Abstract
Nuclear human uracil–DNA glycosylase (hUNG2) initiates base excision repair (BER) of genomic uracils generated through misincorporation of dUMP or through deamination of cytosines. Like many human DNA glycosylases, hUNG2 contains an unstructured N–terminal domain that encodes a nuclear localization signal, protein binding motifs, and sites for post–translational modifications. Although the N–terminal domain has minimal effects on DNA binding and uracil excision kinetics, we report that this domain enhances the ability of hUNG2 to translocate on DNA chains as compared to the catalytic domain alone. The enhancement is most pronounced when physiological ion concentrations and macromolecular crowding agents are used. These data suggest that crowded conditions in the human cell nucleus promote the interaction of the N–terminus with duplex DNA during translocation. The increased contact time with the DNA chain likely contributes to the ability of hUNG2 to locate densely spaced uracils that arise during somatic hypermutation and during fluoropyrimidine chemotherapy.
Graphical abstract
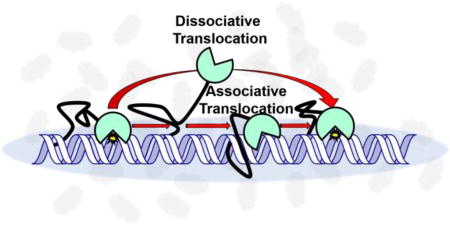
A common attribute of many enzymes that interact with DNA is the ability to track along the DNA chain after an initial encounter event (“DNA translocation”)1. This property arises from the polymeric nature of DNA which provides functionally equivalent binding sites (non–specific) in close proximity, thereby allowing the enzyme to stochastically sample a length of DNA before diffusing to bulk solution. In the case of DNA glycosylases, which need to locate and excise rare damaged bases in large genomes, DNA translocation provides an essential mechanism that allows for the inspection of multiple base pairs during a single encounter event2–4. The efficiency of DNA translocation can be severely impacted by solution properties such as ionic strength, ion composition and molecular crowding2. In particular, crowding has been shown to enhance in vitro DNA translocation by hUNG2 and human 8–oxoguanine DNA glycosylase (hOGG1) by hindering escape of the translocating enzymes to bulk solution5,6. This suggests that similar effects occur in the crowded cell nucleus.
hUNG2 is a highly proficient enzyme that removes uracil from duplex DNA in the context of U/A or U/G base pairs, but also single stranded DNA (ssDNA)6,7. The enzyme consists of a catalytic domain (hUNGcat) that is highly conserved throughout evolution and an unstructured ~90 amino acid N–terminal domain whose length is unique to mammals. The N–terminal domain has been shown to have minor effects on the activity of the enzyme, but contains a portion of the complex nuclear localization signal8, sites for tyrosine, serine and threonine phosphorylation (Y8, T6, T126, T60, S23, S64,)9,10, sites of lysine acetylation, and binding motifs for replication protein A (RPA; res 66–88)11,12, and proliferating cell nuclear antigen (PCNA; res 4–11)11,12. The function for many of the post–translational modifications are not known, but phosphorylation of T60 and S64 serves to target hUNG2 for cell cycle dependent proteolysis13, and phosphorylation within the PCNA and RPA binding motifs disrupts the interactions of the proteins with hUNG29,12.
Here we report that the N–terminal domain of hUNG2 functions to promote DNA translocation and is essential for DNA translocation under physiological conditions where the catalytic domain fails to translocate with high efficiency.
Results and Discussion
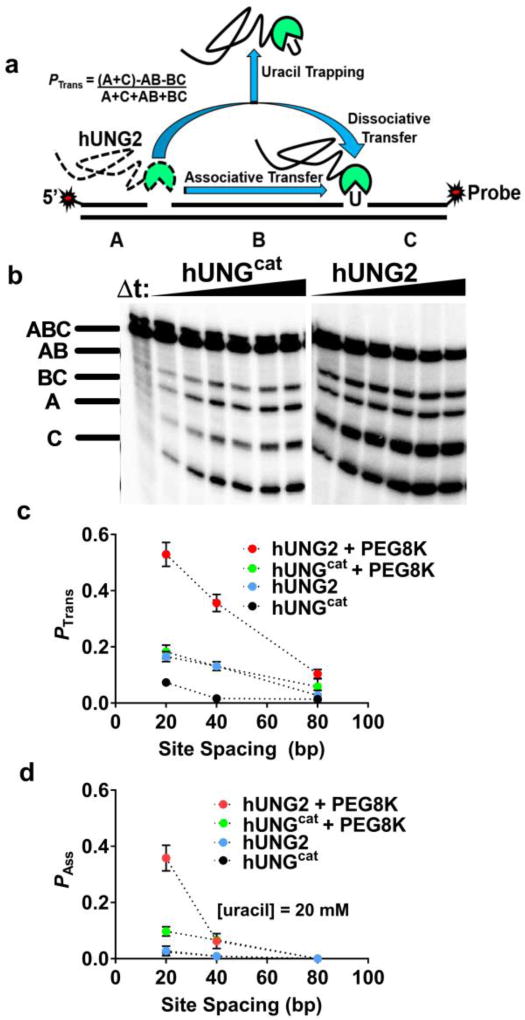
We used our established translocation assay to measure the probability that hUNG2 translocated between two uracil sites separated by a known spacing (Fig. 1a)14. The essence of the assay is that when the enzyme lands on a DNA molecule and excises one of the uracil sites it then has the chance to diffuse away from that DNA and find another new substrate, or translocate to the second uracil site and excise the uracil. Thus, translocation is indicated by higher levels of double site cleavage events as compared to single site cleavage events under initial rate single encounter conditions ([DNA] >> [E]). As indicated in Figure 1a, the single site cleavages generate bands AB and BC (after sample processing and electrophoresis)(Fig. 1b), while the double site cleavages produce bands A and C. The algebraic definition of the DNA translocation efficiency with respect to the fragment band intensities (Ptrans) is shown in Figure 1a.
Figure 1.
The hUNG2 disordered N–terminal domain promotes enhanced DNA translocation. (a) Determination of the DNA translocation efficiency (Ptrans). The DNA duplexes are labeled on the 5′ and 3′ ends of the uracil–containing strands. Performing the reaction in the presence of the uracil trap inhibits dissociative transfers (Pdiss), but associative transfers persist (Pass). (b) hUNG excision of one or both uracils in a single encounter generates distinct DNA fragments after separation (bands AB and BC or A and C). Gels depicting reactions containing hUNGcat or hUNG2 with 40 nM S20 DNA in the presence of 20% PEG8K are shown. (c) Ptrans vs uracil site spacing for hUNG2 and hUNGcat in the presence and absence of 20% PEG8K. Dotted lines are shown through the data to guide the eye. (d) Pass vs uracil site spacing for hUNG2 and hUNGcat in the presence and absence of 20% PEG8K. Reactions were performed in the presence of 20 mM uracil trap.
We investigated the translocation efficiencies of hUNG2 and hUNGcat using substrates with uracil site spacings of 20, 55 and 80 bp using buffer conditions containing 150 mM potassium ions with and without supplementation with the inert crowding agent PEG8K15,16. Previous work has established that PEG8K (but not the ethylene glycol monomer) enhances the translocation behavior hUNGcat in a concentration dependent manner, which is consistent with an excluded volume effect5. In the absence of PEG8K, Ptrans for both enzymes was small at all site spacings (< 0.15), but hUNG2 consistently showed at least a 3–fold larger value than hUNGcat (Fig. 1c). In the presence of 20% PEG8K, Ptrans for hUNGcat remained fairly low for all spacings (<0.2), but the Ptrans values for hUNG2 increased to 0.5 at the 20 bp uracil spacing and a low level of translocation was still measureable at a site spacing of 80 bp (Ptrans = 0.1) (Fig. 1b, c). Thus, the N–terminal extension enhances Ptrans and the effect is increased under the condition of molecular crowding (Fig. S1).
In order to isolate the contribution of associative translocation to site transfer, a high concentration of uracil may be added to the reaction14. Uracil serves to bind to the enzyme active site when hUNG releases from the DNA, effectively capturing all enzyme molecules that translocate by what we call a dissociative or “hopping” mechanism (described by dissociative transfer probability Pdiss). The fraction of the enzyme molecules that transfer without being captured by the uracil trap follow an associative pathway where the enzyme active site is never exposed to the trap (described by associative transfer probability Pass). The overall transfer is the sum of both pathways (Ptrans = Pdiss + Pass)14.
When ten or twenty millimolar uracil is added to the transfer reaction in the absence of PEG8K, all transfers of hUNGcat and hUNG2 are abolished (Fig. 1d, Fig S2). Thus, the dissociative pathway dominates under these conditions. In contrast, in the presence of 20% PEG8K and uracil trap, hUNG2 retained 60% of Ptrans across a 20 bp spacing (Pass = 0.40; Fig. 1d). This result contrasts considerably with hUNGcat, where Pass = 0.1 in the presence of uracil and 20% PEG8K at a 20 bp spacing. At spacings greater than 20 bp, associative transfers diminish to low or undetectable levels for both enzymes. These data establish that under conditions of crowding the disordered tail of hUNG2 confers a significant enhancement to associative site transfers. All of the translocation probabilities are reported in Table S1.
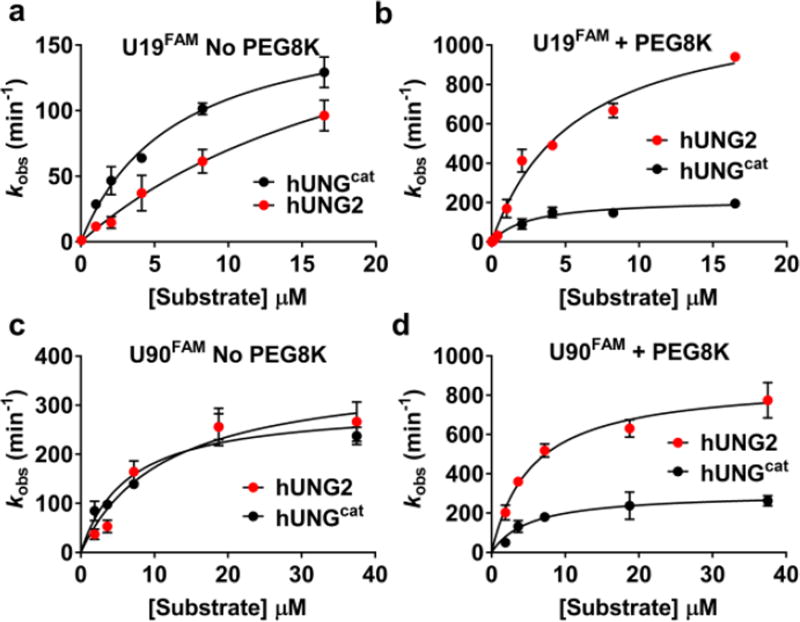
We were curious if molecular crowding influenced other aspects of catalysis by hUNG2 and hUNGcat. We measured the steady–state turnover of both enzymes in the presence and absence of 20% PEG8K using a 19mer duplex (U19FAM) substrate that contained a single uracil (Fig. 2a, b, Fig. S4). Although the kinetic parameters for hUNGcat were only modestly affected upon addition of PEG8K (Table S2), PEG8K introduced a 5–fold increase in kcat and a 4–fold decrease in Km for hUNG2. Due to these effects, hUNG2 is a 2.5–fold more efficient enzyme than hUNGcat in the presence of PEG8K (ratio of kcat/Km values), whereas the opposite is true in the absence of PEG8K.
Figure 2.
Contribution of the N–terminal extension of hUNG2 to steady–state turnover in the absence and presence of 20% PEG8K. (a) Substrate concentration vs velocity plots for hUNG2 and hUNGcat using U19FAM DNA in the absence of 20% PEG8K. (b) Substrate concentration vs velocity plots for hUNG2 and hUNGcat using U19FAM in the presence of 20% PEG8K. (c) [Substrate] vs velocity plot using U90FAM DNA in the absence of PEG8K, and (d) in the presence of 20% PEG8K. All rate measurements were performed in triplicate and the mean and standard deviations are indicated.
We further investigated whether the tail might have a greater effect with a longer DNA substrate. For this purpose, we used a 90mer duplex (U90FAM) with a single central uracil (Fig. 2c, d). The catalytic domain showed only small changes in its kinetic parameters compared to the 19mer, and these were little changed by the presence of PEG8K (Table S2, Fig. S3). For hUNG2, there was a 4–fold increase in kcat/KM with the longer DNA in the absence of PEG8K and a further 4–fold increase upon addition of PEG8K. The overall increase in kcat/KM when hUNG2 reacts with the longer substrate in the presence and absence of PEG8K indicates that the extra DNA flanking the damage site can lead to more efficient catalysis. This is likely manifested in transient contacts between the DNA and the disordered N–terminal domain.
The detailed mechanism by which the N–terminal tail of hUNG2 increases kcat by 5–fold is not known. However, single–turnover kinetic experiments of uracil excision by human hUNG27 have established that the chemical step is ~300 s−1, which is orders of magnitude greater than steady–state turnover, and is therefore not overall rate–limiting. In principle, an increase in kcat brought about by the N–terminus could arise from any mechanism that increases the ratio of productive ES complexes ([ES]prod, defined as any DNA complex that eventually leads to rapid uracil excision) as opposed to non–productive DNA complexes ([ES]non, which do not lead to rapid uracil excision). Alternatively, if product release is rate–limiting for turnover, the tail could in principle accelerate this step. Thus, the tail enhancement could arise from modest contributions from increasing [ES]prod/[ES]non and/or increasing product release.
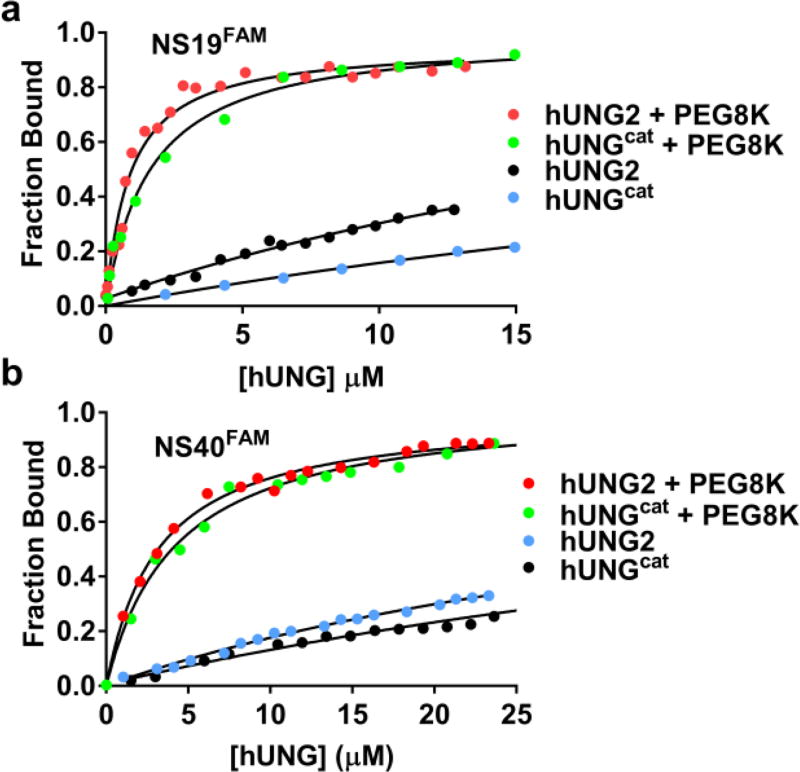
To determine whether the increase in translocation efficiency was mediated by an increase in the equilibrium binding affinity to non–specific DNA, we used fluorescence anisotropy to perform equilibrium DNA binding measurements for both enzymes in the presence and absence of 20% PEG8K (Fig. 3a). The assay employed a 19mer nonspecific DNA duplex (NS19FAM) with a fluorescein group (FAM) on the 5′ end of one strand. Both enzymes showed very weak binding to this duplex in the absence of PEG8K and in the presence of 150 mM [K+], with KD values of 36 µM (hUNG2) and 53 µM (hUNGcat)(Table S3). Strikingly, the KD values for both enzymes dropped about 30–fold to the low micromolar range in the presence of PEG8K. This is the expected effect of a crowding agent which favors the formation of low volume bimolecular complexes over that of the free species17–20. We repeated this experiment with a 5′–FAM labeled 40mer DNA to investigate whether detection of a tail interaction might require additional nucleic acid (Fig. 3b). No significant differences in binding affinity of both enzymes for the 19mer and 40mer DNA duplexes were detected or in the effects of PEG8K (Table S3). These results indicate that the tail extension provides a kinetic benefit and not a change in equilibrium DNA binding affinity (see below).
Figure 3.
Equilibrium binding of hUNG2 and hUNGcat to 19 and 40mer nonspecific DNAs in 150 mM [K+] in the presence and absence of 20% PEG8K (pH 7.5). (a) Changes in the fluorescence anisotropy of the DNA was used to measure binding of hUNG2 and hUNGcat to the 19mer duplex NS19FAM in the presence and absence of 20% PEG8K. (b) Binding of hUNG2 and hUNGcat to a 40mer duplex (NS40FAM) in the presence and absence of 20% PEG8K. The KD values in the presence of 150 mM [K+] and absence of PEG8K were very high (>35 µM). However, these were estimated by fixing the endpoint anisotropy to the value observed at low salt. The KD values for NS19FAM in the presence of PEG8K were 1.6 µM (hUNGcat) and 1.1 µM (hUNG2). The values for NS40FAM were 2.9 µM (hUNG2) and 3.6 µM (hUNGcat). All measurements were performed in triplicate.
The current study establishes how disordered tails can serve to enhance DNA translocation by an enzyme under conditions of molecular crowding. The requirement for crowding suggests that the interaction of the tail with DNA is promoted in restricted volume environments. This requirement may involve DNA assisted folding of the tail, which is further-promoted by the crowded conditions to favor a compact state19. For hUNG2, the tail–DNA interaction is not manifested in the equilibrium binding measurements. Instead, it becomes apparent when the probability of site transfer is measured or in kinetic measurements that include DNA translocation. This apparent anomaly reveals again a unique aspect of DNA translocation1,6,21, where the enzyme is in a short–lived transition state poised for either dissociation or its return to the DNA chain. The transient tail interaction changes this partitioning towards rebinding without affecting a macroscopic property such as equilibrium binding.
Methods
Complete methods are available in the Supporting Information.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by the National Institutes of Health Grants T32 GM8403–23, T32-CA9110, RO1 GM056834 (J.T.S), RO1 CA74305 (P.A.C), F32-GM119230 (B.P.W.).
ABBREVIATIONS
- hUNG
human uracil DNA glycosylase
- hUNG2
full length human uracil DNA glycosylase 2
- hUNGcat
human uracil DNA glycosylase catalytic domain
- BER
Base Excision Repair
- ssDNA
single–stranded DNA
- PEG8K
Polyethylene glycol 8000
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: to be filled in. Methods, DNA sequences, three supporting tables, and 5 Supporting figures (PDF).
Author Contributions
G.R. performed all kinetic and thermodynamic measurements. J.T.S. conceived the approach. B.W. devised and executed the purification of hUNG2. A.E executed the preparation of the FAM site transfer DNA oligonucleotides. J.D.S and P.A.C. edited the manuscript. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
No competing financial interests have been declared.
References
- 1.Halford SE. Biochem. Soc. Trans. 2009;37:343. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 2.Friedman JI, Stivers JT. Biochemistry. 2010;49:4957. doi: 10.1021/bi100593a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedglin M, O’Brien PJ. ACS Chem. Biol. 2010;5(4):427–436. doi: 10.1021/cb1000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, O’Brien PJ. ACS Chem. Biol. 2015;10(11):2606–2615. doi: 10.1021/acschembio.5b00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cravens SL, Schonhoft JD, Rowland MM, Rodriguez AA, Anderson BG, Stivers JT. Nucleic Acids Res. 2015;43:4087–4097. doi: 10.1093/nar/gkv301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schonhoft JD, Kosowicz JG, Stivers JT. Biochemistry. 2013;52:2526–2535. doi: 10.1021/bi301561d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grogan BC, Parker JB, Guminski AF, Stivers JT. Biochemistry. 2011;50:618–627. doi: 10.1021/bi102046h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otterlei M, Haug T, Nagelhus TA, Slupphaug G, Lindmo T, Krokan HE. Nucleic Acids Res. 1998;26:4611–4617. doi: 10.1093/nar/26.20.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liabakk NB, Hagen L, Sousa MM, Torseth K, Slupphaug G, Kavli B, Krokan HE, Jensen ON, Pe a–Diaz J, Otterlei M, Hørning O, Sundheim O. EMBO. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carter RJ, Parsons JL. Mol Cell Biol. 2016;36:1426–1437. doi: 10.1128/MCB.00030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haug T, Otterlei M, Warbrick E, Slupphaug G, Nagelhus TA, Bakke O, Krokan HE, Steinsbekk K, Akbari M, Aas PA. EMBO. 1999;18:3834–3844. doi: 10.1093/emboj/18.13.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiser BP, Stivers JT, Cole PA. Biophys. J. 2017;113(2):393–401. doi: 10.1016/j.bpj.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer JA, Muller–Weeks S, Caradonna S. DNA Repair. 2004;3:505–513. doi: 10.1016/j.dnarep.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Schonhoft JD, Stivers JT. Nat. Chem. Bio. 2012;8:205. doi: 10.1038/nchembio.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillip Y, Sherman E, Haran G, Schreiber G. Biophys. J. 2009;97:875–885. doi: 10.1016/j.bpj.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schreiber G, Haran G, Zhou H. Chemical reviews. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H, Rivas G, Minton AP. Annu. Rev. Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong J, Gierasch LM. J. Am. Chem. Soc. 2010;132:10445. doi: 10.1021/ja103166y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Szasz CS, Alexa A, Toth K, Rakacs M, Langowski J, Tompa P. Biochemistry. 2011;50:5834. doi: 10.1021/bi200365j. [DOI] [PubMed] [Google Scholar]
- 20.Soranno A, Koenig I, Borgia MB, Hofmann H, Zosel F, Nettels D, Schuler B. Proc. Nat. Acad. Sci. 2014;111:4874–4879. doi: 10.1073/pnas.1322611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esadze A, Rodriguez G, Cravens SL, Stivers JT. Biochemistry. 2017;56:1974. doi: 10.1021/acs.biochem.7b00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.