Abstract
Mendelian randomization (MR) is a burgeoning field that involves the use of genetic variants to assess causal relationships between exposures and outcomes. MR studies can be straightforward; for example, genetic variants within or near the encoding locus that is associated with protein concentrations can help to assess their causal role in disease. However, a more complex relationship between the genetic variants and an exposure can make findings from MR more difficult to interpret. In this Review, we describe some of these challenges in interpreting MR analyses, including those from studies using genetic variants to assess causality of multiple traits (such as branched-chain amino acids and risk of diabetes mellitus); studies describing pleiotropic variants (for example, C-reactive protein and its contribution to coronary heart disease); and those investigating variants that disrupt normal function of an exposure (for example, HDL cholesterol or IL-6 and coronary heart disease). Furthermore, MR studies on variants that encode enzymes responsible for the metabolism of an exposure (such as alcohol) are discussed, in addition to those assessing the effects of variants on time-dependent exposures (extracellular superoxide dismutase), cumulative exposures (LDL cholesterol), and overlapping exposures (triglycerides and non-HDL cholesterol). We elaborate on the molecular features of each relationship, and provide explanations for the likely causal associations. In doing so, we hope to contribute towards more reliable evaluations of MR findings.
Introduction
The raison d’être for medical and scientific research is to understand disease aetiologies and identify opportunities for prevention and treatment. Observational epidemiological studies provide a wealth of information on associations between disease exposures and outcomes, but they cannot be interpreted as indicating causality, owing to limitations introduced by confounding and reverse causality1,2. Although randomized, controlled trials (RCTs) remain the gold-standard study design for inferring causality, they are exceedingly expensive and time-consuming efforts with high failure rates (>50% fail owing to lack of efficacy)3,4. In addition, RCTs might involve interventions that are pleiotropic (such as drugs that modify multiple biomarkers), which can challenge causal deductions for any individual biomarker. Finally, RCTs are not always feasible or ethical to conduct5, such as when attempting to clarify the causal role of alcohol in cardiovascular disease6–9.
Mendelian randomization (MR) is an established epidemiological approach that can provide information on causality and prioritize biomarkers for drug-target validation10–14. Grouping individuals in the population according to the possession of genetic variants that modify an exposure allows researchers to infer whether a biomarker is causally related to a disease (Figure 1). This inference is permissible owing to the fundamental nature of the genome: genetic variants should be free from conventional confounding owing to the random independent assortment of DNA at meiotic segregation of alleles. In addition, reverse causality bias would not occur owing to the essentially non-modifiable nature of the transmitted germline genome. Therefore, MR can help to strengthen causal inferences on the role of modifiable exposures, such as circulating biomarkers in disease risk. Genetic variants that are associated with LDL-cholesterol levels have been used to assess the causal role of LDL cholesterol in coronary heart disease (CHD)15–18. Irrespective of the genetic variants used as a proxy for LDL cholesterol, strong dose–response relationships have been identified15. This observation suggests that lowering LDL-cholesterol levels by any means would lead to a reduction in CHD, and further validates the linear dose–response relationship between LDL cholesterol and CHD risk identified from meta-analyses of RCTs assessing statins and other cholesterol-lowering interventions19–21.
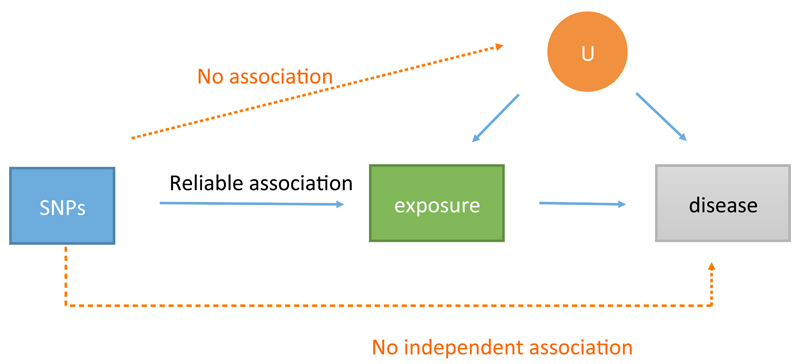
Figure 1. Instrumental variable analysis to generate causal estimates through Mendelian randomization.
The three principles of instrumental variable analysis are: the instrumental variable (in this case a genetic variant either in isolation or in combination with other variants) must associate with the exposure; the instrumental variable must not associate with confounders that are either known or unknown (U); and there is no pathway from the single nucleotide polymorphism (SNP) to disease that does not include the exposure of interest. This figure is a schematic representation and should not be interpreted as a formal directed acyclic graph.
The use of MR analysis had led to several major findings and validations of cause–effect relationships in cardiometabolic disorders. In the absence of any RCTs that have assessed the efficacy of a specific C-reactive protein (CRP)-lowering therapeutic for CHD, MR studies have successfully shown that CRP is unlikely to have a major role in the development of CHD22,23. Similarly, whereas an RCT on the effect of alcohol consumption on cardiovascular health is unfeasible and unethical, MR analyses have shown that moderate alcohol consumption is unlikely to reduce CHD risk24. Increased adiposity was found to increase the risk of CHD in several MR studies25–28, whereas the only RCT investigating this relationship was underpowered for assessing causality29. Furthermore, MR analyses validated findings from RCTs assessing drug targets, including 3-hydroxy- 3-methylglutaryl-coenzyme A reductase (HMG-CoA reductase)30,31, secretory phospholipase A2-IIA32,33, lipoprotein-associated phospholipase A234–38, and Niemann-Pick C1-like protein39. Finally, MR studies have also identified a potential increased risk of diabetes mellitus with pharmacological inhibition of PCSK940–42. These studies and other notable examples of MR analyses that have progressed our understanding of the aetiology of cardiovascular and metabolic diseases are outlined in Table 1.
Table 1. Notable Mendelian randomization studies in cardiometabolic disease.
| Exposure | Outcome | Interpretation | Importance | Refs |
|---|---|---|---|---|
| Biomarkers and drug targets | ||||
| BMI | Metabolites | BMI causally influences many circulating metabolites | Supports the interpretation that BMI may influence cardiometabolic disease through its influence on metabolites | 139 |
| HMGCR/Statins | Metabolites | Casual | Shows consistency of observational data on statins vs predicted MR effects on metabolites | 30 |
| Adiposity (BMI and waist-hip ratio) | CHD | BMI and waist–hip ratio (adjusted for BMI) causally increases risk of CHD | No trial yet to show this causal relationship29 | 25–28 |
| C-reactive protein | CHD | No causal relationship | No trial of a therapy specific to CRP for CVD events has been conducted | 22,23,50 |
| LDL-C | CHD | Dose-response relationship irrespective of locus | Suggests LDL-cholesterol lowering by any means is beneficial, consistent with trials involving statin and other cholesterol-lowering interventions19,21,108 | 15 |
| HDL-C | CHD | No causal effect | Contradicts observational data140, but supports findings from recent RCTs61–63 | 16–18 |
| TGs | CHD | Causal | Precedes trial data of a TG-lowering agent | 16,18,141 |
| sPLA2-IIA | CHD | Non-causal | Published at a similar time to the VISTA-16 trial33 of a sPLA2- IIA-lowering drug that did not have beneficial effects on CVD | 32 |
| Lp-PLA-IIA | CHD | Non-causal | Many resources were spent on trials35,62 that showed therapeutic lowering of Lp-PLA2 level does not lower risk of CVD; some MR studies were published before the reporting of RCT results | 34,36,37, 142 |
| NPC1L1/Ezetimibe | CHD | Causal | MR studies preceded RCT data143, that showed lowering of LDL-cholesterol level via inhibition of NPC1L1 results in reduced risk of CVD | 39,144 |
| PCSK9, Lipoprotein (a) and ANGPTL4 | CHD | Causal | Drugs developed for CVD prevention on basis of genetic findings, some of which have since shown cardiovascular benefit in phase III RCTs145 | 146–148 |
| LDL-C | Diabetes | Causal | Suggests LDL-cholesterol lowering might generally lead to increased risk of diabetes mellitus, and has potential ramifications for drugs that lower LDL-cholesterol level | 18 |
| HMGCR/Statins | Diabetes | Causal | Indicates that the (albeit small) diabetogenic effects of statins (that are outweighed by the cardiovascular benefits149) seen in RCTs are on-target150 | 31 |
| PCSK9/PCSK9 inhibitors | Diabetes | Causal | Suggests PCSK9 inhibition might increase risk of diabetes | 40–42 |
| Exogenous exposures | ||||
| Alcohol | Cardiovascular diseases (including blood pressure, coronary artery calcification and CHD) | Causal | Suggests alcohol is harmful to cardiovascular health at all doses of consumption, contrary to decades of observational data6; findings are important for public-health policy151 | 24,83,152 |
ANGPTL4, angiopoietin-related protein 4; CHD, coronary heart disease; CRP, C-reactive protein; CVD, cardiovascular disease; Lp-PLA2, lipoprotein-associated phospholipase A2; MR, Mendelian randomization; NPC1L1, Niemann–Pick C1-like protein 1; RCT, randomized, controlled trial; sPLA2-IIA, secretory phospholipase A2-IIA.
As with conventional observational, epidemiological, and interventional trials, MR analyses typically yield a quantitative association between a biomarker and disease risk. This approach is most robust when a variant is in or near a gene that is responsible for the synthesis of the protein under investigation, and associates with concentrations of the same protein without disrupting protein function (for example, using genetic variants around the CRP gene that are associated with concentrations of circulating CRP)22. However, there are various scenarios in which the accuracy of data derived from MR analyses becomes compromised. In this Review, we highlight scenarios in which the interpretation of MR analyses is not straightforward and, in each case, we elaborate on the molecular details and provide what we consider to be a more accurate interpretation (Figure 2).
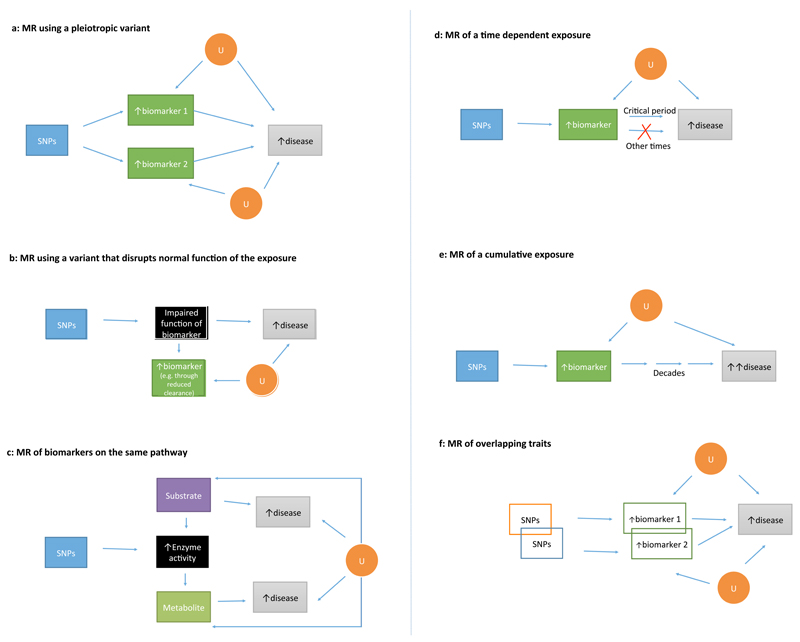
Figure 2. Paradoxical scenarios in Mendelian randomization.
a | An example of MR using a pleiotropic variant. The genetic variant associates with multiple biomarkers on separate biological pathways. Generating separate causal estimates for biomarkers 1 and 2 is invalid as they ascribe the same single nucleotide polymorphism (SNP)–disease effect to each biomarker. Furthermore, if only one of the biomarkers is causal, then using the SNP to make causal inferences on the non-causal biomarker can generate an erroneous conclusion. b | An example of MR using a variant that disrupts normal function of the exposure. Possession of the genetic variant might lead to increased concentration of the exposure (for example, owing to impaired clearance), but paradoxically lead to an increased risk of disease (if the normal function of the biomarker would be protective of disease), or vice versa if the normal function of the biomarker increases risk of the disease. c | An example of MR involving biomarkers on the same pathway, in which the genetic variant encodes an enzyme that metabolizes a substrate into a metabolite. If the substrate and metabolite have contrasting roles in the development of diseases, this discrepancy might lead to complexity in the interpretation of findings. d | An example of MR involving a time-dependent exposure. If the biomarker is causal for disease only during a critical time period, MR might show evidence of a protective effect. However, intervening on the biomarker during the noncritical time period will not alter risk of disease. e | An example of MR of a cumulative exposure, in which the exposure is causal for disease, but has a long latency. For example, the disease might typically present after decades of exposure-induced subclinical disease development. f | An example of MR involving overlapping traits. MR of overlapping biomarkers can lead to paradoxical findings as the overlapping nature of the traits might lead to a diminution of their apparent causal effect on multivariate analyses. These figures are schematic representations and should not be interpreted as formal directed acyclic graphs. U, unknown.
Multiple biomarkers on separate pathways
In general, a genetic variant used for MR should affect only a single pathway on which the exposure of interest lies (Box 1). For example, the use of a genetic variant in MR that, through its association with the exposure of interest, associates with multiple biomarkers on a single biological pathway is valid and is termed vertical pleiotropy. By contrast, horizontal pleiotropy — when a genetic variant employed in MR associates with multiple biomarkers on discrete pathways (Box 1; Figure 2A) — can yield invalid causal estimates. Two related examples are discussed below: the first example involves the same genetic variant used incorrectly to assess causal relationships of multiple traits on discrete pathways, and the second example relates to the use of pleiotropic single nucleotide polymorphisms (SNPs) to assess the causal role of CRP in CHD without applying appropriate analytical checks.
Box 1. Pleiotropy in MR and implications for causal deduction.
Single nucleotide polymorphisms (SNPs) can be used in isolation or combination as genetic instruments to assess the causal role of a biomarker with disease risk. Vertical pleiotropy refers to when genetic variants associate with multiple biomarkers that are on the same pathway from exposure through to disease, and does not invalidate the findings as long as the primary phenotype (most proximal to the genotype) that is influenced by the genetic variant is understood. Horizontal pleiotropy refers to when a genetic variant associates with traits on discrete pathways that are also causal in disease. When using multiple genetic variants in combination, horizontal pleiotropy can ‘balance out’ and have no net effect on the association of the exposure and disease risk. Balanced horizontal pleiotropy should not bias the causal effect derived from Mendelian randomization (MR), even when using conventional approaches; however, it does lead to increased variance in the effect estimation, and thus less-precise confidence intervals. By contrast, unbalanced horizontal pleiotropy distorts the association between the exposure and the outcome, and the effect estimate from conventional MR approaches can be exaggerated or diminished, depending on the direction of the pleiotropy. The presence of unbalanced horizontal pleiotropy can be formally assessed by using MR-Egger53, provided certain assumptions are satisfied. MR-Egger provides a valid MR estimate that takes into account presence of unbalanced horizontal pleiotropy. Other approaches include median and weighted median MR54, which provide a valid MR estimate as long as the majority of SNPs (or the majority of the statistical weight contributed by the SNPs) in the instrument are valid, and the modal estimate122, which assumes that the most common effect estimate among a set of instruments is the one most likely to be valid. Each of these approaches has their own assumptions54. When possible, investigators should perform these sensitivity analyses when conducting conventional (inverse variance-weighted) MR analyses. Figure adapted from White, J. et al. Association of lipid fractions with risks for coronary artery disease and diabetes. JAMA Cardiol. 1, 692–699 (2016), with permission from the American Heart Association.
Branched-chain amino acids and diabetes
Observational studies have reported an association between the branched-chain amino acids (leucine, isoleucine, and valine) and risk of incident diabetes43–48. In a genome-wide association study (GWAS) published in 2016, a genetic variant associated with these three amino acids was identified and used to conduct MR analyses49. At the PPM1K locus, the rs1440581 SNP was used to generate causal estimates for both leucine and valine, whereas a SNP in the same locus (rs7678928, in linkage disequilibrium with rs1440581 at r2 = 0.79 and Dʹ = 1) was used in combination with other SNPs for isoleucine. According to the investigators, “the association of genetic variants appeared highly specific”, and they subsequently generated MR estimates for each of the three amino acids, and used these to implicate the role of branched-chain amino acid metabolism in the development of diabetes49.
As shown in Figure 3A, the PPM1K locus encodes mitochondrial phosphatase that activates branched-chain α-ketoacid dehydrogenase (BCKD), which is responsible for the metabolism of leucine, isoleucine, and valine49. Therefore, by activating the enzyme responsible for the metabolism of these three amino acids, SNPs in the PPM1K locus associate with the concentrations of these amino acids. In MR analyses, the association of the SNPs with risk of disease is scaled to the association of the SNP with the exposure to generate a single causal estimate for the relationship between the exposure and disease (Figure 1). When the same SNP is used to make causal deductions on multiple exposures, there is only a single association of the SNP with risk of disease; therefore, this SNP-to-disease association is used multiple times, each time assuming that the exposure individually accounts for the disease association. In this scenario, the association of PPM1K at rs1440581 (or a SNP in linkage disequilibrium with rs7678928) with diabetes is used to generate individual MR estimates for each of the three branched-chain amino acids, ascribing a causal estimate that is scaled to the effect of the SNP on the amino acid49 (Table 2). Crucially, this analysis makes the invalid assumption that each of the three amino acids in isolation would be causal, and is in violation of one of the principal rules of MR: that the instrument acts only on the outcome through the exposure of interest. Furthermore, the genetic variant association with diabetes is triple-counted, in that the full effect is attributed to three different exposures, which is incoherent. Using this single locus, the MR analyses cannot clarify which, if any, of the three amino acids is actually driving the causal relationship with diabetes. Given that the researchers concluded that their findings are “consistent with a causal role of [branched-chain amino acid] metabolism in the aetiology of type 2 diabetes”, readers might incorrectly interpret the quantitative MR estimates reported for each of the three branched-chain amino acids as evidence that each branched-chain amino acid is individually and independently related to type 2 diabetes49. The only information that can be inferred from this study is that the PPM1K locus activates an enzyme (BCKD) that has a range of substrates (which might not be limited to the three amino acids studied), and that one or more of these pathways leads to diabetes. Therefore, although we agree with the researchers regarding their results being consistent with the proposed causal role of branched-chain amino acid metabolism in diabetes, the presentation of the data could lead to misinterpretation49.
Figure 3. Mendelian randomization using a genetic variant that associates with multiple biomarkers on separate pathways.
a | Using single nucleotide polymorphisms (SNPs) in PPM1K (which encodes a mitochondrial phosphatase that activates branched-chain α-keto acid dehydrogenase [BCKD], responsible for the rate-limiting step of metabolism of the branched-chain amino acids) to infer causality of three separate amino acids yields an erroneous conclusion, as this inference ascribes a causal estimate to each amino acid from the same PPM1K– diabetes association that is scaled to the PPM1K–amino acid estimate (TABLE 2). The three amino acids are initially catabolized prior to the enzymatic action of BCKD. b | Using SNPs in APOE to infer causality of C-reactive protein (CRP) yields an erroneous conclusion, as the SNP is pleiotropic for CRP and LDL cholesterol (LDL-C). These figures are schematic representations and should not be interpreted as formal directed acyclic graphs. CHD, coronary heart disease.
Table 2. Mendelian randomization of discrete biomarkers when using identical or highly correlated genetic variants.
| Branched chain amino acid (BCAA) | SNP in PPM1K | Linkage disequilibrium with rs1440581 | Effect/other allele | EAF | Standardized Beta (SE) of Metabolite Level per Allele | P-value for BCAA | OR (95% CI) for T2DM per Allele | P-value for T2DM | MR estimate of T2DM per 1-SD higher BCAA | P-value for MR estimate |
|---|---|---|---|---|---|---|---|---|---|---|
| Leucine | rs1440581 | 1.0 | C/T | 53% | 0.08 (0.013) | 3.9×10−25 | 1.04 (1.02–1.07) | 0.00034 | 1.85 (1.41–2.42) | 7.3×10−6 |
| Isoleucine | rs7678928 | 0.8 | T/C | 46% | 0.09 (0.013) | 5.6×10−19 | 1.03 (1.01–1.05) | 0.0055 | 1.40 (1.10–1.78) | 5.5×10−3 |
| Valine | rs1440581 | 1.0 | C/T | 53% | 0.10 (0.013) | 4.4×10−24 | 1.04 (1.02–1.07) | 0.00034 | 1.54 (1.28–1.84) | 4.2×10−6 |
Legend: Branched-chain amino acids (BCAAs) are arranged according to the effect size of the single nucleotide polymorphism (SNP) on each trait (sorted from smallest to largest). Given that the association of each SNP with type 2 diabetes mellitus (T2DM) is identical (or near-identical when using rs7678928, in linkage disequilibrium with rs1440581 at r2 = 0.79), the MR estimate is scaled to this effect. Therefore, on Mendelian randomization (MR) analysis, leucine has the greatest magnitude of association with risk of T2DM, and valine has the lowest magnitude of association. This analysis reflects how the MR estimates are generated by dividing the SNP–T2DM estimate by SNP–BCAA. When the SNP–BCAA estimate is the smallest (as for leucine), the MR point estimate is the greatest. Critically, none of the MR estimates for the three BCAAs is valid because they all assume that each BCAA individually is causal. EAF, effect allele frequency. Adapted from Lotta, L. A. et al. Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a Mendelian randomisation analysis. PLoS Med. 13, e1002179 (2016).
APOE, C-reactive protein, and risk of CHD
In MR studies, incorrect analysis of a genetic variant in APOE that associates with circulating levels of CRP might erroneously imply a causal relationship between CRP levels and CHD risk50. However, this relationship is actually driven by the association of the APOE genotype with multiple biomarkers on discrete pathways, including LDL cholesterol, which is causally related to CHD (Figure 3B). Use of such pleiotropic variants in isolation is, therefore, likely to lead to incorrect causal interpretations. An alternative approach would be to combine multiple SNPs across the genome into a genetic variant score for CRP51. Although pleiotropy in a gene score might ‘balance out’ in certain settings (so-called ‘balanced horizontal pleiotropy’; Box 1), the horizontal pleiotropy of the CRP gene score is likely to persist, resulting in potential biased estimates using conventional MR approaches (Boxes 2, 3). Furthermore, when using a multilocus genetic variant score for CRP and removing SNPs on the basis of tests for heterogeneity, this approach can still yield biased results, because multiple SNPs that have pleiotropic effects might remain. Therefore, causal associations for disease that arise from horizontal pleiotropy when analysed using conventional MR methods might be biased (known as unbalanced horizontal pleiotropy)52. This approach could be improved by using SNPs that are either confined to the CRP locus (which are more likely to show specificity for the protein trait), or using SNPs across the genome identified from GWAS in combination with use of more novel approaches, such as MR-Egger (Box 4), which allows relaxation of the instrumental variable assumption that there is no unbalanced horizontal pleiotropy53,54. Therefore, our recommendation would be to utilize a range of applicable methodologies as sensitivity analyses to test the robustness of the MR estimates against potential violations, including unbalanced horizontal pleiotropy.
Box 2. Conventional MR approaches.
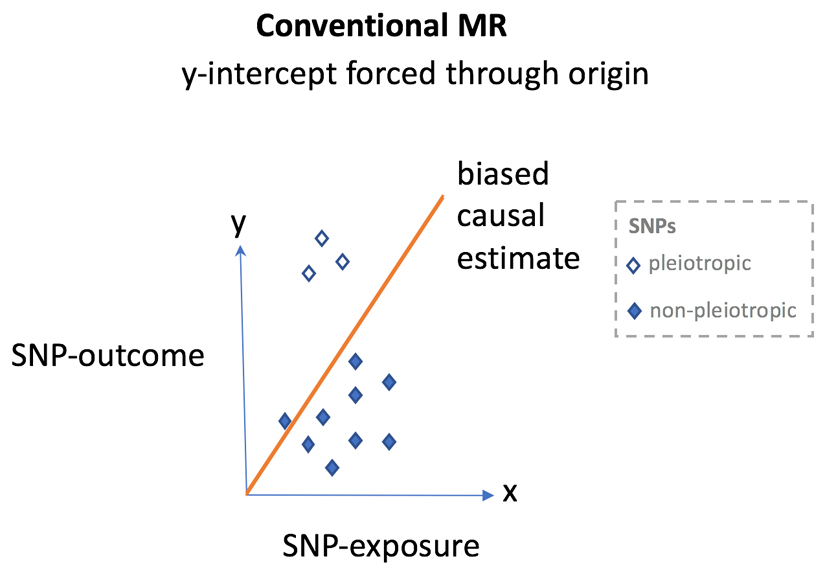
When using multiple single nucleotide polymorphisms (SNPs) for Mendelian randomization (MR) in summary-level data, inverse variance-weighted analysis was, until recently, the most common analytical approach. In this form of regression, each SNP contributes a data point on the x-axis (SNP-to-exposure association) and y-axis (SNP-to-disease association). However, inverse-variance-weighted MR forces the y-intercept through the origin. In the setting of balanced pleiotropy, bias should not be evident and the slope of the regression line can be reliably interpreted as the causal effect of the exposure on the outcome. However, in the presence of unbalanced horizontal pleiotropy, conventional MR analysis can lead to bias as the y-intercept, being forced through the origin, means that the directional bias influences the regression slope. To overcome this issue, investigators previously relied on approaches such as ‘manual pruning’ of SNPs that they considered pleiotropic. However, this approach relies on the availability and precision of SNP estimates with multiple biomarkers to inform on the presence of such pleiotropy; many SNP associations that do not meet conventional significance thresholds in genome-wide association studies (GWAS) are false-negatives, owing to the stringent α values used to avoid false-positives133,134. Second, this approach can be subjective as one investigator might consider a trait to be indicative of vertical pleiotropy and retain SNPs associated with the trait, whereas another could consider the same trait to be evidence of horizontal pleiotropy and remove SNPs showing association with the trait. This approach would lead to different genetic instruments and potentially inconsistent results between studies. Furthermore, reasons for exclusion can be nontransparent and differ by study, further reducing the degree of objectivity. Finally, manual pruning can lead to a genetic instrument that is no longer biologically meaningful (Box 3).
Each diamond represents a single SNP plotted so that the SNP to exposure estimate is on the x-axis and the SNP to outcome estimate is on the y-axis. Filled diamonds = non-pleiotropic variants and open diamonds = pleiotropic variants. In MR using summary level data, the regression slope provides an estimation of the causal effect of the exposure on the outcome.
Box 3. Effects of manual pruning on interpretation of MR analysis.
As mentioned in Box 2, manual pruning of single nucleotide polymorphisms (SNPs) that are considered pleiotropic might result in a genetic instrument that is no longer biologically meaningful, as SNPs that are retained might not be indicative of any tangible entity. For example, if we have SNPs in 70 loci associated with HDL cholesterol from genome-wide association studies (GWAS) using conventional P-value thresholds (that is, P <5 × 10-8)135, and manually prune out the SNPs associated with LDL cholesterol or triglycerides (associated with either LDL cholesterol or triglycerides at P <5 × 10-8, or by using a more relaxed P-value threshold) in order to obtain an instrument that is more ‘specific’ for HDL cholesterol, this approach will retain a smaller number of SNPs in the genetic instrument. However, by removing SNPs that account for the underlying genetic architecture of HDL cholesterol acquired in a hypothesis-free GWAS study, the remaining SNPs might not be representative of features that are biologically meaningful of HDL cholesterol. Furthermore, manual pruning can exacerbate the problem when removal of SNPs occurs on the basis of vertical pleiotropy. For example, pruning SNPs associated with fasting glucose for their association with type 2 diabetes mellitus to evaluate whether fasting glucose is associated with coronary heart disease independently of diabetes might inadvertently lead to SNPs being selected on the basis of pleiotropy (as a fasting glucose SNP that is not associated with diabetes might associate with pathways that counterbalance the glucose association, such that the overall association of the SNP with diabetes is null)136. Therefore, an MR analysis using manually pruned SNPs in a genetic instrument can lead to findings that are very challenging to interpret.
Box 4. New MR approaches.
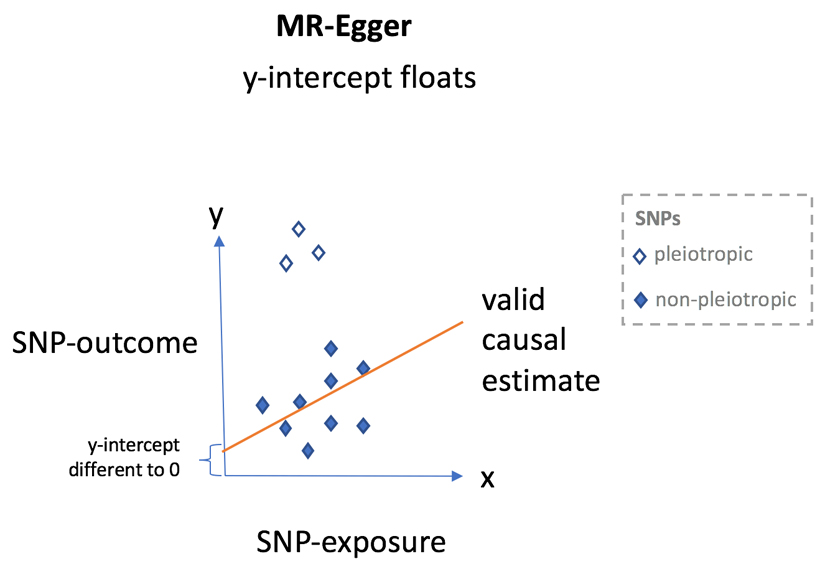
In contrast to conventional Mendelian randomization (MR), MR-Egger53 takes the approach of Egger regression137 (introduced in the context of small study bias evaluation in clinical trials) and allows the y-intercept to float, which achieves two outcomes. First, this flotation provides a statistical test for the presence of unbalanced horizontal pleiotropy; evidence that y is different to 0 when x = 0 is suggestive of the presence of unbalanced horizontal pleiotropy. Second, by absorbing the pleiotropic effects into the y-intercept, MR-Egger can provide a reliable estimate for the underlying causal effect (when certain additional assumptions are satisfied53) from the slope of the regression line. Therefore, the advantages of MR-Egger are manifold, as it obviates the need for manual pruning of SNPs, the intercept can inform on the presence of unbalanced pleiotropy, and the slope can provide a valid causal estimate even in the presence of such pleiotropy (as long as certain conditions are met). One disadvantage of MR-Egger is that, for a given sample size, power is reduced compared with inverse variance-weighted MR, although newer extensions to MR-Egger, such as MR-Egger with SIMEX138, seek to increase power.
Each diamond represents a single SNP plotted so that the SNP to exposure estimate is on the x-axis and the SNP to outcome estimate is on the y-axis. Filled diamonds = non-pleiotropic variants and open diamonds = pleiotropic variants. In MR using summary level data, the regression slope provides an estimation of the causal effect of the exposure on the outcome.
Disrupting the function of the exposure
A paradoxical association between the levels of the biomarker and disease risk in MR (Figure 2B) can take place if a SNP disrupts the normal function of a causal exposure (such as binding of the exposure to its target receptor). In this setting, the difference in biomarker concentrations from the genetic variant might be used to infer inaccurate directionality of causation. However, by perturbing the normal function of the biomarker, the genetic variant can paradoxically be linked with a biomarker that confers an opposite risk of disease in MR analysis to that observed in epidemiological studies.
SCARB1, HDL cholesterol, and CHD risk
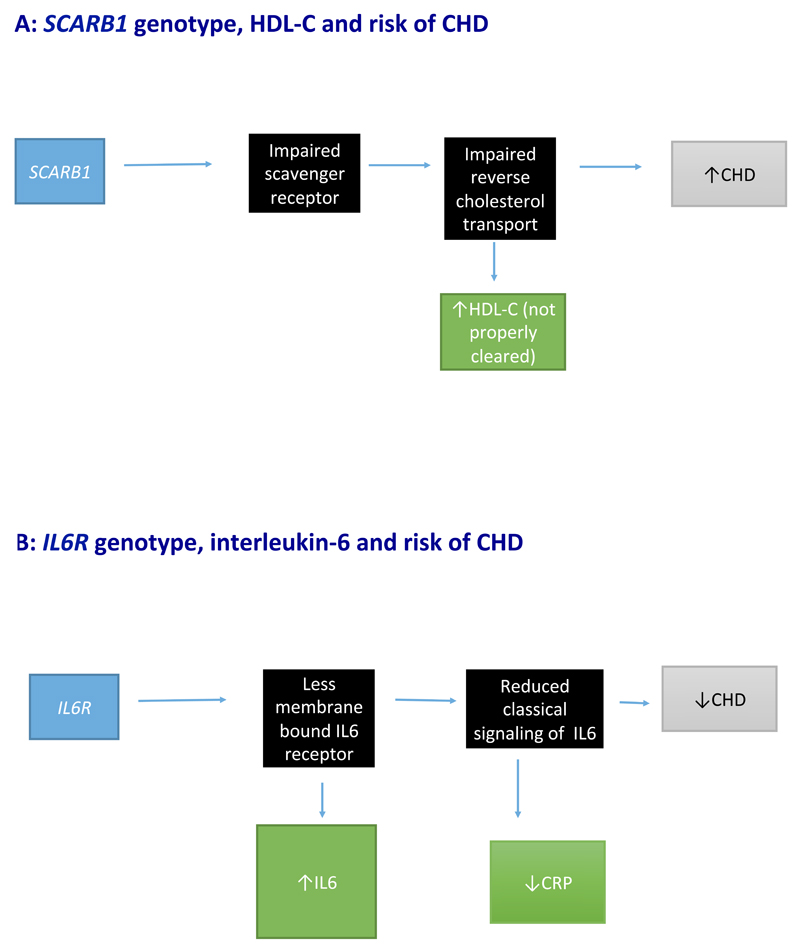
In 2016, a rare variant in SCARB1 was identified that results in the loss of function of the scavenger receptor B1 (SR-B1)55. Individuals carrying this variant have higher levels of circulating HDL-cholesterol levels and an elevated risk of CHD. The study investigators proposed that reduced hepatic SR-B1 function in humans causes impaired reverse cholesterol transport, which in turn leads to increased risk of CHD despite elevated HDL-cholesterol levels55. These findings have been widely interpreted to mean that high levels of HDL cholesterol can actually contribute to the development of CHD, which is inconsistent with the well-established putative protective role of HDL cholesterol against cardiovascular risk56,57.
The HDL-mediated transport of cholesterol from peripheral tissues to the liver (reverse cholesterol transport) is thought to result in a decrease in atheroma burden, and a commensurate reduction in the risk of CHD58,59. However, a critical component of HDL-mediated reverse cholesterol transport is the selective uptake of circulating HDL particles by the liver. After hepatic uptake, cholesterol is excreted in bile; the binding and uptake of HDL particles into the liver occurs principally through SR-B160. Therefore, the increased risk of CHD associated with a genetic variant that disrupts normal function of SR-B1 potentially provides new evidence that HDL-mediated reverse cholesterol transport (through SR-B1) might have a role in preventing the development of CHD (Figure 4A). However, to temper enthusiasm, whole-genome sequencing studies that investigated sequence variants within the SCARB1 locus have identified additional links with other biomarkers, including lipoprotein-associated phospholipase A2 and vitamin E, suggesting the potential for horizontal pleiotropy (Box 1). Furthermore, both RCTs61–63 and MR studies16–18 have failed to show that increasing HDL-cholesterol levels can reduce cardiovascular disease risk. Therefore, whether HDL metabolism has a causal role in the aetiology of CHD remains speculative, and studies are now evaluating the function of HDL particles, as opposed to measuring only circulating HDL-cholesterol concentrations64–67.
Figure 4. Mendelian randomization using a variant that disrupts normal function of the exposure.
a | Reduced hepatic uptake of HDL particles through the scavenger receptors leads to the accumulation of circulating HDL cholesterol (HDL-C) and increased risk of coronary heart disease (CHD). However, this observation does not indicate that HDL-C is harmful, but provides some support for the notion that appropriate function of reverse cholesterol transport might be beneficial to cardiovascular health. b | A variant in IL6R leads to reduced membrane-bound IL-6, which in turn results in increased levels of circulating IL-6, disruption of classical IL-6 signalling, reduced C-reactive protein (CRP) levels, and a reduction in risk of CHD. These figures are schematic representations and should not be interpreted as formal directed acyclic graphs.
IL-6 signalling and risk of CHD
IL-6 is a proinflammatory cytokine produced by stromal and immune cells that circulates in the blood and binds to plasma membrane receptor complexes. IL-6 can exert its biological effect via two signalling mechanisms: classical signalling and trans-signalling. Classical signalling involves the binding of IL-6 to cellular membranes that express both the IL-6 receptor (IL6R) and glycoprotein-130 68. Most cells express glycoprotein-130, but only a limited number express IL6R. In trans-signalling, a soluble form of the IL6R binds to circulating IL-6 in the blood, and this IL-6/IL6R complex can then bind to any cell expressing glycoprotein-130. Since glycoprotein-130 is ubiquitous, trans-signalling can involve many more cell types than classical signalling. Classical IL-6 signalling is thought to have a more prominent role in the development of systemic diseases, whereas trans-IL-6 signalling might be more involved in local tissue inflammation69.
Using a nonsynonymous SNP (rs8192284) in the gene encoding IL6R, two studies have reported that variants associated with increased concentrations of circulating IL-6 are related to a reduction in CHD risk70,71. A naive interpretation, and one that the researchers explain is not the case, would be that IL-6 signalling is protective against CHD. However, this deduction would be at odds with the well-established role of IL-6 as an inflammatory cytokine72.
A genetic variant in IL6R (rs8192284) has also been shown to increase generation of soluble IL6R through increased proteolytic cleavage of membrane-bound IL6R, leading to a subsequent reduction in membrane-bound IL6R73,74. This reduction in turn leads to diminished IL-6-mediated classical signalling, and a shift from classical signalling to trans-signalling, effectively attenuating downstream classical signalling of IL-6. Consequently, decreased classical IL-6 signalling increases circulating IL-6 (owing to a reduction in membrane-bound IL6R and an increase in the circulating IL-6/IL6R complex), but a reduction in CRP levels (as classical IL-6 signalling is impaired; Figure 4B)75,76. Of note, the association of SNPs in IL6R with CRP concentrations in this setting reflects vertical pleiotropy. Although in no way does this observation indicate that CRP is causal for CHD, the association of SNPs in IL6R with CRP does not invalidate the use of IL6R in MR because, unlike in Figures 3A, B, CRP is downstream of the same pathway as IL-6. One way to dissect these relationships would be to use separate genetic instruments for IL-6 and CRP, and construct a causal framework involving molecular intermediates using mediation analysis77.
Multiple biomarkers on the same pathway
If a variant is associated with multiple dependent traits on the same pathway, and if those biomarkers have different roles in disease, paradoxical situations can arise (Figure 2C).
ALDH2 and alcohol consumption
Alcohol is metabolized in tissues and in the liver by the enzymes alcohol dehydrogenase 1B (ADH1B) and acetaldehyde dehydrogenase 2 (ALDH2). Metabolism of alcohol by ADH1B yields acetaldehyde, a group 1 human carcinogen78, which is rapidly metabolized by ALDH2 into acetate. When ADH1B and ALDH2 function normally, systemic and tissue concentrations of acetaldehyde are low. However, when ADH1B enzymatic function is increased, or when ALDH2 enzymatic function is impaired, acetaldehyde concentrations rise, resulting in symptoms such as flushing, nausea, and headache upon consumption of alcohol. A naturally occurring genetic variation in ALDH2 (rs671, in which carriage of the variant that influences enzyme activity is referred to as *2) that is present in East-Asian individuals, but not in white individuals (ALDH2*1*1) results in loss-of-function of ALDH2 in a dose-dependent fashion. Individuals who are homozygous for the normal variant can consume alcohol in the usual way, whereas those who are heterozygous can still consume alcohol, although they experience symptoms of acetaldehyde toxicity. However, individuals who are homozygous for the ALDH2*2 variant tend to consume almost no alcohol79, given the symptoms experienced by this population. Individuals who carry one copy of the ALDH2*2 variant experience a less severe flushing response as compared with individuals who carry two copies of the *2 variant80, and this is evidenced by differences in blood acetaldehyde levels for a given dose of alcohol81.
ALDH2 and hypertension
Given that alcohol consumption is associated with hypertension82, the ALDH2*2 variant can be used as a genetic instrument to assess the causal role of alcohol consumption on blood pressure levels83. As each additional ALDH2*2 allele reduces alcohol consumption in a dose-dependent manner (Figure 5), a conventional MR study using a per-allele genetic model for the *2 allele of ALDH2 will produce a valid causal estimate83. Among East-Asian individuals, women generally consume considerably lower amounts of alcohol compared with men; therefore, stratifying the MR analysis by sex can test one of the fundamental principles of MR: that the genetic instrument is acting through the exposure of interest84,85. Given that the genetic variant should associate with blood pressure only in the presence of alcohol, a larger association of the genetic variant with blood pressure should be seen in men versus women (because men consume more alcohol); this pattern has been reported in several studies83,86. The interaction between sex and ALDH2 genotype can be used as the instrumental variable to estimate the causal effect of alcohol on outcomes, and is a robust analytical strategy that circumvents the instrumental variables assumption of no pleiotropy79.
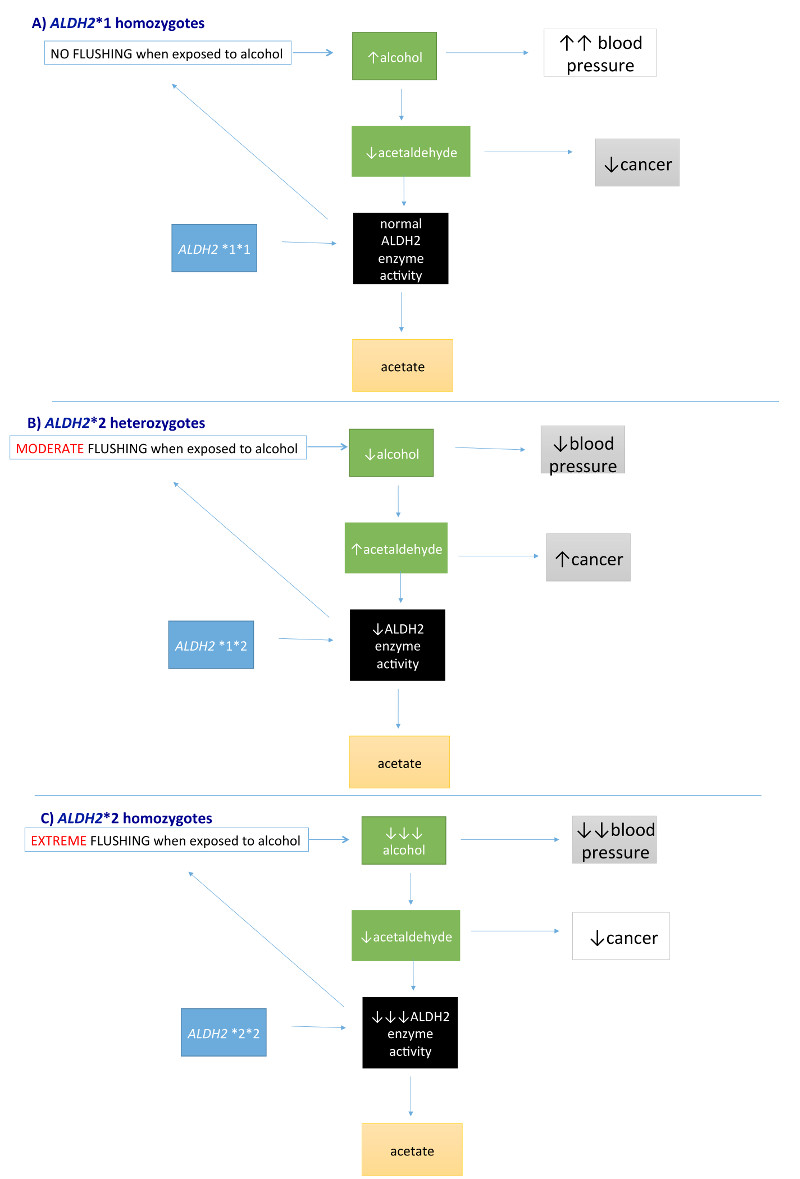
Figure 5. Mendelian randomization of biomarkers on the same pathway.
a | Individuals who are homozygous for the ALDH2*1 variant can consume normal amounts of alcohol without symptoms of flushing and nausea, which can lead to increased alcohol intake and subsequent high blood pressure. However, because acetaldehyde is efficiently cleared by aldehyde dehydrogenase (ALDH2), the risk of oesophageal cancer is low. b | Individuals who are heterozygous for ALDH2*2 are likely to consume lower amounts of alcohol than those who are homozygous for the ALDH2*1 variant, given their symptoms of moderate flushing. This lower alcohol consumption leads to lower blood pressure. Reduced functioning of ALDH2 leads to increased acetaldehyde levels, which in turn results in increased risk of oesophageal cancer. c | Individuals homozygous for ALDH2*2 consume almost no alcohol, given the severe symptoms and, therefore, blood pressure levels are expected to be lower than in those who are heterozygous for ALDH2*2 or homozygous for the ALDH2*1 variant. Similarly, acetaldehyde levels in individuals homozygous for ALDH2*2 are also expected to be lower than in individuals heterozygous for the ALDH2*2 variant, resulting in lower risk of oesophageal cancer (and similar to or lower than carriers of ALDH2*1*1, depending on alcohol consumption). These figures are schematic representations and should not be interpreted as formal directed acyclic graphs.
ALDH2 and oesophageal carcinoma
When investigating the association between ALDH2 and oesophageal carcinoma, a different interpretive framework is required. Individuals who drink the most (that is, those who are homozygous for the wild-type gene) might be expected to have the highest risk of oesophageal carcinoma; however, individuals who carry one copy of the *2 allele actually have the highest risk87. Although it might be tempting to interpret this observation as meaning that moderate drinkers having the highest risk of oesophageal carcinoma (a paradoxical scenario that is at odds with dose-dependent increase in risk with alcohol intake), this relationship can be explained by the effect of ALDH2 on both alcohol consumption and circulating concentrations of acetaldehyde81 (Figure 5).
Individuals who are heterozygous for ADLH2 rs671 (that is, carriers of the *1*2 allele) have the highest circulating concentrations of acetaldehyde (Figure 5B), despite the fact that they do not consume the greatest amount of alcohol (ALDH2 *1 homozygotes consume the greatest amounts; Figure 5A), nor do they have the genetic variant (like that carried by ALDH2 *2 homozygotes81) that conveys highest concentrations of acetaldehyde for a given amount of alcohol (Figure 5C). The highest absolute concentration of circulating and tissue acetaldehyde by genotype results in an increased risk of oesophageal carcinoma in *1*2 allele group compared with either the *1*1 allele or *2*2 allele groups. In this setting, the genetic variant influences both alcohol consumption and, among alcohol drinkers, acetaldehyde levels. For these reasons, a per-allele analytical model would be inappropriate for evaluation of causality88.
To summarize, the way in which a genetic variant might serve as an exposure measure for different traits depends on the outcome under investigation. For blood pressure, which seems not to be influenced in the long term by acetaldehyde levels, the variant is an instrument for causal inferences with respect to alcohol intake. For oesophageal cancer, the variant is an additional instrument for assessing the causal effects of acetaldehyde among consumers of alcohol. Without a good understanding of the biological basis of the effects of the genetic variant, misleading interpretations could be drawn from MR analyses.
Time-dependent exposures
If an exposure is time-dependent (for example, if an exposure is influential only during a period of development, such as during adolescence), then despite MR results suggesting a causal effect, modification of the exposure in later life will not necessarily alter the risk of disease (Figure 2D).
Vitamin D and multiple sclerosis
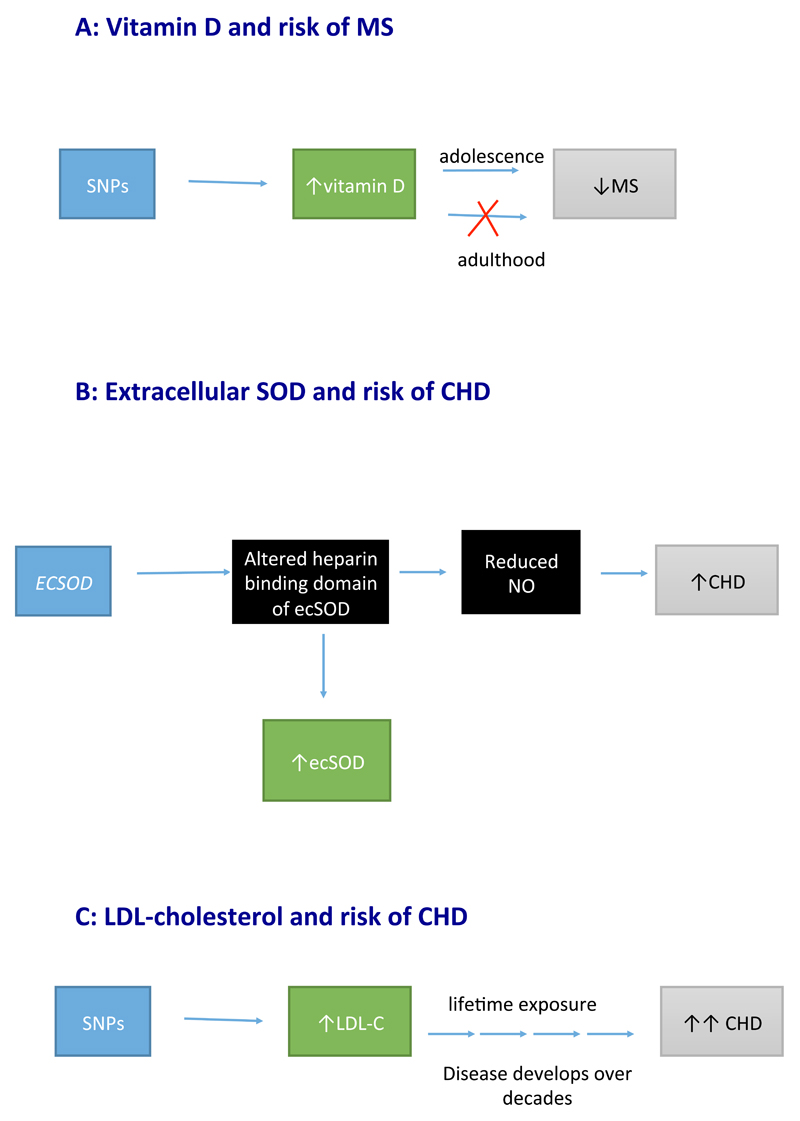
A MR study published in 2015 suggested a causal role of vitamin D in the aetiology of multiple sclerosis89. Although this example does not pertain directly to cardiovascular disease, we use it to introduce a general issue in MR analysis that might also apply for studies in cardiovascular disease. Previous observational studies of migration and risk of multiple sclerosis have consistently identified a time-dependent relationship between these two variables, indicating that sunlight exposure (and perhaps vitamin D levels) during early life, but not during adulthood, is associated with the risk of multiple sclerosis90–93. In this scenario, a MR study utilizing genetic variants associated with circulating levels of vitamin D would yield evidence in support of a protective role of vitamin D in the aetiology of multiple sclerosis (Figure 6A). However, modifying vitamin D levels after the critical time period (infanthood up to early adulthood) would not reduce the risk of multiple sclerosis. If this time period were proven to be critical for disease development, RCTs would need to be commenced in at-risk individuals (perhaps identified through family history or genetic risk scores) during childhood, as interventions commenced after this critical period would not be expected to influence disease risk.
Figure 6. Mendelian randomization of a time dependent and cumulative exposure.
a | The genetic variant-to-disease (multiple sclerosis) association reflects lifetime associations (including causal effects mediated through vitamin D that only occur during adolescence). Therefore, Mendelian randomization might provide evidence of a causal effect when this effect actually only occurs during a critical time period. b | The genetic variant alters the heparin-binding domain of extracellular superoxide dismutase (ecSOD), meaning that it cannot bind to the external membrane of endothelial cells, and cannot prevent nitric oxide (NO) from being degraded by superoxide anions. Less NO results in vasoconstriction and increased risk of coronary heart disease (CHD). c | Genetic variants instrumenting LDL cholesterol (LDL-C) have large effects on risk of CHD. Given that CHD is a disease that develops over decades, the effect estimates are equivalent to the estimates that would be derived from lifelong lowering of LDL-cholesterol levels. These figures are schematic representations and should not be interpreted as formal directed acyclic graphs.
Extracellular superoxide dismutase and CHD
Antioxidants have long been hypothesized to prevent CHD94; however, evidence from major RCTs conducted in adults have yielded largely neutral findings95,96. Extracellular superoxide dismutase (ecSOD) protects the nitric oxide released from smooth muscle cells from degradation by superoxide97,98. In preserving the function of nitric oxide, ecSOD facilitates the vasodilatation of arterioles, allowing maintenance of normotension99. Therefore, ecSOD is considered an endogenous antioxidant that has an important role in the development of vascular disease. A large population-based cohort study assessing a genetic variant that encodes a missense mutation (R231G in the ECSOD gene) reported that, contrary to expectations, the variant was associated with elevated levels of circulating ecSOD and linked to an increase in CHD risk100. The genetic variant altered the heparin- binding domain of ecSOD, affecting the binding capacity of ecSOD to the external membrane of endothelial cells (Figure 6B). As a result, ecSOD plasma levels increase, but the ecSOD cannot protect nitric oxide from degradation by superoxide anions98. Reduced bioavailability of nitric oxide can lead to hypertension and increased risk of CHD. This scenario is, therefore, another example of a seemingly paradoxical association between the concentration of the biomarker (ecSOD) in relation to its purported role in disease development, and supports the protective role of antioxidants in CHD.
Interestingly, other studies have shown that this same genetic variant associated with higher CHD risk is also associated with lower risk of lung disease101–103, albeit with some mixed findings104. However, this observation can be explained by findings from animal studies; the R231G SNP in the ECSOD gene has been shown to reduce ecSOD concentration in blood vessels, while concomitantly increasing ecSOD levels in alveolar fluids. Therefore, this variant might be associated with detrimental effects on the vasculature, but beneficial effects for lung function105. Notably, antioxidants might be important only at critical times during the development of vascular disease (unlike LDL cholesterol, which we discuss below). This critical time effect is consistent with other MR studies that have provided some, admittedly weak, evidence for a causal role for vitamin C deficiency in CHD development106, and might explain the possible discrepancy between these findings and the null findings from RCTs assessing vitamin C supplementation and risk of vascular disease in later life107.
Cumulative exposures
If an exposure accumulates to cause disease over many years, MR analyses might generate a causal estimate that is larger than that from a RCT (which alters exposure for only a limited time period) or from observational epidemiology (which generally captures the effect of the exposure for only a particular time period). In general, MR findings should be interpreted to reflect a lifelong exposure to a biomarker (Figure 2E), although this inference changes if an exposure occurs only after a certain age (for example, with alcohol or smoking, in which case the genetic instrument influences exposure only after the habit has been taken up)11. Furthermore, MR studies should demonstrate that the SNP associates with the biomarker across the lifetime, to allow appropriate interpretations to be made.
LDL cholesterol and CHD risk
An MR study using multiple independent genetic loci that influence concentrations of LDL cholesterol reported that a decrease in LDL-cholesterol level of 1 mmol/l results in >50% reduction in the risk of CHD39. This projection is approximately double the estimated effect reported in RCTs for a similar reduction in LDL-cholesterol level (25% reduction in the risk of a major coronary event for each 1 mmol/l reduction with statins)108. As such, this magnitude of effect from the MR study could be considered an overestimation109. However, the causal estimate from MR depicts lifelong exposure to a harmful trait (Figure 6C). Given that atherosclerosis is a disease that accumulates over a lifetime110,111 and the clinical effects of CHD generally present at advanced ages112, genetic variants provide an insight into the expected effect sizes if interventions were initiated from early childhood to reduce circulating LDL-cholesterol levels. Therefore, effect sizes from MR analyses should not be considered equivalent to those from an RCT of a short-term intervention. Differences in estimates from MR and RCTs can inform about disease latency and critical exposure periods.
Overlapping exposures
An emerging approach in MR analyses is the combination of multiple traits and genetic instruments into one model to try to tease out independent causal effects (so-called ‘multivariable MR’113. However, multivariable MR for discrete traits (for example, BMI and blood pressure in relation to cardiovascular disease) has several limitations, such as collider bias, in which the conditioning of a mediator between the exposure and outcome can induce new confounding by factors that are not related to the MR instrument before such conditioning114,115. This situation becomes more complex if the traits themselves overlap (that is, they contain the same element in their total value; Figure 2F).
Non-HDL cholesterol and triglycerides
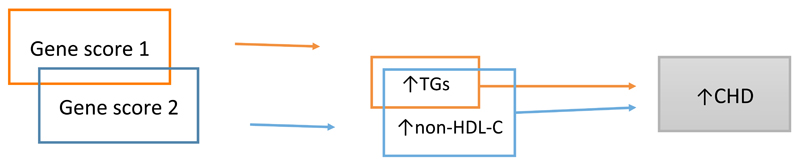
In a study by Helgadottir and colleagues, gene scores for non-HDL cholesterol and triglycerides were used to determine whether triglycerides have an independent causal role in CHD116. The researchers suggested that although LDL cholesterol (calculated using the Friedewald equation117) does not include cholesterol from triglyceride-rich lipoproteins (TRLs), non-HDL cholesterol does include this class of lipoproteins. However, estimation of LDL cholesterol via the Friedewald equation also includes intermediate-density lipoproteins118, and given that intermediate-density lipoprotein particles are semi-enriched in triglycerides, LDL cholesterol estimated via the Friedewald equation should also contain TRL-related cholesterol. A MR analysis that included both non-HDL cholesterol and triglycerides showed that although the association of non-HDL cholesterol with risk of CHD remained largely unaltered, the association of triglycerides with CHD after adjusting for non-HDL cholesterol diminished towards the null116.
In this scenario, non-HDL cholesterol and triglycerides should not be considered as discrete entities, but as overlapping entities (Figure 7). Therefore, adjustment of the triglyceride genetic instrument for non-HDL cholesterol adjusts for overlapping components, and the diminution of the triglyceride score should not be interpreted as meaning that triglycerides have no causal effect (that is, in effect, the multivariable analysis has adjusted triglycerides for a component of triglycerides contained within non-HDL cholesterol). By contrast, although non-HDL cholesterol consists of cholesterol in TRLs, it also contains LDL cholesterol, which has a triglyceride-independent effect on CHD18. Therefore, a lack of diminution of the association of the non-HDL cholesterol gene score with risk of CHD after adjustment for triglycerides provides no additional information beyond what is already understood about the causal role of LDL cholesterol in CHD, and is not useful for assessing the causality of triglycerides in CHD. By contrast, MR for correlated, but non-overlapping traits can be highly informative. For example, separate genetic instruments (albeit not using a multivariable MR framework) for CRP and IL-6 show that whereas IL-6 upregulates CRP119, causality for CHD is limited to IL-6 22,23,50,70,71.
Figure 7. Mendelian randomization of overlapping exposures.
As triglycerides (TGs) overlap with non-HDL cholesterol (non-HDL-C), adjusting the association of TGs with risk of coronary heart disease (CHD) for non-HDL-C diminishes the causal effect of TGs to null. By contrast, non-HDL-C contains the entire cascade of apolipoprotein B-containing lipoproteins, including intermediate-density lipoprotein cholesterol and LDL cholesterol, meaning that an association persists between non-HDL-C and CHD on adjustment for TGs. The attenuation of the TG–CHD association does not provide any information about the causality of TG, because it adjusts for an overlapping trait. This figure is a schematic representation and should not be interpreted as a formal directed acyclic graph.
Solutions for rigorous interpretations of MR
Although each example highlighted above is unique, they can be categorized into general themes, each of which has potential solutions to aid interpretation. First, when a single genetic variant associates with multiple traits on discrete pathways (horizontal pleiotropy), the use of this individual genetic variant to generate causal estimates for each individual trait is invalid, because it makes assumptions that each trait alone accounts for the causal effect. Furthermore, ascribing causal effects when using a single genetic variant to instrument a complex phenotype (such as FTO for BMI120) should be undertaken cautiously owing to the high likelihood of horizontal pleiotropy. This notion is especially true for non-protein (complex) traits, because no single genetic variant will tag a direct pathway to the exposure under investigation.
When an MR analysis generates associations that directly contradict what has been established from observational, epidemiological studies, investigators should consider whether the genetic variants used in the instrument disrupts the normal function of an exposure. If so, the association might arise owing to the biomarker not exerting its normal biological effect, and levels of this biomarker might be elevated despite this lower functional effect. An alternative explanation in this setting might be negative bias owing to unbalanced horizontal pleiotropy. By contrast, if the magnitude of effect from MR is directionally consistent but larger in magnitude than that seen in observational studies, an alternative explanation might be cumulative exposure, given that the genetic variant proxies a lifetime exposure. Alternative explanations include measurement error in the observational analysis (leading to regression dilution bias, from which MR analysis is protected) or a positive bias induced by horizontal pleiotropy.
In these examples, pleiotropy can seriously perturb estimates derived from MR analyses. Uncovering the presence of horizontal pleiotropy when using a single genetic variant is challenging. Detailed knowledge about the function of the variable, or access to large cohorts in which a phenome-wide association scan can reveal associations that might be indicative of unknown pleiotropy can guide interpretation. By contrast, when multiple SNPs are used in combination as genetic instruments, approaches now exist (such as MR-Egger; Box 4) to quantify and assess the presence of pleiotropy, and can provide valid causal estimates even in the presence of pleiotropy (although additional assumptions are required)53,54,79,113,121–123. These tests for pleiotropy should be used as sensitivity analyses in addition to conventional MR approaches.
When an MR study provides evidence of causality that has not been recapitulated in RCTs, one explanation is that the biomarker might be causal only during a particular time period of life. Therefore, the evidence obtained from MR studies might not translate into equivalent benefit if the intervention to modify the biomarker is at a different period of the life course from when it has its causal effect.
Finally, when assessing multiple traits in combination, if the traits are overlapping (that is, they contain elements of each other in their individual measures), then MR might not allow reliable individual assessment of which trait is causal. In this scenario, MR analyses are fraught with the same issues as conventional observational, epidemiological studies whereby adjusting for an overlapping trait diminishes associations of traits in the model with risk of disease.
Conclusions
In this Review, we have sought to illustrate and provide explanations for potentially paradoxical and implausible findings from MR analyses. As MR studies are increasingly performed to clarify causal relationships of the expanding number of traits that are measurable (such as ‘-omics’ analyses, including metabolomics44,124, lipidomics125, and proteomics126), these scenarios are likely to become more commonplace, highlighting the need for careful application and critical appraisal of MR findings. Indeed, as the relative ease of performing two-sample MR studies utilizing readily available data increases, the reliability of such studies are likely to decrease, through both methodological errors and through publication bias influencing which results are deemed ‘of interest’127. Despite these caveats, with increasing large-scale genetic data becoming available to facilitate two-sample MR128, together with resources such as MR-Base129, LD Hub130, and PhenoScanner131, MR promises to provide an efficient and pragmatic means to identify traits that are likely to be causal in cardiometabolic and other diseases, and to help to prioritize drug targets to take forward into therapeutic clinical trials132. Such drug-target prioritization might result in fewer failed multibillion dollar clinical trials and can contribute to improving the drug-development pipeline.
Key points.
Mendelian randomization (MR) is a powerful tool that utilizes genetic information to inform about the likely causal relevance of an exposure to an outcome
When performed rigorously, MR findings should be free from reverse causality bias, and only minimally affected by confounding
The number of MR studies has been increasing in the past decade, providing important new insights into disease aetiology
However, as MR studies become more common, and as increasingly complex gene-to-exposure and exposure-to-outcome relationships are investigated, reliable conduct and interpretation of MR analyses can be challenging
Potential solutions to aid the conduct and interpretation of MR studies can be derived, for example, through use of emerging statistical approaches to investigate potential genetic pleiotropy that can distort the findings
Acknowledgements
M.A.-K. is supported by Biocenter Oulu and Sigrid Juselius Foundation, Finland. M.A.-K. and G.D.S. work in a unit that receives funding from the University of Bristol and UK Medical Research Council (MC_UU_12013/1). M.V.H. works in a unit that receives funds from the University of Oxford and the UK Medical Research Council. These funding bodies did not have a role in the study design, decision to publish, or preparation of the manuscript.
Glossary
- Randomized, controlled trials (RCTs)
An interventional study in which individuals are randomized to an ‘exposed’ arm (for example, an active drug) or to a comparator control arm (for example, placebo) to test the efficacy of an intervention on an outcome (such as disease risk). Such trials are considered the gold standard for asserting causal relationships of an exposure to disease risk.
- Mendelian randomization
A genetic epidemiological approach that aims to quantify the causal relationship between an exposure and an outcome, by using the properties of the genome (randomized owing to Mendel’s second law, and invariant) to minimize the issues of confounding and reverse causality that can undermine traditional observational epidemiology.
- Vertical pleiotropy
The association of a genetic marker (for example, a single nucleotide polymorphism [SNP] in isolation, or a genetic instrument consisting of multiple SNPs) with more than one phenotype on the same biological pathway.
- Horizontal pleiotropy
The association of a genetic marker with more than one phenotype on discrete biological pathways.
- Genome-wide association study (GWAS)
The hypothesis-free investigation of hundreds of thousands to millions of genetic variants (typically single nucleotide polymorphisms) for their association with a phenotype to characterize the underlying genetic architecture of the phenotype.
- Linkage disequilibrium
The nonrandom assortment of genetic variants, meaning that when linkage disequilibrium between a pair of variants is high (for example, as measured by a r2 value of >0.80), a single nucleotide polymorphism (SNP) can be used as a ‘proxy’ for another SNP in the absence of this second SNP being directly genotyped.
- Unbalanced horizontal pleiotropy
When horizontal pleiotropy is such that alternative pathways from the genetic marker to disease can lead to distortion of the association of the exposure under investigation. Unbalanced horizontal pleiotrophy is a violation of the exclusion restriction assumption of an instrumental variable.
- Instrumental variable
A variable used as a proxy for an exposure of interest that is not associated with confounders and only associates with an outcome through the exposure of interest
- Friedewald equation
The estimation of LDL-cholesterol levels in the absence of its direct measurement, from total cholesterol, high-density lipoprotein cholesterol, and triglycerides
- Two-sample Mendelian randomization
The single nucleotide polymorphism (SNP)-to-exposure estimate is obtained from a separate dataset to that of the SNP-to-outcome estimate.
Biographies
Michael V. Holmes is a Senior Clinical Research Fellow in Cardiovascular Medicine at the University of Oxford, UK. After medical school and practising medicine within the NHS, he completed a Master's in Epidemiology at London School of Hygiene & Tropical Medicine and a PhD in Genetic Epidemiology from University College London, UK. He has led large-scale international collaborations to investigate alcohol, lipids, and drug targets for their causal role in cardiovascular disease using Mendelian randomization. Presently, he investigates physiological traits (including metabolomics) and environmental exposures for their causal role in cardiovascular and metabolic disease within the China Kadoorie Biobank.
Mika Ala-Korpela is a Professor of Computational Medicine at the Medical Faculty, University of Oulu, Finland and at the School of Social and Community Medicine, University of Bristol, UK. He received his PhD from the University of Oulu for a multidisciplinary thesis, shared by the Departments of Physics and Internal Medicine, for applications of NMR spectroscopy to human blood plasma and lipoprotein research. His current research is mainly in the area of systems epidemiology, focusing on the development and utilization of -omics technologies to improve molecular understanding of metabolic diseases, advancing metabolic phenotyping, and improving disease risk assessment.
George Davey Smith is Professor of Clinical Epidemiology at the University of Bristol, Director of the MRC Integrative Epidemiology Unit, and Scientific Director of the Avon Longitudinal Study of Parents and Children. As an epidemiologist, he has researched socioeconomic inequalities in health, the methodologies (and application) of meta-analysis, sexually transmitted disease, and HIV/AIDS risk in Nicaragua and India, and has contributed to the development of Mendelian randomization approaches and applied epigenetic epidemiology.
Footnotes
Competing interests statement
The authors declare no competing interests.
Author contributions
All authors researched data for the article, and made substantial contributions to discussion of content, writing, reviewing, and editing the manuscript before submission.
References
- 1.Fewell Z, Davey Smith G, Sterne JA. The impact of residual and unmeasured confounding in epidemiologic studies: a simulation study. Am J Epidemiol. 2007;166:646–655. doi: 10.1093/aje/kwm165. [DOI] [PubMed] [Google Scholar]
- 2.Davey Smith G, et al. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007;4:e352. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison RK. Phase II and phase III failures: 2013–2015. Nat Rev Drug Discov. 2016;15:817–818. doi: 10.1038/nrd.2016.184. [DOI] [PubMed] [Google Scholar]
- 4.Fordyce CB, et al. Cardiovascular drug development: is it dead or just hibernating? J Am Coll Cardiol. 2015;65:1567–1582. doi: 10.1016/j.jacc.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 5.Wang Q, et al. Effects of hormonal contraception on systemic metabolism: cross-sectional and longitudinal evidence. Int J Epidemiol. 2016;45:1445–1457. doi: 10.1093/ije/dyw147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corrao G, Rubbiati L, Bagnardi V, Zambon A, Poikolainen K. Alcohol and coronary heart disease: a meta-analysis. Addiction. 2000;95:1505–1523. doi: 10.1046/j.1360-0443.2000.951015056.x. [DOI] [PubMed] [Google Scholar]
- 7.Marmot M, Brunner E. Alcohol and cardiovascular disease: the status of the U shaped curve. BMJ. 1991;303:565–568. doi: 10.1136/bmj.303.6802.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kloner RA, Rezkalla SH. To drink or not to drink? That is the question. Circulation. 2007;116:1306–1317. doi: 10.1161/CIRCULATIONAHA.106.678375. [DOI] [PubMed] [Google Scholar]
- 9.Mukamal KJ, Rimm EB. Alcohol’s effects on the risk for coronary heart disease. Alcohol Res Health. 2001;25:255–261. [PMC free article] [PubMed] [Google Scholar]
- 10.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23:R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 12.Swerdlow DI, et al. Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. Int J Epidemiol. 2016;45:1600–1616. doi: 10.1093/ije/dyw088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans DM, Davey Smith G. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genomics Hum Genet. 2015;16:327–350. doi: 10.1146/annurev-genom-090314-050016. [DOI] [PubMed] [Google Scholar]
- 14.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 15.Ference BA, et al. Effect of long-term exposure to lower low-density lipoprotein cholesterol beginning early in life on the risk of coronary heart disease: a Mendelian randomization analysis. J Am Coll Cardiol. 2012;60:2631–2639. doi: 10.1016/j.jacc.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 16.Holmes MV, et al. Mendelian randomization of blood lipids for coronary heart disease. Eur Heart J. 2015;36:539–550. doi: 10.1093/eurheartj/eht571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voight BF, et al. Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet. 2012;380:572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White J, et al. Association of lipid fractions with risks for coronary artery disease and diabetes. JAMA Cardiol. 2016;1:692–699. doi: 10.1001/jamacardio.2016.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cholesterol Treatment Trialists’ Collaborators et al. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet. 2010;376:1670–1681. doi: 10.1016/S0140-6736(10)61350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins R, et al. Interpretation of the evidence for the efficacy and safety of statin therapy. Lancet. 2016;388:2532–2561. doi: 10.1016/S0140-6736(16)31357-5. [DOI] [PubMed] [Google Scholar]
- 21.Silverman MG, et al. Association between lowering LDL-C and cardiovascular risk reduction among different therapeutic interventions: a systematic review and meta-analysis. JAMA. 2016;316:1289–1297. doi: 10.1001/jama.2016.13985. [DOI] [PubMed] [Google Scholar]
- 22.C Reactive Protein Coronary Heart Disease Genetics Collaboration et al. Association between C reactive protein and coronary heart disease: Mendelian randomisation analysis based on individual participant data. BMJ. 2011;342:d548. doi: 10.1136/bmj.d548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zacho J, et al. Genetically elevated C-reactive protein and ischemic vascular disease. N Engl J Med. 2008;359:1897–1908. doi: 10.1056/NEJMoa0707402. [DOI] [PubMed] [Google Scholar]
- 24.Holmes MV, et al. Association between alcohol and cardiovascular disease: Mendelian randomisation analysis based on individual participant data. BMJ. 2014;349:g4164. doi: 10.1136/bmj.g4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagg S, et al. Adiposity as a cause of cardiovascular disease: a Mendelian randomization study. Int J Epidemiol. 2015;44:578–586. doi: 10.1093/ije/dyv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nordestgaard BG, et al. The effect of elevated body mass index on ischemic heart disease risk: causal estimates from a Mendelian randomisation approach. PLoS Med. 2012;9:e1001212. doi: 10.1371/journal.pmed.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emdin CA, et al. Genetic association of waist-to-hip ratio with cardiometabolic traits, type 2 diabetes, and coronary heart disease. JAMA. 2017;317:626–634. doi: 10.1001/jama.2016.21042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dale C, et al. Causal associations of adiposity and body fat distribution with coronary heart disease, stroke subtypes and type 2 diabetes: a Mendelian randomization analysis. Circulation. doi: 10.1161/CIRCULATIONAHA.116.026560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Look AHEAD Research Group et al. Cardiovascular effects of intensive lifestyle intervention in type 2 diabetes. N Engl J Med. 2013;369:145–154. doi: 10.1056/NEJMoa1212914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wurtz P, et al. Metabolomic profiling of statin use and genetic inhibition of HMG-CoA reductase. J Am Coll Cardiol. 2016;67:1200–1210. doi: 10.1016/j.jacc.2015.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swerdlow DI, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2014;385:351–361. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmes MV, et al. Secretory phospholipase A2-IIA and cardiovascular disease: a Mendelian randomization study. J Am Coll Cardiol. 2013;62:1966–1976. doi: 10.1016/j.jacc.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholls SJ, et al. Varespladib and cardiovascular events in patients with an acute coronary syndrome: the VISTA-16 randomized clinical trial. JAMA. 2014;311:252–262. doi: 10.1001/jama.2013.282836. [DOI] [PubMed] [Google Scholar]
- 34.Polfus LM, Gibbs RA, Boerwinkle E. Coronary heart disease and genetic variants with low phospholipase A2 activity. N Engl J Med. 2015;372:295–296. doi: 10.1056/NEJMc1409673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.STABILITY Investigators et al. Darapladib for preventing ischemic events in stable coronary heart disease. N Engl J Med. 2014;370:1702–1711. doi: 10.1056/NEJMoa1315878. [DOI] [PubMed] [Google Scholar]
- 36.Millwood IY, et al. Lipoprotein-associated phospholipase A2 loss-of-function variant and risk of vascular diseases in 90,000 Chinese adults. J Am Coll Cardiol. 2016;67:230–231. doi: 10.1016/j.jacc.2015.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Millwood IY, et al. A phenome-wide association study of a lipoprotein-associated phospholipase A2 loss-of-function variant in 90 000 Chinese adults. Int J Epidemiol. 2016;45:1588–1599. doi: 10.1093/ije/dyw087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talmud PJ, Holmes MV. Deciphering the causal role of sPLA2s and Lp-PLA2 in coronary heart disease. Arterioscler Thromb Vasc Biol. 2015;35:2281–2289. doi: 10.1161/ATVBAHA.115.305234. [DOI] [PubMed] [Google Scholar]
- 39.Ference BA, Majeed F, Penumetcha R, Flack JM, Brook RD. Effect of naturally random allocation to lower low-density lipoprotein cholesterol on the risk of coronary heart disease mediated by polymorphisms in NPC1L1, HMGCR, or both: a 2 × 2 factorial Mendelian randomization study. J Am Coll Cardiol. 2015;65:1552–1561. doi: 10.1016/j.jacc.2015.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt AF, et al. PCSK9 genetic variants and risk of type 2 diabetes: a Mendelian randomisation study. Lancet Diabetes Endocrinol. 2017;5:97–105. doi: 10.1016/S2213-8587(16)30396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ference BA, et al. Variation in PCSK9 and HMGCR and risk of cardiovascular disease and diabetes. N Engl J Med. 2016;375:2144–2153. doi: 10.1056/NEJMoa1604304. [DOI] [PubMed] [Google Scholar]
- 42.Lotta LA, et al. Association between low-density lipoprotein cholesterol-lowering genetic variants and risk of type 2 diabetes: a meta-analysis. JAMA. 2016;316:1383–1391. doi: 10.1001/jama.2016.14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guasch-Ferre M, et al. Metabolomics in prediabetes and diabetes: a systematic review and meta-analysis. Diabetes Care. 2016;39:833–846. doi: 10.2337/dc15-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soininen P, Kangas AJ, Wurtz P, Suna T, Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circ Cardiovasc Genet. 2015;8:192–206. doi: 10.1161/CIRCGENETICS.114.000216. [DOI] [PubMed] [Google Scholar]
- 45.Wurtz P, et al. Branched-chain and aromatic amino acids are predictors of insulin resistance in young adults. Diabetes Care. 2013;36:648–655. doi: 10.2337/dc12-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wurtz P, et al. Circulating metabolite predictors of glycemia in middle-aged men and women. Diabetes Care. 2012;35:1749–1756. doi: 10.2337/dc11-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wurtz P, et al. Metabolic signatures of insulin resistance in 7,098 young adults. Diabetes. 2012;61:1372–1380. doi: 10.2337/db11-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang TJ, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lotta LA, et al. Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: a Mendelian randomisation analysis. PLoS Med. 2016;13:e1002179. doi: 10.1371/journal.pmed.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elliott P, et al. Genetic loci associated with C-reactive protein levels and risk of coronary heart disease. JAMA. 2009;302:37–48. doi: 10.1001/jama.2009.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans DM, et al. Mining the human phenome using allelic scores that index biological intermediates. PLoS Genet. 2013;9:e1003919. doi: 10.1371/journal.pgen.1003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prins BP, et al. Investigating the causal relationship of C-reactive protein with 32 complex somatic and psychiatric outcomes: a large-scale cross-consortium Mendelian randomization study. PLoS Med. 2016;13:e1001976. doi: 10.1371/journal.pmed.1001976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–314. doi: 10.1002/gepi.21965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zanoni P, et al. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science. 2016;351:1166–1171. doi: 10.1126/science.aad3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ledford H. ‘Good’ cholesterol mutation linked to heart disease. Nature. 2016 http://www.nature.com/news/good-cholesterol-mutation-linked-to-heart-disease-1.19543?WT.mc_id=FBK_NA_1603_NEWSCHOLESTEROLMUTATION_PORTFOLIO.
- 57.Harb J. Why having too much ‘good’ cholesterol can actually be BAD for you. 2016 Daily Mail http://www.dailymail.co.uk/health/article-3384473/Why-having-good-cholesterol-actually-BAD-you.html.
- 58.Tall AR. An overview of reverse cholesterol transport. Eur Heart J. 1998;19(Suppl. A):A31–A35. [PubMed] [Google Scholar]
- 59.Rosenson RS, et al. Cholesterol efflux and atheroprotection: advancing the concept of reverse cholesterol transport. Circulation. 2012;125:1905–1919. doi: 10.1161/CIRCULATIONAHA.111.066589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, et al. Hepatic expression of scavenger receptor class B type I (SR-BI) is a positive regulator of macrophage reverse cholesterol transport in vivo. J Clin Invest. 2005;115:2870–2874. doi: 10.1172/JCI25327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barter PJ, et al. Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med. 2007;357:2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 62.Schwartz GG, et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N Engl J Med. 2012;367:2089–2099. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 63.Eli Lilly and Company. Lilly to discontinue development of evacetrapib for high-risk atherosclerotic cardiovascular disease. 2015 Lilly https://investor.lilly.com/releasedetail.cfm?ReleaseID=936130.
- 64.Rohatgi A, et al. HDL cholesterol efflux capacity and incident cardiovascular events. N Engl J Med. 2014;371:2383–2393. doi: 10.1056/NEJMoa1409065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anastasius M, et al. Cholesterol efflux capacity: an introduction for clinicians. Am Heart J. 2016;180:54–63. doi: 10.1016/j.ahj.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 66.Rosenson RS, et al. Dysfunctional HDL and atherosclerotic cardiovascular disease. Nat Rev Cardiol. 2016;13:48–60. doi: 10.1038/nrcardio.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kingwell BA, Chapman MJ, Kontush A, Miller NE. HDL-targeted therapies: progress, failures and future. Nat Rev Drug Discov. 2014;13:445–464. doi: 10.1038/nrd4279. [DOI] [PubMed] [Google Scholar]
- 68.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 69.Lissilaa R, et al. Although IL-6 trans-signaling is sufficient to drive local immune responses, classical IL-6 signaling is obligate for the induction of T cell-mediated autoimmunity. J Immunol. 2010;185:5512–5521. doi: 10.4049/jimmunol.1002015. [DOI] [PubMed] [Google Scholar]
- 70.IL6R Genetics Consortium Emerging Risk Factors Collaboration et al. Interleukin-6 receptor pathways in coronary heart disease: a collaborative meta-analysis of 82 studies. Lancet. 2012;379:1205–1213. doi: 10.1016/S0140-6736(11)61931-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Interleukin-6 Receptor Mendelian Randomisation Analysis Consortium. The interleukin-6 receptor as a target for prevention of coronary heart disease: a Mendelian randomisation analysis. Lancet. 2012;379:1214–1224. doi: 10.1016/S0140-6736(12)60110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med. 2005;352:1685–1695. doi: 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- 73.Rafiq S, et al. A common variant of the interleukin 6 receptor (IL-6r) gene increases IL-6r and IL-6 levels, without other inflammatory effects. Genes Immun. 2007;8:552–559. doi: 10.1038/sj.gene.6364414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mullberg J, et al. The soluble human IL-6 receptor. Mutational characterization of the proteolytic cleavage site. J Immunol. 1994;152:4958–4968. [PubMed] [Google Scholar]
- 75.Rose-John S. IL-6 trans-signaling via the soluble IL-6 receptor: importance for the pro-inflammatory activities of IL-6. Int J Biol Sci. 2012;8:1237–1247. doi: 10.7150/ijbs.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kleveland O, et al. Effect of a single dose of the interleukin-6 receptor antagonist tocilizumab on inflammation and troponin T release in patients with non-ST-elevation myocardial infarction: a double-blind, randomized, placebo-controlled phase 2 trial. Eur Heart J. 2016;37:2406–2413. doi: 10.1093/eurheartj/ehw171. [DOI] [PubMed] [Google Scholar]
- 77.Richmond RC, Hemani G, Tilling K, Davey Smith G, Relton CL. Challenges and novel approaches for investigating molecular mediation. Hum Mol Genet. 2016;25:R149–R156. doi: 10.1093/hmg/ddw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Secretan B, et al. A review of human carcinogens — part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol. 2009;10:1033–1034. doi: 10.1016/s1470-2045(09)70326-2. [DOI] [PubMed] [Google Scholar]
- 79.Cho Y, et al. Alcohol intake and cardiovascular risk factors: a Mendelian randomisation study. Sci Rep. 2015;5:18422. doi: 10.1038/srep18422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6:e50. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peng GS, Yin SJ. Effect of the allelic variants of aldehyde dehydrogenase ALDH2*2 and alcohol dehydrogenase ADH1B*2 on blood acetaldehyde concentrations. Hum Genomics. 2009;3:121–127. doi: 10.1186/1479-7364-3-2-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Marmot MG, et al. Alcohol and blood pressure: the INTERSALT study. BMJ. 1994;308:1263–1267. doi: 10.1136/bmj.308.6939.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen L, Davey Smith G, Harbord RM, Lewis SJ. Alcohol intake and blood pressure: a systematic review implementing a Mendelian randomization approach. PLoS Med. 2008;5:e52. doi: 10.1371/journal.pmed.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 85.Davey Smith G. Use of genetic markers and gene-diet interactions for interrogating population-level causal influences of diet on health. Genes Nutr. 2011;6:27–43. doi: 10.1007/s12263-010-0181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tabara Y, et al. Mendelian randomization analysis in three Japanese populations supports a causal role of alcohol consumption in lowering low-density lipid cholesterol levels and particle numbers. Atherosclerosis. 2016;254:242–248. doi: 10.1016/j.atherosclerosis.2016.08.021. [DOI] [PubMed] [Google Scholar]
- 87.Lewis SJ, Davey Smith G. Alcohol, ALDH2, and esophageal cancer: a meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev. 2005;14:1967–1971. doi: 10.1158/1055-9965.EPI-05-0196. [DOI] [PubMed] [Google Scholar]
- 88.Zuccolo L, Holmes MV. Commentary: Mendelian randomization-inspired causal inference in the absence of genetic data. Int J Epidemiol. 2016 doi: 10.1093/ije/dyw327. [DOI] [PubMed] [Google Scholar]
- 89.Mokry LE, et al. Vitamin D and risk of multiple sclerosis: a Mendelian randomization study. PLoS Med. 2015;12:e1001866. doi: 10.1371/journal.pmed.1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cabre P. Migration and multiple sclerosis: the French West Indies experience. J Neurol Sci. 2007;262:117–121. doi: 10.1016/j.jns.2007.06.044. [DOI] [PubMed] [Google Scholar]
- 91.Dean G, Elian M. Age at immigration to England of Asian and Caribbean immigrants and the risk of developing multiple sclerosis. J Neurol Neurosurg Psychiatry. 1997;63:565–568. doi: 10.1136/jnnp.63.5.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Elian M, Nightingale S, Dean G. Multiple sclerosis among United Kingdom-born children of immigrants from the Indian subcontinent, Africa and the West Indies. J Neurol Neurosurg Psychiatry. 1990;53:906–911. doi: 10.1136/jnnp.53.10.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dean G, Kurtzke JF. On the risk of multiple sclerosis according to age at immigration to South Africa. Br Med J. 1971;3:725–729. doi: 10.1136/bmj.3.5777.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Navab M, et al. The Yin and Yang of oxidation in the development of the fatty streak. A review based on the 1994 George Lyman Duff Memorial Lecture. Arterioscler Thromb Vasc Biol. 1996;16:831–842. doi: 10.1161/01.atv.16.7.831. [DOI] [PubMed] [Google Scholar]
- 95.Myung SK, et al. Efficacy of vitamin and antioxidant supplements in prevention of cardiovascular disease: systematic review and meta-analysis of randomised controlled trials. BMJ. 2013;346:f10. doi: 10.1136/bmj.f10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vivekananthan DP, Penn MS, Sapp SK, Hsu A, Topol EJ. Use of antioxidant vitamins for the prevention of cardiovascular disease: meta-analysis of randomised trials. Lancet. 2003;361:2017–2023. doi: 10.1016/S0140-6736(03)13637-9. [DOI] [PubMed] [Google Scholar]
- 97.Powers KM, Oberley LW, Domann FE. In: Oxidants in Biology: A Question of Balance. Valacchi G, Davis PA, editors. Springer; 2008. pp. 183–201. [Google Scholar]
- 98.Jung O, et al. Extracellular superoxide dismutase is a major determinant of nitric oxide bioavailability: in vivo and ex vivo evidence from ecSOD-deficient mice. Circ Res. 2003;93:622–629. doi: 10.1161/01.RES.0000092140.81594.A8. [DOI] [PubMed] [Google Scholar]
- 99.Gongora MC, et al. Role of extracellular superoxide dismutase in hypertension. Hypertension. 2006;48:473–481. doi: 10.1161/01.HYP.0000235682.47673.ab. [DOI] [PubMed] [Google Scholar]
- 100.Juul K, et al. Genetically reduced antioxidative protection and increased ischemic heart disease risk: The Copenhagen City Heart Study. Circulation. 2004;109:59–65. doi: 10.1161/01.CIR.0000105720.28086.6C. [DOI] [PubMed] [Google Scholar]
- 101.Siedlinski M, van Diemen CC, Postma DS, Vonk JM, Boezen HM. Superoxide dismutases, lung function and bronchial responsiveness in a general population. Eur Respir J. 2009;33:986–992. doi: 10.1183/09031936.00171507. [DOI] [PubMed] [Google Scholar]
- 102.Young RP, et al. Functional variants of antioxidant genes in smokers with COPD and in those with normal lung function. Thorax. 2006;61:394–399. doi: 10.1136/thx.2005.048512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Juul K, Tybjaerg-Hansen A, Marklund S, Lange P, Nordestgaard BG. Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2006;173:858–864. doi: 10.1164/rccm.200509-1387OC. [DOI] [PubMed] [Google Scholar]
- 104.Kobylecki CJ, Afzal S, Nordestgaard BG. Does SOD3 R213G homozygosity influence morbidity, mortality, and lung function in the general population? Antioxid Redox Signal. 2016;24:884–891. doi: 10.1089/ars.2016.6629. [DOI] [PubMed] [Google Scholar]
- 105.Hartney JM, et al. A common polymorphism in extracellular superoxide dismutase affects cardiopulmonary disease risk by altering protein distribution. Circ Cardiovasc Genet. 2014;7:659–666. doi: 10.1161/CIRCGENETICS.113.000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kobylecki CJ, Afzal S, Davey Smith G, Nordestgaard BG. Genetically high plasma vitamin C, intake of fruit and vegetables, and risk of ischemic heart disease and all-cause mortality: a Mendelian randomization study. Am J Clin Nutr. 2015;101:1135–1143. doi: 10.3945/ajcn.114.104497. [DOI] [PubMed] [Google Scholar]
- 107.Cook NR, et al. A randomized factorial trial of vitamins C and E and beta carotene in the secondary prevention of cardiovascular events in women: results from the Women’s Antioxidant Cardiovascular Study. Arch Intern Med. 2007;167:1610–1618. doi: 10.1001/archinte.167.15.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cholesterol Treatment Trialists’ Collaborators et al. The effects of lowering LDL cholesterol with statin therapy in people at low risk of vascular disease: meta-analysis of individual data from 27 randomised trials. Lancet. 2012;380:581–590. doi: 10.1016/S0140-6736(12)60367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Davey Smith G, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol. 2004;33:30–42. doi: 10.1093/ije/dyh132. [DOI] [PubMed] [Google Scholar]
- 110.McGill HC, Jr, et al. Origin of atherosclerosis in childhood and adolescence. Am J Clin Nutr. 2000;72:1307S–1315S. doi: 10.1093/ajcn/72.5.1307s. [DOI] [PubMed] [Google Scholar]
- 111.Strong JP, Malcom GT, Newman WP, III, Oalmann MC. Early lesions of atherosclerosis in childhood and youth: natural history and risk factors. J Am Coll Nutr. 1992;11(Suppl):51S–54S. doi: 10.1080/07315724.1992.10737984. [DOI] [PubMed] [Google Scholar]
- 112.Rosengren A, et al. Age, clinical presentation, and outcome of acute coronary syndromes in the Euroheart acute coronary syndrome survey. Eur Heart J. 2006;27:789–795. doi: 10.1093/eurheartj/ehi774. [DOI] [PubMed] [Google Scholar]
- 113.Burgess S, Thompson SG. Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects. Am J Epidemiol. 2015;181:251–260. doi: 10.1093/aje/kwu283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aschard H, Vilhjalmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am J Hum Genet. 2015;96:329–339. doi: 10.1016/j.ajhg.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Richiardi L, Bellocco R, Zugna D. Mediation analysis in epidemiology: methods, interpretation and bias. Int J Epidemiol. 2013;42:1511–1519. doi: 10.1093/ije/dyt127. [DOI] [PubMed] [Google Scholar]
- 116.Helgadottir A, et al. Variants with large effects on blood lipids and the role of cholesterol and triglycerides in coronary disease. Nat Genet. 2016;48:634–639. doi: 10.1038/ng.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- 118.Niemi J, et al. Estimation of VLDL, IDL, LDL, HDL2, apoA-I, and apoB from the Friedewald inputs — apoB and IDL, but not LDL, are associated with mortality in type 1 diabetes. Ann Med. 2009;41:451–461. doi: 10.1080/07853890902893392. [DOI] [PubMed] [Google Scholar]
- 119.Shah T, et al. Gene-centric analysis identifies variants associated with interleukin-6 levels and shared pathways with other inflammation markers. Circ Cardiovasc Genet. 2013;6:163–170. doi: 10.1161/CIRCGENETICS.112.964254. [DOI] [PubMed] [Google Scholar]
- 120.Fall T, et al. The role of adiposity in cardiometabolic traits: a Mendelian randomization analysis. PLoS Med. 2013;10:e1001474. doi: 10.1371/journal.pmed.1001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Burgess S, Dudbridge F, Thompson SG. Re: “Multivariable Mendelian randomization: the use of pleiotropic genetic variants to estimate causal effects”. Am J Epidemiol. 2015;181:290–291. doi: 10.1093/aje/kwv017. [DOI] [PubMed] [Google Scholar]
- 122.Hartwig FP, Davey Smith G, Bowden J. Robust inference in two-sample Mendelian randomisation via the zero modal pleiotropy assumption. Int J Epidemiol. doi: 10.1093/ije/dyx102. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bowden J, et al. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med. 2017;36:1783–1802. doi: 10.1002/sim.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ala-Korpela M, Davey Smith G. Metabolic profiling-multitude of technologies with great research potential, but (when) will translation emerge? Int J Epidemiol. 2016;45:1311–1318. doi: 10.1093/ije/dyw305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mundra PA, Shaw JE, Meikle PJ. Lipidomic analyses in epidemiology. Int J Epidemiol. 2016;45:1329–1338. doi: 10.1093/ije/dyw112. [DOI] [PubMed] [Google Scholar]
- 126.Ganz P, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. 2016;315:2532–2541. doi: 10.1001/jama.2016.5951. [DOI] [PubMed] [Google Scholar]
- 127.Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomisation: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 2016;6:1717–1726. doi: 10.1093/ije/dyx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Khera AV, Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet. 2017 doi: 10.1038/nrg.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hemani G, et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. bioRxiv. 2016 doi: 10.1101/078972. [DOI] [Google Scholar]
- 130.Zheng J, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Staley JR, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Davey Smith G, Paternoster L, Relton C. When will Mendelian randomization become relevant for clinical practice and public health? JAMA. 2017;317:589–591. doi: 10.1001/jama.2016.21189. [DOI] [PubMed] [Google Scholar]
- 133.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bush WS, Moore JH. Chapter 11: genome-wide association studies. PLoS Comput Biol. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Global Lipids Genetics Consortium et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Merino J, et al. Genetically driven hyperglycemia increases risk of coronary artery disease separately from type 2 diabetes. Diabetes Care. 2017;40:687–693. doi: 10.2337/dc16-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bowden J, et al. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol. 2016;45:1961–1974. doi: 10.1093/ije/dyw220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wurtz P, et al. Metabolic signatures of adiposity in young adults: Mendelian randomization analysis and effects of weight change. PLoS Med. 2014;11:e1001765. doi: 10.1371/journal.pmed.1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Emerging Risk Factors Collaboration et al. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302:1993–2000. doi: 10.1001/jama.2009.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Do R, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Casas JP, et al. PLA2G7 genotype, lipoprotein-associated phospholipase A2 activity, and coronary heart disease risk in 10 494 cases and 15 624 controls of European Ancestry. Circulation. 2010;121:2284–2293. doi: 10.1161/CIRCULATIONAHA.109.923383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cannon CP, et al. Ezetimibe added to statin therapy after acute coronary syndromes. N Engl J Med. 2015;372:2387–2397. doi: 10.1056/NEJMoa1410489. [DOI] [PubMed] [Google Scholar]
- 144.Myocardial Infarction Genetics Consortium Investigators et al. Inactivating mutations in NPC1L1 and protection from coronary heart disease. N Engl J Med. 2014;371:2072–2082. doi: 10.1056/NEJMoa1405386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sabatine MS, et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 146.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 147.Clarke R, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 148.Dewey FE, et al. Inactivating variants in ANGPTL4 and risk of coronary artery disease. N Engl J Med. 2016;374:1123–1133. doi: 10.1056/NEJMoa1510926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ridker PM, Pradhan A, MacFadyen JG, Libby P, Glynn RJ. Cardiovascular benefits and diabetes risks of statin therapy in primary prevention: an analysis from the JUPITER trial. Lancet. 2012;380:565–571. doi: 10.1016/S0140-6736(12)61190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sattar N, et al. Statins and risk of incident diabetes: a collaborative meta-analysis of randomised statin trials. Lancet. 2010;375:735–742. doi: 10.1016/S0140-6736(09)61965-6. [DOI] [PubMed] [Google Scholar]
- 151.Piepoli MF, et al. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts). Developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR) Eur Heart J. 2016;37:2315–2381. doi: 10.1093/eurheartj/ehw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yun KE, et al. Alcohol and coronary artery calcification: an investigation using alcohol flushing as an instrumental variable. Int J Epidemiol. 2017 doi: 10.1093/ije/dyw237. [DOI] [PubMed] [Google Scholar]