathogen,37, 39 and distinct metabolite profiles identify specific end-organ damage resulting from sepsis.39, 45 More recently, the long-term detrimental consequences of sepsis have become apparent in survivors.46 Specifically, patients display impaired cognition and functionality.47 Certainly, prognostic and predictive biomarkers of long-term outcome of sepsis survival would be of great value, particularly if targets for drug therapy could be identified. These findings may lie, in part, in the new insights into sepsis pathophysiology that metabolomics offers, which may in turn provide new areas for targeted therapy.
Metabolomics has identified differences in energy metabolism between healthy subjects and patients with sepsis, and between survivors and nonsurvivors of sepsis. Metabolites differentiating these groups are primarily amino acids and their derivatives, and demonstrate the role of energy metabolism in the pathophysiology of sepsis (Table 2). Specific metabolites are identified when comparing adults with both induced endotoxemia and community-acquired sepsis with healthy controls; the direction of change in metabolites is similar as well.41 When survivors and nonsurvivors were compared, alterations were seen in free fatty acid metabolism, and there was a suggestion of a profound defect in mitochondrial fatty acid beta-oxidation in nonsurvivors; differences in glycolysis, gluconeogenesis, and citric acid cycle were also noted.48-50 In addition, kynurenine, a by-product of tryptophan metabolism, which also differentiates SIRS (Table 1) from sepsis, is elevated in nonsurvivors compared to survivors (Table 2).37, 48 The principle of a sepsis-induced energy disruption is evidenced in the accompanying case.18 We have also demonstrated this in adult septic shock patients in which L-carnitine supplementation appears to be less effective in patients with elevated pretreatment levels of ketone bodies, implying that patients who present with evidence of a disruption in energy metabolism are less likely to survive sepsis.51
Ultimately, the strength and “added value” of metabolomics in sepsis for both children and adults may be when it is combined with clinical data and other measurements to improve prediction of outcomes and demonstrate enhanced performance when compared with currently available diagnostic tests including procalcitonin, C-reactive protein, and lactate levels.43, 52, 53 In an example of this strategy, profiling children early after presentation in the emergency department using metabolomics and inflammatory protein mediators differentiated children who did and did not require ICU care. This application of metabolomics may aid in triage decisions and risk stratification in sepsis, particularly in clinical environments without pediatric expertise.54
Precision Medicine and Drug Targets for Sepsis
One of the more intriguing aspects of metabolomics is the potential to provide early, clinically relevant information on an individual's eligibility and response to treatment.14, 35, 36, 55 With the current focus on resistant pathogens and antibiotic stewardship, pharmacometabolomics could permit customization and targeting of therapy early, thereby decreasing the prevalence of multidrug-resistant organisms. In the case of most infections, results of diagnostic tests for pathogen detection are not available for days to weeks after treatment is initiated. Importantly, specific information to guide antimicrobial therapy choices is not available until the organism has been cultured and sensitivities obtained. Often, particularly in children, the specific pathogen is not identified or cultures are negative. In the case of fungal infections, definitive results may not be available for weeks. In a study of neonates with fungal infections, the amino acid, serine, was elevated compared to healthy controls, and levels gradually declined in response to antifungal therapy, providing treatment-specific feedback prior to the time when culture results would be finalized.56 In animal studies, expressed metabolites differed in mice receiving effective versus ineffective antibiotic treatment against the bacterium Staphylococcus aureus within 2 hours after initiation of therapy.57 The same authors were able to demonstrate similar changes in response to therapy in vivo, in both S. aureus and Escherichia coli infections.57
As new therapies are developed for sepsis, metabolomics may be used as a tool to elucidate the impacted biochemical pathways, monitor for adverse events, and predict and track treatment responsiveness (pharmacometabolomics)35, 36 as well as facilitate a precision medicine approach for sepsis.58 Furthermore, integration of pharmacometabolomics with other systems pharmacology sciences (Figure 1B), such as pharmacokinetics and pharmacogenomics, could lead to further refining of drug response phenotypes.36 As identification of viable drug targets is critical for drug discovery, metabolomics could be particularly useful in this regard. For example, in an animal study, erythropoietin was found to reduce end-organ damage in sepsis, and distinct metabolic profiles were found between the treated and untreated groups.59 In addition, based on the metabolites identified, the specific pathways affected by erythropoietin were identified. This insight may lead to further investigation of the experimental therapy as well provide new hypothesis-generating data for the development of other therapies. In a phase I trial of supplemental L-carnitine for the treatment of adults with septic shock, pretreatment metabolite profiles differed between those patients who did and did not respond to the therapy.51 In addition, the metabolites that differentiated L-carnitine responders and nonresponders highlighted affected biochemical pathways that could aid in identifying drug target opportunities for L-carnitine–nonresponsive patients. Importantly, there were no evident clinical differences between responders and nonresponders prior to treatment. This finding provides a direction for further study in that the metabolomics profile may assist in the early identification of patients most likely to respond to specific therapies who are not readily clinically differentiated using conventional means. Identifying the metabolic phenotypes of responders versus nonresponders to specific therapies could again provide insight into the differences in sepsis physiology and guide more focused precision therapies, thereby providing an approach to overcome what is considered to be one of the major impediments responsible for the repeatedly negative clinical trials, namely a largely heterogeneous patient population. For example, many metabolomics studies have identified mitochondrial beta-oxidation as a key pathway upon which interventions may be targeted (Table 2).42, 43, 60 Knowledge of these metabolic differences could also be highly valuable for the design of clinical trials in which patients would be enrolled and stratified based on their pretreatment metabolic profiles. Use of metabolomics as an inclusion criterion could result in reduced study patient heterogeneity and improve the likelihood of clinical trial success.55
Future Directions
Metabolomics has shown significant potential as a diagnostic tool to differentiate patients with sepsis and sterile inflammation (e.g., SIRS) and for predicting mortality. Metabolomics may also be useful in predicting illness severity in sepsis in both adults and children, with the latter exemplified in our case report,18 differentiating pathogens to guide appropriate antimicrobial therapy, and identifying the optimal timing for assessing response to therapy. In contrast to the other systems biology approaches, metabolomics comes closest to accounting for the direct interplay between individual, environment, and pathogen. It is precisely these characteristics that lend to its utility in the evaluation of sepsis given its heterogeneity as a disease. As the field evolves, pharmacometabolomics—the application of metabolomics to drug response prediction and phenotyping—will likely emerge as an increasingly important member of systems pharmacology (Figure 2). Integration of these sciences could be particularly informative of drug response phenotypes and prediction of adverse drug reactions.
One challenge in the clinical application of metabolomics to both adult and pediatric patients is having timely and accessible results. Presently, the metabolomics work flow, which is lengthy, is not conducive to generating real-time data to be used for clinical decision making. Importantly, as key differentiating metabolites of drug response, diagnosis, and prognosis are identified, confirmation of metabolite identification and quantification will be essential. In addition, the findings of metabolomics studies will require validation in prospective studies in order to achieve robust biomarker credentialing.33 They will also be needed for the development of accurate point-of-care tests that are optimal for the care of critically ill patients. New technologies are being developed, including programmable nano-bio-chips 61 and metabolomics-on-a-chip technology. As metabolomics is applied in future investigations of therapeutic interventions, metabolic phenotyping may be a useful tool in clinical trials for benchmarking or risk stratification of subjects.
Acknowledgments
Sources of support: This work was supported in part by a grant from the National Institute of General Medical Sciences (NIGMS; GM111400 to KAS; K23 GM113041 to MAP) and the National Heart Lung and Blood Institute (NHBLI; R01 HL123515 to TJS). The content is solely the responsibility of the authors and does not necessarily present the official views of the NIGMS, NHLBI, or the National Institutes of Health.
References
- 1.Fleischmann C, Scherag A, Adhikari NK, et al. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am J Respir Crit Care Med. 2016;3:259–72. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- 2.Vincent JL, Marshall JC, Namendys-Silva SA, et al. Assessment of the worldwide burden of critical illness: the intensive care over nations (ICON) audit. Lancet Respir Med. 2014;5:380–6. doi: 10.1016/S2213-2600(14)70061-X. [DOI] [PubMed] [Google Scholar]
- 3.Watson RS, Carcillo JA. Scope and epidemiology of pediatric sepsis. Pediatr Crit Care Med. 2005;3(1):S3–5. doi: 10.1097/01.PCC.0000161289.22464.C3. [DOI] [PubMed] [Google Scholar]
- 4.Bryce J, Boschi-Pinto C, Shibuya K, Black RE Group WCHER. WHO estimates of the causes of death in children. Lancet. 2005;9465:1147–52. doi: 10.1016/S0140-6736(05)71877-8. [DOI] [PubMed] [Google Scholar]
- 5.Kadri SS, Rhee C, Strich JR, et al. Estimating Ten-Year Trends in Septic Shock Incidence and Mortality in United States Academic Medical Centers Using Clinical Data. Chest. 2016 doi: 10.1016/j.chest.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartman ME, Linde-Zwirble WT, Angus DC, Watson RS. Trends in the epidemiology of pediatric severe sepsis*. Pediatr Crit Care Med. 2013;7:686–93. doi: 10.1097/PCC.0b013e3182917fad. [DOI] [PubMed] [Google Scholar]
- 7.Martin GS, Mannino DM, Moss M. The effect of age on the development and outcome of adult sepsis. Crit Care Med. 2006;1:15–21. doi: 10.1097/01.ccm.0000194535.82812.ba. [DOI] [PubMed] [Google Scholar]
- 8.Vincent JL, Opal SM, Marshall JC, Tracey KJ. Sepsis definitions: time for change. Lancet. 2013;9868:774–5. doi: 10.1016/S0140-6736(12)61815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;8:762–74. doi: 10.1001/jama.2016.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shankar-Hari M, Phillips GS, Levy ML, et al. Developing a New Definition and Assessing New Clinical Criteria for Septic Shock: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;8:775–87. doi: 10.1001/jama.2016.0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) JAMA. 2016;8:801–10. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Randolph AG, McCulloh RJ. Pediatric sepsis: important considerations for diagnosing and managing severe infections in infants, children, and adolescents. Virulence. 2014;1:179–89. doi: 10.4161/viru.27045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen J, Vincent JL, Adhikari NK, et al. Sepsis: a roadmap for future research. The Lancet Infectious diseases. 2015;5:581–614. doi: 10.1016/S1473-3099(15)70112-X. [DOI] [PubMed] [Google Scholar]
- 14.Sims CR, Nguyen TC, Mayeux PR. Could Biomarkers Direct Therapy for the Septic Patient? The Journal of pharmacology and experimental therapeutics. 2016;2:228–39. doi: 10.1124/jpet.115.230797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein B, Giroir B, Randolph A International Consensus Conference on Pediatric S. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr Crit Care Med. 2005;1:2–8. doi: 10.1097/01.PCC.0000149131.72248.E6. [DOI] [PubMed] [Google Scholar]
- 16.Leteurtre S, Martinot A, Duhamel A, et al. Development of a pediatric multiple organ dysfunction score: use of two strategies. Med Decis Making. 1999;4:399–410. doi: 10.1177/0272989X9901900408. [DOI] [PubMed] [Google Scholar]
- 17.Calfee CS. Opening the Debate on the New Sepsis Definition. Precision Medicine: An Opportunity to Improve Outcomes of Patients with Sepsis. Am J Respir Crit Care Med. 2016;2:137–9. doi: 10.1164/rccm.201604-0697ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ambroggio L, Florin T, Shah S, et al. Urine metabolomics of sepsis and necrotizing methicillin-resistant staphylococcus aureus pneumonia: A case report of early recognition of severe illness in the emergency department. Pharmacotherapy submitted. doi: 10.1002/phar.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clish CB. Metabolomics: an emerging but powerful tool for precision medicine. Molecular Case Studies. 2015;1:a000588. doi: 10.1101/mcs.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serkova NJ, Standiford TJ, Stringer KA. The emerging field of quantitative blood metabolomics for biomarker discovery in critical illnesses. Am J Respir Crit Care Med. 2011;6:647–55. doi: 10.1164/rccm.201103-0474CI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carraro S, Giordano G, Reniero F, Perilongo G, Baraldi E. Metabolomics: a new frontier for research in pediatrics. J Pediatr. 2009;5:638–44. doi: 10.1016/j.jpeds.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 22.Wishart DS, Tzur D, Knox C, et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;(Database issue):D521–6. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emwas AH, Luchinat C, Turano P, et al. Standardizing the experimental conditions for using urine in NMR-based metabolomic studies with a particular focus on diagnostic studies: a review. Metabolomics : Official journal of the Metabolomic Society. 2015;4:872–94. doi: 10.1007/s11306-014-0746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serkova NJ, Glunde K. Metabolomics of cancer. In: Tainsky M, editor. Tumor Biomarker Discovery. New York, New York: Humana Press; 2009. pp. 273–96. [Google Scholar]
- 25.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;1:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sas KM, Karnovsky A, Michailidis G, Pennathur S. Metabolomics and diabetes: analytical and computational approaches. Diabetes. 2015;3:718–32. doi: 10.2337/db14-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stringer KA, McKay RT, Karnovsky A, Quemerais B, Lacy P. Metabolomics and Its Application to Acute Lung Diseases. Front Immunol. 2016;44 doi: 10.3389/fimmu.2016.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lacy P, McKay RT, Finkel M, et al. Signal intensities derived from different NMR probes and parameters contribute to variations in quantification of metabolites. PloS one. 2014;1:e85732. doi: 10.1371/journal.pone.0085732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren S, Hinzman AA, Kang EL, Szczesniak RD, Lu LJ. Computational and statistical analysis of metabolomics data. Metabolomics : Official journal of the Metabolomic Society. 2015;6:1492–513. [Google Scholar]
- 30.Storey JD. A direct approach to false discovery rates. J Roy Stat Soc B. 2002:479–98. [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;1:289–300. [Google Scholar]
- 32.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;W1:W251–7. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koulman A, Lane GA, Harrison SJ, Volmer DA. From differentiating metabolites to biomarkers. Anal Bioanal Chem. 2009;3:663–70. doi: 10.1007/s00216-009-2690-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karnovsky A, Weymouth T, Hull T, et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics. 2012;3:373–80. doi: 10.1093/bioinformatics/btr661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Everett JR. From Metabonomics to Pharmacometabonomics: The Role of Metabolic Profiling in Personalized Medicine. Frontiers in pharmacology. 2016;297 doi: 10.3389/fphar.2016.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaddurah-Daouk R, Weinshilboum RM. Pharmacometabolomics: implications for clinical pharmacology and systems pharmacology. Clinical pharmacology and therapeutics. 2014;2:154–67. doi: 10.1038/clpt.2013.217. [DOI] [PubMed] [Google Scholar]
- 37.Ferrario M, Cambiaghi A, Brunelli L, et al. Mortality prediction in patients with severe septic shock: a pilot study using a target metabolomics approach. Sci Rep. 2016;20391 doi: 10.1038/srep20391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin ZY, Xu PB, Yan SK, et al. A metabonomic approach to early prognostic evaluation of experimental sepsis by (1)H NMR and pattern recognition. NMR in biomedicine. 2009;6:601–8. doi: 10.1002/nbm.1373. [DOI] [PubMed] [Google Scholar]
- 39.Langley RJ, Tipper JL, Bruse S, et al. Integrative “omic” analysis of experimental bacteremia identifies a metabolic signature that distinguishes human sepsis from systemic inflammatory response syndromes. Am J Respir Crit Care Med. 2014;4:445–55. doi: 10.1164/rccm.201404-0624OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blaise BJ, Gouel-Cheron A, Floccard B, Monneret G, Allaouchiche B. Metabolic phenotyping of traumatized patients reveals a susceptibility to sepsis. Analytical chemistry. 2013;22:10850–5. doi: 10.1021/ac402235q. [DOI] [PubMed] [Google Scholar]
- 41.Mickiewicz B, Duggan GE, Winston BW, Doig C, Kubes P, Vogel HJ. Metabolic profiling of serum samples by 1H nuclear magnetic resonance spectroscopy as a potential diagnostic approach for septic shock. Crit Care Med. 2014;5:1140–9. doi: 10.1097/CCM.0000000000000142. [DOI] [PubMed] [Google Scholar]
- 42.Fanos V, Caboni P, Corsello G, et al. Urinary 1H-NMR and GC-MS metabolomics predicts early and late onset neonatal sepsis. Early Human Development. 2014:S78–S83. doi: 10.1016/S0378-3782(14)70024-6. [DOI] [PubMed] [Google Scholar]
- 43.Mickiewicz B, Vogel HJ, Wong HR, Winston BW. Metabolomics as a novel approach for early diagnosis of pediatric septic shock and its mortality. Am J Respir Crit Care Med. 2013;9:967–76. doi: 10.1164/rccm.201209-1726OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kamisoglu K, Haimovich B, Calvano SE, et al. Human metabolic response to systemic inflammation: assessment of the concordance between experimental endotoxemia and clinical cases of sepsis/SIRS. Critical care (London, England) 2015;71 doi: 10.1186/s13054-015-0783-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stringer KA, Serkova NJ, Karnovsky A, Guire K, Paine R, 3rd, Standiford TJ. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma (1)H-nuclear magnetic resonance quantitative metabolomics and computational analysis. American journal of physiology Lung cellular and molecular physiology. 2011;1:L4–L11. doi: 10.1152/ajplung.00231.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iwashyna TJ, Ely EW, Smith DM, Langa KM. Long-term cognitive impairment and functional disability among survivors of severe sepsis. Jama. 2010;16:1787–94. doi: 10.1001/jama.2010.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farris RW, Weiss NS, Zimmerman JJ. Functional outcomes in pediatric severe sepsis: further analysis of the researching severe sepsis and organ dysfunction in children: a global perspective trial. Pediatr Crit Care Med. 2013;9:835–42. doi: 10.1097/PCC.0b013e3182a551c8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langley RJ, Tsalik EL, van Velkinburgh JC, et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci Transl Med. 2013;195:195ra95. doi: 10.1126/scitranslmed.3005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogers AJ, McGeachie M, Baron RM, et al. Metabolomic derangements are associated with mortality in critically ill adult patients. PloS one. 2014;1:e87538. doi: 10.1371/journal.pone.0087538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Yin P, Amathieu R, Savarin P, Xu G. Application of LC-MS-based metabolomics method in differentiating septic survivors from non-survivors. Anal Bioanal Chem. 2016 doi: 10.1007/s00216-016-9845-9. [DOI] [PubMed] [Google Scholar]
- 51.Puskarich MA, Finkel MA, Karnovsky A, et al. Pharmacometabolomics of l-carnitine treatment response phenotypes in patients with septic shock. Ann Am Thorac Soc. 2015;1:46–56. doi: 10.1513/AnnalsATS.201409-415OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garcia-Simon M, Morales JM, Modesto-Alapont V, et al. Prognosis Biomarkers of Severe Sepsis and Septic Shock by 1H NMR Urine Metabolomics in the Intensive Care Unit. PloS one. 2015;11:e0140993. doi: 10.1371/journal.pone.0140993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kauppi AM, Edin A, Ziegler I, et al. Metabolites in Blood for Prediction of Bacteremic Sepsis in the Emergency Room. PloS one. 2016;1:e0147670. doi: 10.1371/journal.pone.0147670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mickiewicz B, Thompson GC, Blackwood J, et al. Development of metabolic and inflammatory mediator biomarker phenotyping for early diagnosis and triage of pediatric sepsis. Critical care (London, England) 2015;320 doi: 10.1186/s13054-015-1026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burt T, Nandal S. Pharmacometabolomics in Early-Phase Clinical Development. Clinical and translational science. 2016;3:128–38. doi: 10.1111/cts.12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dessi A, Liori B, Caboni P, et al. Monitoring neonatal fungal infection with metabolomics. The journal of maternal-fetal & neonatal medicine : the official journal of the European Association of Perinatal Medicine, the Federation of Asia and Oceania Perinatal Societies, the International Society of Perinatal Obstet. 2014:34–8. doi: 10.3109/14767058.2014.954787. [DOI] [PubMed] [Google Scholar]
- 57.Antti H, Fahlgren A, Nasstrom E, et al. Metabolic profiling for detection of Staphylococcus aureus infection and antibiotic resistance. PloS one. 2013;2:e56971. doi: 10.1371/journal.pone.0056971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beger RD, Dunn W, Schmidt MA, et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics : Official journal of the Metabolomic Society. 2016;10:149. doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi X, Yang F, Zheng YN, et al. Metabolomic approach for the identification of therapeutic targets of erythropoietin against sepsis in rat models. European review for medical and pharmacological sciences. 2016;3:537–46. [PubMed] [Google Scholar]
- 60.Izquierdo-García JL, Nin N, Ruíz-Cabello J, et al. A metabolomic approach for diagnosis of experimental sepsis. Intensive Care Med. 2011;12:2023–32. doi: 10.1007/s00134-011-2359-1. [DOI] [PubMed] [Google Scholar]
- 61.Jokerst JV, McDevitt JT. Programmable nano-bio-chips: multifunctional clinical tools for use at the point-of-care. Nanomedicine (London, England) 2010;1:143–55. doi: 10.2217/nnm.09.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamisoglu K, Calvano SE, Coyle SM, Corbett SA, Androulakis IP. Integrated transcriptional and metabolic profiling in human endotoxemia. Shock. 2014;6:499–508. doi: 10.1097/SHK.0000000000000248. [DOI] [PubMed] [Google Scholar]