Summary paragraph
Nitrification, the oxidation of ammonia (NH3) via nitrite (NO2-) to nitrate (NO3-), is a key process of the biogeochemical nitrogen cycle. For decades, ammonia and nitrite oxidation were thought to be separately catalyzed by ammonia-oxidizing bacteria (AOB) and archaea (AOA), and by nitrite-oxidizing bacteria (NOB). The recent discovery of complete ammonia oxidizers (comammox) in the NOB genus Nitrospira1,2, which alone convert ammonia to nitrate, raised questions about the ecological niches where comammox Nitrospira successfully compete with canonical nitrifiers. Here we isolated the first pure culture of a comammox bacterium, Nitrospira inopinata, and show that it is adapted to slow growth in oligotrophic and dynamic habitats based on a high affinity for ammonia, low maximum rate of ammonia oxidation, high growth yield compared to canonical nitrifiers, and genomic potential for alternative metabolisms. The nitrification kinetics of four AOA from soil and hot springs were determined for comparison. Their surprisingly poor substrate affinities and lower growth yields reveal that, in contrast to earlier assumptions, not all AOA are most competitive in strongly oligotrophic environments and that N. inopinata has the highest substrate affinity of all analyzed ammonia oxidizer isolates except the marine AOA Nitrosopumilus maritimus SCM13. These results suggest a role of comammox organisms for nitrification under oligotrophic and dynamic conditions.
Nitrification links reduced and oxidized pools of inorganic nitrogen, is a pivotal step of wastewater treatment, reduces nitrogen fertilization efficiency in agriculture4, and produces the greenhouse gas nitrous oxide (N2O)5. A century after the discovery of AOB and NOB, landmark studies demonstrated that ammonia-oxidizing chemolithoautotrophic Thaumarchaeota are widespread and abundant in oxic environments6,7. Substrate affinity measurements on the first isolated AOA strain, Nitrosopumilus maritimus SCM16, revealed a half-saturation constant for NH3 two orders of magnitude below that of any cultured AOB and suggested that ammonia oxidation kinetics are a major driver of niche separation between AOA and AOB3. Comammox Nitrospira comprise a previously overlooked third group of aerobic ammonia oxidizers1,2. They appear to be environmentally widespread1,2, but the lack of pure comammox cultures has hampered their physiological characterization.
As previously reported, four years of enrichment yielded a culture containing the comammox organism Nitrospira inopinata (~60% of the community) and a betaproteobacterium (~40%)1. Sub-cultivation resulted in the isolation of N. inopinata (Supplementary Information and Extended Data Fig. 1a), which appeared mostly in cell aggregates with some individual cells and had a coiled spiral morphology (Extended Data Fig. 2a). Isolated N. inopinata oxidized NH3 stoichiometrically to NO3- with a transient accumulation of NO2- (Extended Data Fig. 2b) and showed optimal activity at 37 °C (Extended Data Fig 3a).
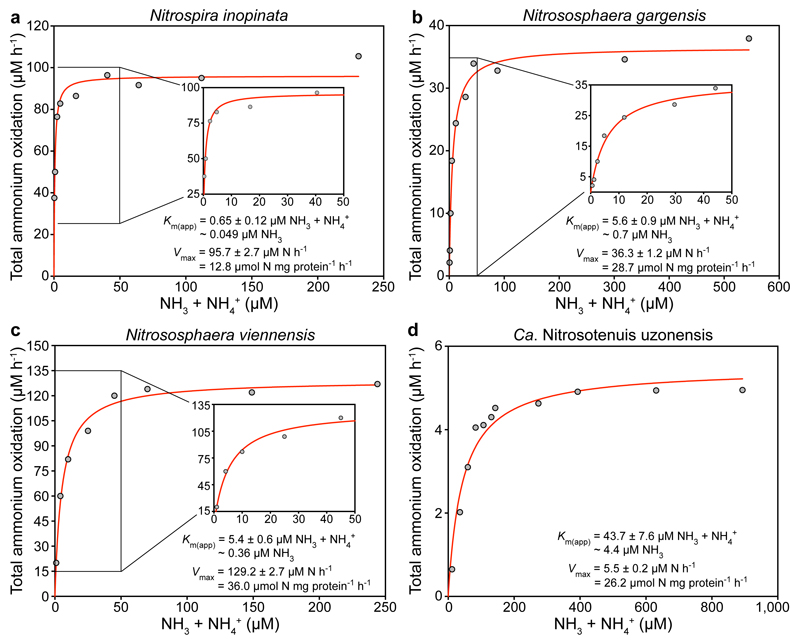
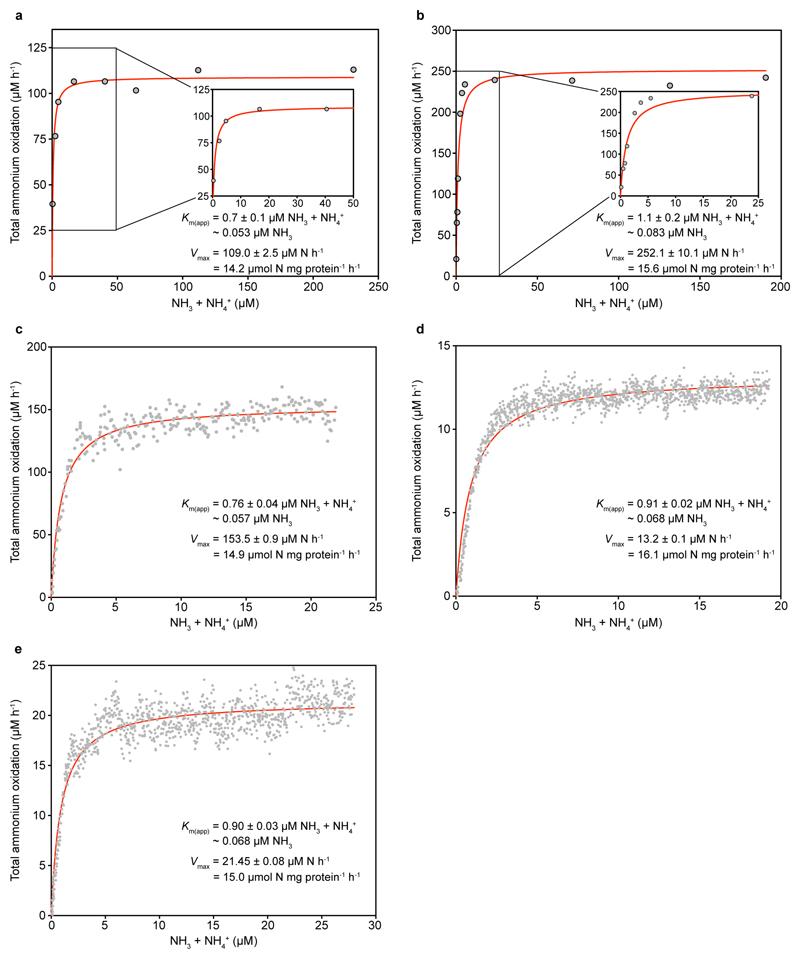
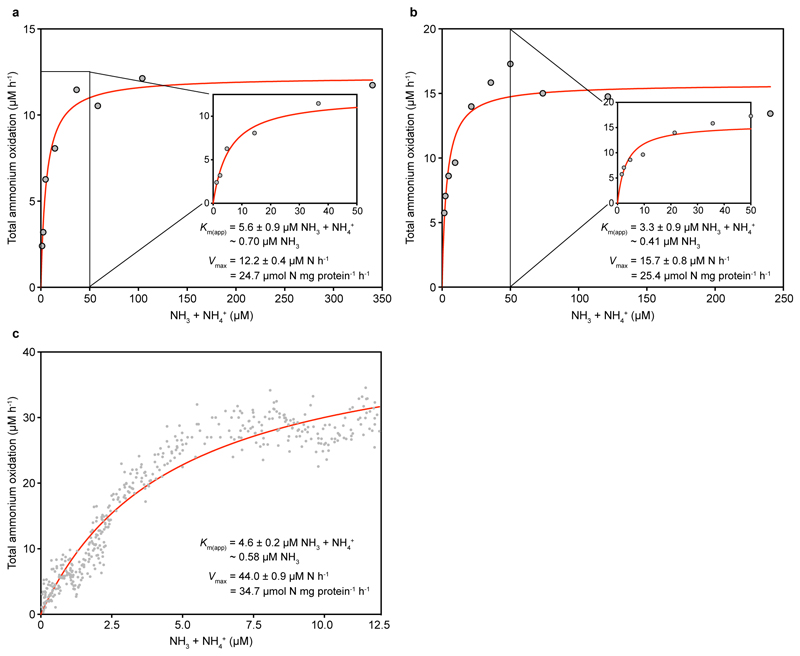
The pure N. inopinata culture enabled us to elucidate the nitrification kinetics of this first isolated comammox organism without interference by other microorganisms. We determined the rates of total ammonium (NH3 + NH4+) and NO2- oxidation from substrate-dependent oxygen (O2) consumption rates of early stationary phase batch cultures, which had depleted both total ammonium and NO2-. Ammonium-dependent O2 consumption reached a maximum rate within seconds of ammonium addition, and total ammonium and O2 were consumed with a stoichiometry of 1:2 (mean 1:2.03, s.d. 0.06, n = 3) as expected for complete nitrification. The calculated mean maximum oxidation rate of total ammonium (Vmax) was 14.8 μmol N mg protein-1 h-1 (s.d. 1.2, n = 6) (Fig. 1a and Extended Data Fig. 4). Remarkably, N. inopinata reached Vmax at total ammonium concentrations as low as 5 μM. Ammonia oxidation by N. inopinata followed Michaelis-Menten type kinetics, with a mean apparent half-saturation constant of Km(app) = 0.84 μM total ammonium (s.d. 0.17, n = 6; equivalent to ~63 ± 10 nM NH3) (Fig. 1a and Extended Data Fig. 4). This high affinity of N. inopinata for ammonia was confirmed using two techniques to determine Km(app), which consistently yielded Km(app) values of 0.65 to 1.1 μM total ammonium (~49 to 83 nM NH3) (Fig. 1a and Extended Data Fig. 4). Since substrate diffusion into the (albeit small) cell aggregates of N. inopinata may be rate limiting, we consider the measured Km(app) values as an upper estimate for the half-saturation constant.
Figure 1. Ammonia oxidation kinetics of N. inopinata and three ammonia-oxidizing Thaumarchaeota.
a-d, Michaelis-Menten plots for N. inopinata, N. gargensis, N. viennensis, and Ca. N. uzonensis. Total ammonium oxidation rates were calculated from microsensor measurements of NH3 dependent O2 consumption and discrete slopes over many substrate concentrations. Apparent half-saturation constants (Km(app)) and maximum oxidation rates (Vmax) for total ammonium were calculated by fitting the data to the Michaelis-Menten kinetic equation. The red curve indicates the best fit of the data. Standard errors of the estimates based on non-linear regression are reported. Data from biological replicates are shown in Extended Data Figures 4, 6, 7 and 8.
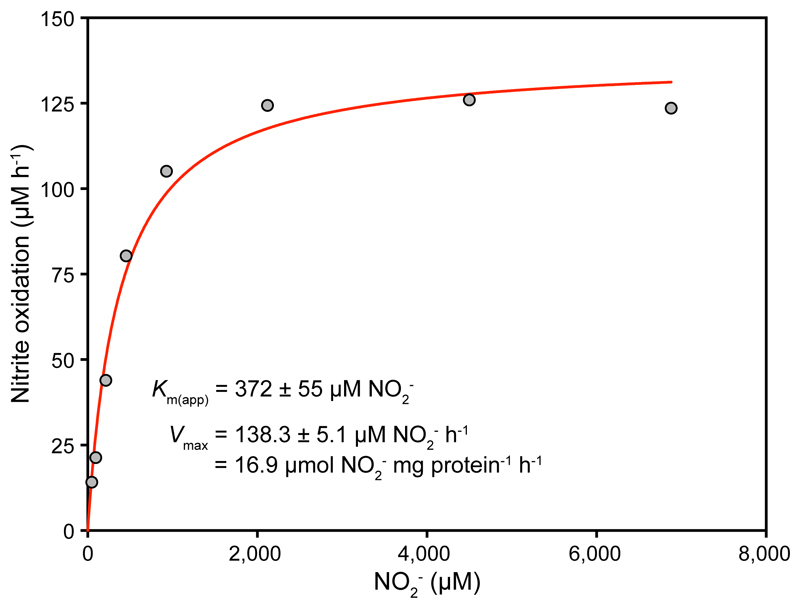
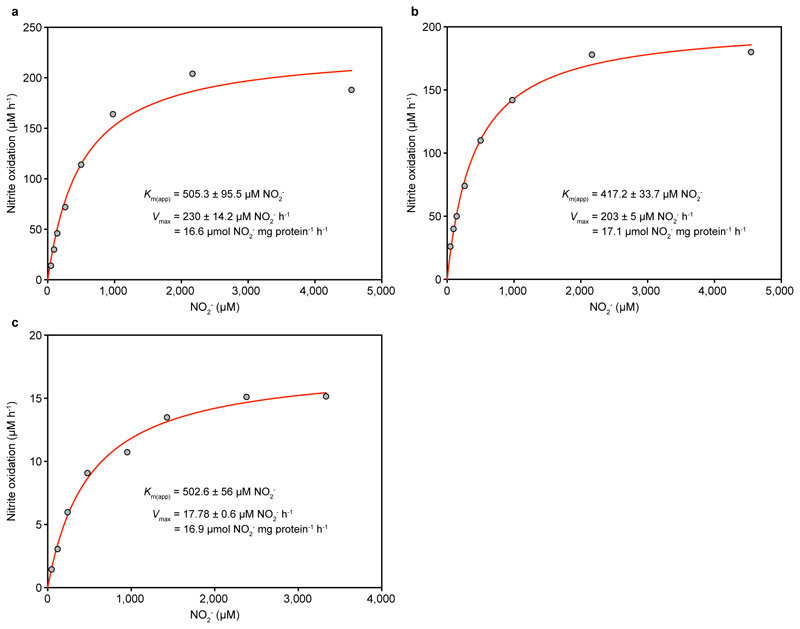
Nitrite oxidation by N. inopinata also followed Michaelis-Menten kinetics, and the maximum rate of nitrite-dependent O2 uptake was reached at high concentrations >2 mM NO2-. The stoichiometry of NO2- and O2 consumption was 2:1 as in canonical NOB8 (mean = 1.96:1, s.d. 0.03, n = 3). Interestingly, the mean Km(app) for NO2- (449.2 μM NO2-, s.d. 65.8, n = 4, Fig. 2 and Extended Data Fig. 5) was up to 50-fold larger than the known Km(app) values (9 to 27 µM NO2-) of canonical nitrite-oxidizing Nitrospira8,9. The poorer affinity for NO2- of N. inopinata resembles that of Nitrobacter hamburgensis, a nitrite oxidizer adapted to eutrophic conditions (Fig. 3), and is consistent with the transient accumulation of nitrite observed during complete nitrification (Extended Data Fig. 2b).
Figure 2. Nitrite oxidation kinetics of N. inopinata.
Rates of NO2- oxidation were calculated from microsensor measurements of NO2- dependent O2 consumption and discrete slopes over many substrate concentrations. The apparent half-saturation constant (Km(app)) and maximum oxidation rate (Vmax) for NO2- were calculated by fitting the data to the Michaelis-Menten kinetic equation. The red curve indicates the best fit of the data. Standard errors of the estimates based on non-linear regression are reported. Data from biological replicates are shown in Extended Data Figure 5.
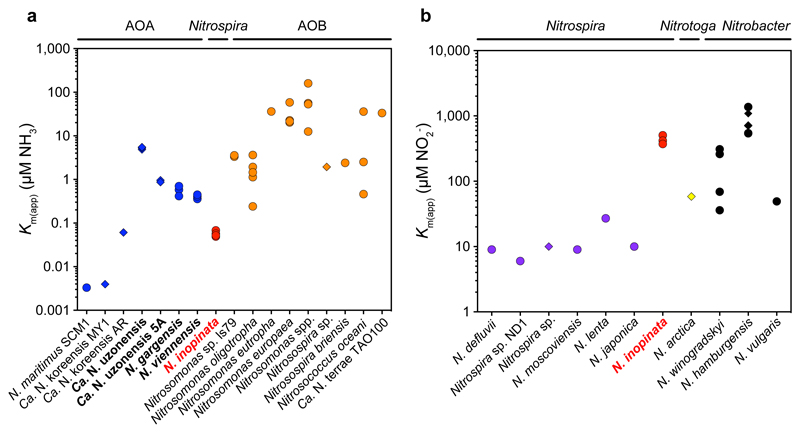
Figure 3. Comparison of the affinities for NH3 and NO2- between the comammox organism N. inopinata and canonical ammonia- and nitrite-oxidizing microbes.
a, Km(app) for NH3 of N. inopinata (red), AOA (blue), and AOB (orange). Km(app) values for NH3 were used when given, or calculated from reported total ammonium concentrations using the pH and temperature values provided from the respective studies. b, Km(app) for NO2- of N. inopinata (red), canonical Nitrospira (purple), Nitrotoga (yellow), and Nitrobacter (black). Km(app) values shown in a and b were derived from measurements with either pure cultures (circles) or enrichments (diamonds). Species for which affinities were determined in this study are depicted in bold. See Methods for references.
The Km(app) for NH3 of N. inopinata is 4 to 2,500 fold below values reported for AOB (Fig. 3a). Only the reported affinities for ammonia of N. maritimus SCM1 (Km(app) ~3 nM NH3), which is closely related to AOA living in oxic seawater containing <10 nM ammonium3, and of two AOA enrichment cultures of Candidatus (Ca.) Nitrosoarchaeum koreensis from marine sediments and agricultural soil10,11 exceed that of N. inopinata (Fig. 3a). However, substrate affinity determinations by microrespirometry in nitrifier enrichments can lead to overestimation (but not underestimation) of actual affinities, because of additional oxygen consumption by contaminating species, if the NH4+:O2 consumption stoichiometry is not determined. Therefore, N. maritimus remains the only nitrifier with a documented higher affinity for ammonia than N. inopinata. However, N. inopinata was cultured from biofilm covered by hot water1 and belongs to Nitrospira lineage II, which is usually not found in seawater1,12. Therefore, the ammonia oxidation kinetics of N. inopinata were compared to cultured AOA from soil and thermal habitats, which represent habitat types colonized by lineage II Nitrospira including comammox organisms1,12. We carried out the first kinetic analyses on two pure cultures of non-marine AOA: Nitrososphaera viennensis from soil13 and Nitrososphaera gargensis from a hot spring14 (both group I.1b Thaumarchaeota). The highly enriched AOA Ca. Nitrosotenuis uzonensis (group I.1a Thaumarchaeota) that was obtained from a thermal spring15 and enrichment ‘5A’ of a Ca. Nitrosotenuis uzonensis-related AOA (Extended Data Fig. 1b) recently obtained from the same source as N. inopinata were also analyzed.
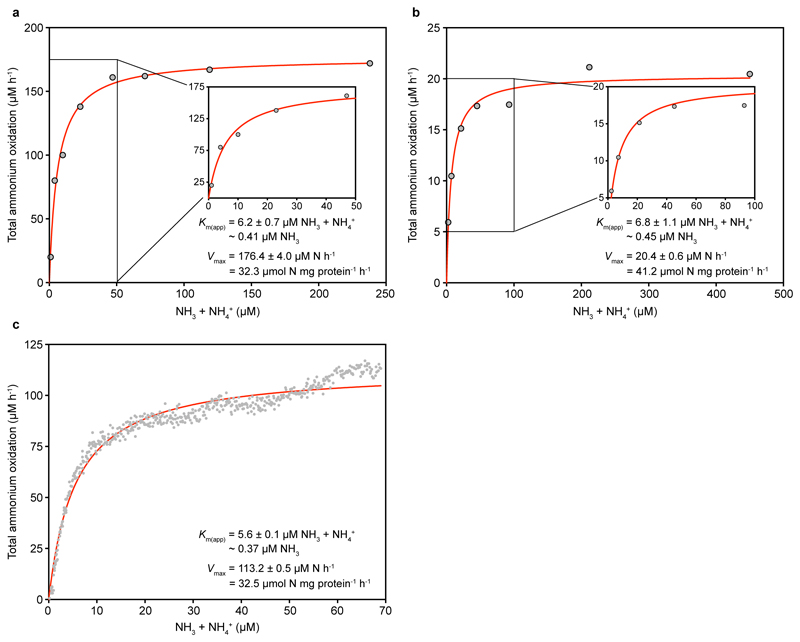
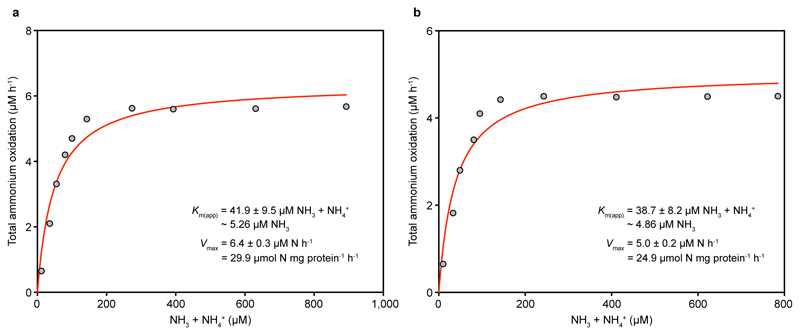
The mean Km(app) values of the two group I.1b AOA isolates of the genus Nitrososphaera (3.3 to 6.8 μM total ammonium, equivalent to ~410 to 703 nM NH3, n = 4 each) were one or two orders of magnitude larger than those of N. inopinata (P < 0.0001) and the group I.1a member N. maritimus SCM1, respectively (Fig. 1b and c, Fig. 3a, Extended Data Figures 6 and 7). For Ca. N. uzonensis (41.4 µM total ammonium, ~5.2 µM NH3, n = 3) (Fig. 1d, Fig. 3a, Extended Data Fig. 8) and the closely related strain 5A (8.6 to 9.5 µM total ammonium, ~870 to 960 nM NH3, n = 2), even higher mean Km(app) values were measured, showing variability of substrate affinity within group I.1a AOA. A comparison of the amoA, amoB, amoC, and amoX sequences from the N. gargensis pure culture (sequenced in 2014) to sequences from its first enrichment in 2008 (only amoA and amoB)16 and the first complete genome17 sequence obtained in 2010 revealed no differences, demonstrating that the relatively poor substrate affinity was not caused by mutations in the ammonia monooxygenase during this period of cultivation. The Km(app) values determined with enrichment cultures of Ca. N. uzonensis from 201315 and strain 5A, which has been cultured only since May 2016, also show that the poorer affinities of the AOA investigated here compared to N. maritimus SCM1 are not caused by adaptation during long-term cultivation.
Previous work has regarded differences in ammonia oxidation kinetics as a major driver of niche differentiation and competition between AOA and AOB18,19, with AOA assumed to all have an extremely high affinity for NH3 similar to that of N. maritimus SCM13. In contrast, our results reveal that the affinity of AOA for ammonia is not consistently orders of magnitude higher than the affinity of AOB. Instead, the known Km(app) values (0.3 to 4.0 μM NH3) of oligotrophic AOB belonging to Nitrosomonas cluster 6a are in the range we report for non-marine AOA (Fig. 3a). This finding demonstrates that predictions on the ecological niche separation of AOA and oligotrophic AOB are quite different than previously assumed.
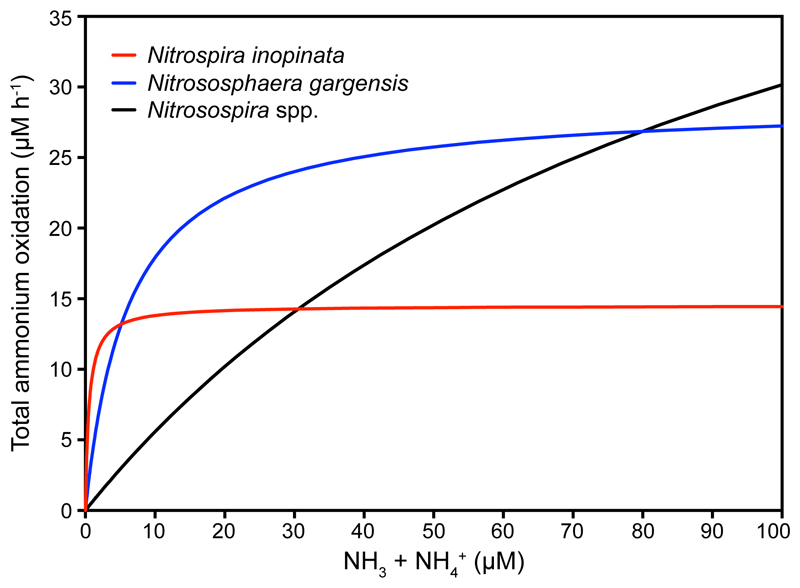
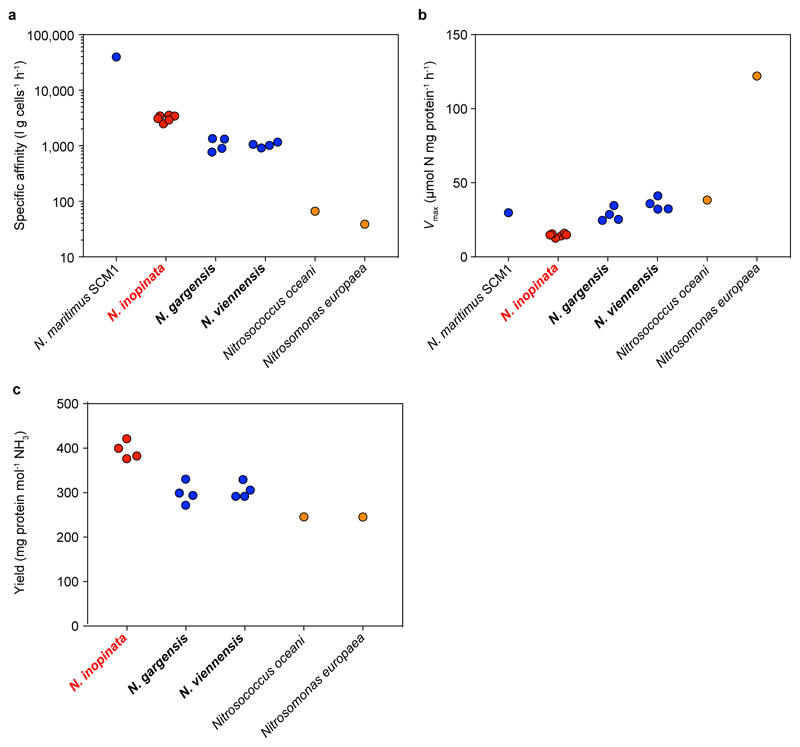
Intriguingly, the comammox organism N. inopinata has a lower Km(app) for NH3 than all isolated and kinetically characterized non-marine ammonia oxidizers (Fig. 3a). The ability of an organism to collect substrate from a dilute solution is best described by its specific affinity (ao)20. As shown by comparing the specific affinities for NH4+ of ammonia oxidizers (Extended Data Fig. 9a) and by simulating the competition among these organisms (Fig. 4), N. inopinata (mean ao = 3,162 l g cells-1 h-1, s.d. 409, n = 6) might outcompete all so-far investigated AOB and the tested non-marine AOA at the lowest ammonia concentrations (< 5 μM total ammonium).
Figure 4. Influence of total ammonium concentration on the rate of ammonia oxidation by N. inopinata, N. gargensis, and Nitrosospira spp. as described by ammonia oxidation kinetics.
For N. inopinata and N. gargensis, calculations were based on the kinetic parameters as determined in this study. For Nitrosospira spp., Km(app) and Vmax were inferred from the literature assuming a linear relationship between the rate of ammonia oxidation and the maximum specific growth rate. See Methods for references.
Theory predicts that complete ammonia oxidizers have a higher growth yield per mol ammonia oxidized than incomplete oxidizers, because their longer pathway offers additional ATP production from nitrite oxidation. A more efficient coupling between catabolic and anabolic reactions could also increase the growth yield. We demonstrated that N. inopinata has a growth yield per mol NH3 oxidized that is 32.3% and 29.7% higher than that of the AOA N. gargensis (P = 0.0003) and N. viennensis (P = 0.0004) (Extended Data Fig. 9c), respectively. Published yields of AOB are even lower than those of both AOA (Extended Data Figure 9c), likely reflecting the use of a more efficient CO2 fixation pathway by AOA compared to AOB21. The high yield, high substrate affinity, and comparatively low maximum oxidation rate (Extended Data Fig. 9b) suggest a niche for N. inopinata that is characterized by slow growth under highly oligotrophic conditions. These results lend support to an earlier hypothesis, which was based entirely on theoretical considerations and predicted that slow growth in ammonia-depleted biofilms, soils or sediments represents the fundamental niche of comammox organisms22.
Uncultured comammox Nitrospira are highly abundant in biofilms from groundwater wells, drinking water treatment systems, and freshwater biofilters exposed to bulk concentrations of NH4+ from ~4 to 60 µM1,23–26. Furthermore, recent amoA qPCR data showed that comammox Nitrospira were the most abundant ammonia oxidizers in a groundwater well containing on average 2 µM ammonium and they represented 12 to 30% of all detected ammonia oxidizers in a rice paddy soil, a forest soil and an activated sludge sample, providing further support for the environmental relevance of these ammonia oxidizers27. The mean Km(app) of in situ ammonia oxidation reported for grassland soils (12 nM NH3)28 is close to the substrate affinity of N. inopinata and would be consistent with an important role of comammox Nitrospira for nitrification in these soils, but comparisons of the Km(app) from heterogeneous soil environments with the Km(app) of a pure culture have to be interpreted with caution. However, like AOB and also AOA (Fig. 3a), the diverse1,2,27 comammox Nitrospira might span a broad range of adaptations to different substrate levels and it is impossible to extrapolate substrate affinities from a single or a few representatives of a group of ammonia oxidizers to all members of that group. Multiple environmental factors18, the ability to utilize additional sources of ammonia like cyanate and urea14,29, and alternative energy metabolisms30 also influence the niche partitioning and specialization of nitrifiers. Consistently, annotation of the whole genome sequence of N. inopinata suggested extensive metabolic versatility, including hydrogen and sulfide oxidation and fermentative metabolism (Supplementary Information). An encompassing understanding of the ecophysiology of both comammox Nitrospira and canonical nitrifiers will be essential for our perception of nitrogen cycling on our planet and for optimizing nitrification management strategies in drinking water treatment, wastewater treatment, and agriculture.
Methods
Studied ammonia-oxidizing microbes
The complete nitrifier N. inopinata1 isolated in this study; N. gargensis, an AOA strain isolated from a hot spring14,16; Ca. N. uzonensis, an AOA strain enriched from a hot spring15; Ca. N. uzonensis-related strain 5A enriched from hot well water (this study); and N. viennensis, an AOA strain isolated from soil13 were used in this study. N. gargensis (JCM 31473, DSM-103042) has been deposited in both the Japan Collection of Microorganisms (JCM) and the German Collection of Microorganisms and Cell Cultures (DSMZ). N. inopinata has been deposited in the JCM (JCM 31988).
Isolation of N. inopinata
A pure culture of the completely nitrifying bacterium N. inopinata was isolated from a previously described enrichment culture, ENR6, which contained N. inopinata and a betaproteobacterium affiliated with the family Hydrogenophilaceae1. Culture ENR6 had been obtained from a microbial biofilm that grew on the surface of a pipe and was covered by hot water (56 °C, pH 7.5), which was raised from a 1,200 m deep oil exploration well. Details of the sampling site and the procedure of enrichment have been provided elsewhere1. Successful isolation of N. inopinata was achieved through continued serial dilution to extinction as described previously1, with the only exception that the cultures were incubated at 42 instead of 46 °C. Purity of the culture was initially indicated by phase contrast microscopy showing a uniform cell morphology resembling that of N. inopinata1, and by the absence of heterotrophic growth in organic media (0.1× lysogeny broth, 0.1× R2A medium, or 0.1× tryptic soy broth) and in AO medium (see below) supplemented with either 20 mM thiosulfate or 5 mM sodium methanesulfonate (both substrates support the growth of the betaproteobacterium). In addition, fluorescence in situ hybridization (FISH) and quantitative PCR (qPCR) analyses of the culture were performed to confirm absence of the betaproteobacterial contaminant. FISH with 16S rRNA-targeted oligonucleotide probes was performed as described elsewhere31 using probes Ntspa662 and Ntspa712, which are specific for the genus and phylum Nitrospira32, and probe Nmir1009, which is specific for the betaproteobacterial contaminant1 in ENR6. Cells were counterstained by incubation for 5 min in a DAPI (4’,6-diamidino-2-phenylindole, 0.1 µg ml-1) solution. The qPCR tests were performed using primers Nino_amoA_19F and Nino_amoA_252R, which target the amoA gene of N. inopinata, and primers soxB_F1 and soxB_R1, which target the soxB gene of the betaproteobacterium, as described previously1. For qPCR, cell lysis for 10 min at 95 °C was followed by 40 PCR cycles of 30 s at 95 °C, 30 s at 53 °C, and 30 s at 72 °C. All qPCR reactions were performed using the iQ SYBR Green Supermix kit on a C1000 CFX96 real-Time PCR system (Bio-Rad). Finally, the purity of the N. inopinata culture was confirmed by deep Illumina sequencing (see below).
Enrichment of Ca. N. uzonensis-related strain 5A
The Ca. N. uzonensis-related AOA strain 5A was enriched from the same sampling site as N. inopinata in Aushiger, North Caucasus. Details of the site have been provided elsewhere1. One liter of hot water (52 °C, pH 7.6), which was raised from a 1,200 m deep oil exploration well, was filtered using membrane filters (0.2 µm pore size) on April 27, 2016. The retained particles were suspended in 40 ml of the sampling water and the sample was kept at 4 °C prior to cultivation. On May 10, 2016, 5 ml of the suspension was inoculated into a 100 ml Schott bottle containing 45 ml AO medium (see below) and 0.5 mM NH4Cl. The culture was incubated at 46 °C and fed with 0.5 to 1 mM NH4Cl when all ammonium had been consumed. Serial dilutions were performed 66 and 108 days after the initial inoculation. An aliquot of 5 ml was taken from the most diluted (10-7) ammonia-oxidizing culture from the second dilution series. To select for very small cells, this aliquot was filtered through a cellulose acetate filter (0.2 µm pore size, Thermo Scientific) into a 100 ml Schott bottle containing 20 ml AO medium. This filtered culture was propagated at 42 °C and supplied with 0.5 mM NH4Cl when all ammonium had been consumed. Observation by light microscopy revealed cells whose morphology resembled that of Ca. N. uzonensis15. Deep Illumina sequencing of the culture confirmed a high degree of enrichment of Thaumarchaeota (Extended Data Fig. 1b) closely related to Ca. N. uzonensis (99.93% 16S rRNA and 99.54% amoA gene sequence identity). The enriched AOA was tentatively named ‘strain 5A’.
Cultivation of N. inopinata and AOA
A medium for ammonia oxidizers (AO medium), whose composition and preparation are detailed elsewhere1, was used for the cultivation of ENR6, the isolation and further cultivation of N. inopinata, and the cultivation of all investigated AOA except N. viennensis. The desired concentration of NH4+ was adjusted by adding an autoclaved NH4Cl stock solution to autoclaved AO medium. N. viennensis was cultured in a freshwater medium (FWM)13 amended with 1 ml of sterile filtered (0.2 µm) non-chelated trace element mixture33, 1 ml of vitamin solution33, and 1 ml of Fe-NaEDTA solution13. Sterile filtered (0.2 μm pore size) sodium bicarbonate and sodium pyruvate were added to autoclaved FWM to final concentrations of 2 mM and 1 mM, respectively. The pH was adjusted to 7.6 by adding 10 ml l-1 HEPES buffer (1 M HEPES, 0.6 M NaOH)13,34. Unless otherwise stated, all nitrifier cultures were grown chemolithoautotrophically in the respective medium containing 1 mM NH4+ at 37 °C (N. inopinata), 42 °C (N. viennensis, Ca. Nitrosotenuis uzonensis and the Ca. Nitrosotenuis uzonensis-related enrichment 5A), or 46 °C (N. gargensis). N. inopinata and N. gargensis were cultured in 250 to 300 ml of AO medium in 1 l Schott bottles. The enrichment cultures of Ca. N. uzonensis and the Ca. N. uzonensis-related strain 5A were cultured in 20 to 50 ml of AO medium in 100 ml Schott bottles. N. viennensis was cultured in 50 ml of FWM in 250 ml Schott bottles. All cultures were grown in the dark and without agitation.
(Meta)genome sequencing and bioinformatics analyses of the N. inopinata pure culture and of the enriched Ca. N. uzonensis-related strain 5A
DNA was extracted from N. inopinata using the PowerSoil DNA Isolation Kit (MoBio) and from strain 5A using the FastDNA SPIN Kit for Soil (MP Biomedicals). For N. inopinata, sequencing libraries were prepared using the Nextera DNA Sample Preparation Kit (Illumina Inc.) following the manufacturer’s recommendation using 50 ng of input DNA. The prepared libraries were sequenced using an Illumina MiSeq with MiSeq Reagent Kit v3 (2×301 bp; Illumina Inc.). For Ca. N. uzonensis-related strain 5A, sequencing libraries were prepared using the NEBNext Ultra DNA Library Prep Kit (Illumina Inc.) following the manufacturer’s recommendation with 95.4 ng of input DNA. The prepared libraries were sequenced using an Illumina HiSeq with HiSeq Reagent Kit v4 (2×125 bp; Illumina Inc). Paired-end Illumina reads in the FASTQ format were imported into CLC Genomics Workbench v. 10.0 (CLCBio, Qiagen) and trimmed using a minimum phred score of 20 and a minimum length of 50 bp, allowing no ambiguous nucleotides and trimming off Illumina sequencing adaptors if found. All trimmed reads were co-assembled using CLCs de novo assembly algorithm, using a kmer of 63 and a minimum scaffold length of 1 kbp. The trimmed reads were mapped to the de novo assembled scaffolds using CLCs map reads to reference algorithm, with a minimum similarity of 90% over 80% of the read length. The metagenomic data set obtained from the Ca. N. uzonensis-related enrichment 5A was binned using coverage and the G+C content of DNA. Furthermore, for the N. inopinata pure culture the trimmed reads were mapped, by using similar settings, to an earlier metagenome assembly from N. inopinata enrichment culture ENR6 containing the previous betaproteobacterial contaminant1. The data was analyzed using mmgenome35.
Chemical analyses
Concentrations of ammonium were determined photometrically using a previously described assay36,37. Nitrite concentrations were measured photometrically by the acidic Griess reaction38. Nitrate was reduced to nitrite with vanadium chloride, measured as NOx by the Griess assay, and nitrate concentrations were calculated from the NOx measurements as described elsewhere39. Standards for the photometric assays were prepared in AO or FWM medium. All photometric analyses were performed using an Infinite 200 Pro spectrophotometer (Tecan Group).
Determination of temperature optima of activity
Specific ammonia oxidation rates of N. inopinata and N. gargensis were measured over ranges of incubation temperatures (30 to 50 °C for N. inopinata and 37 to 50 °C for N. gargensis). Biomass was harvested by centrifugation (9,000g, 15 min, 20 °C), washed with NH4+-free AO medium, and resuspended in AO medium containing 1 mM NH4+. Samples (0.5 ml) were taken regularly from triplicate cultures, centrifuged (21,000g, 15 min, 4 °C), and the supernatants were stored at -20 °C until chemical analysis (see above). Specific ammonia oxidation rates were determined by calculating the slope of the log-transformed NO2- (N. gargensis) or inverse NH4+ (N. inopinata) concentrations against time.
Measurements of substrate-dependent oxygen uptake
The ammonium concentrations in the respective nitrifier cultures were monitored daily during a period of 7 days prior to the instantaneous oxygen uptake measurements. To ensure that the organisms were analyzed during the early stationary phase, the cultures were sampled upon substrate depletion, which was predictable to within 2 to 3 h of stationary phase onset. Experiments with N. inopinata and N. viennensis were performed using concentrated or un-concentrated biomass to collect data across a range of cell concentrations. Cells were concentrated by gentle centrifugation (6 min, N. inopinata: 3,000g, N. viennensis: 6,000g). Since filtration or centrifugation of N. gargensis cells and filtration of Ca. N. uzonensis cells resulted in a complete loss of activity, experiments with these organisms were carried out using un-concentrated biomass. Oxygen uptake measurements were performed with a microrespiration (MR) system that consisted of a 2 ml glass MR chamber fitted with a MR injection lid, a glass coated stir bar, a microsensor multimeter (Unisense), and an OX-MR oxygen microsensor with 500 μm tip diameter (Unisense). Oxygen sensors were polarized continuously for >7 days before use, and all measurements were performed in a recirculating water bath at the optimum temperature for activity of each studied organism3. The optimum temperatures were 37 °C for N. inopinata (Extended Data Fig. 3a), 46 °C for N. gargensis (Extended Data Fig. 3b), 42 °C for N. viennensis34, and 46 °C for Ca. N. uzonensis15. Aliquots (~50 ml) of the cultures were incubated in a recirculating water bath before being sub-sampled into a 2 ml MR chamber with a stir bar. MR chambers were overflown with culture and closed with MR injection lids to ensure that no headspace was left inside the chambers. Full chambers were immersed in a recirculating water bath, and initial sensor equilibration (1 to 2 h) and stirring (200 to 400 rpm) were started. The sensor signal drift was stable for at least 10 min prior to initial substrate injections. A concentrated stock solution of substrate was injected into MR chambers with a Hamilton syringe through the port in the MR injection lid. For single trace oxygen uptake measurements, ammonium was injected once into a MR chamber and oxygen uptake was recorded until ammonium depletion. For multiple trace oxygen uptake measurements, discrete slopes of oxygen uptake were measured for 5 to 10 min for each individual injection of ammonium (the injections led to different start concentrations of ammonium in the MR chamber). Multiple trace oxygen uptake measurements with nitrite as substrate were performed as described for ammonium. After the oxygen uptake measurements, the total protein content in each MR chamber was determined photometrically using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific) according to the manufacturer’s ‘Enhanced Test-tube’ procedure. Cells were lysed with Bacterial Protein Extraction Reagent (Thermo Scientific) according to the manufacturer’s instructions prior to BCA quantification. Concentrations of ammonium, nitrite and nitrate were measured photometrically (see above) to confirm injected substrate concentrations and their oxidation to product.
Calculation of kinetic constants
The kinetic constants (Km(app) and Vmax) of N. inopinata, N. gargensis, and N. viennensis for ammonium were estimated from both single and multiple trace oxygen uptake measurements for comparison. Total numbers of biological replicates were n = 6 for N. inopinata, n = 4 for N. gargensis, and n = 4 for N. viennensis. The kinetics of Ca. N. uzonensis (biological triplicates) and of the closely related strain 5A (biological duplicates) were determined only from multiple trace oxygen uptake measurements, because single trace analyses were hampered by slow rates of ammonia oxidation. The kinetics of N. inopinata with nitrite were also analyzed only by multiple trace oxygen uptake experiments (four biological replicates). Single trace measurements with nitrite were not feasible because of the high Km(app) of N. inopinata for NO2- and the low solubility of O2 in water at 37 °C. Substrate oxidation rates were calculated from the measured oxygen uptake data according to a substrate to oxygen consumption stoichiometry of 1:1.5 for AOA, 1:2 for N. inopinata using ammonium, and 2:1 for N. inopinata using nitrite. For all analyzed organisms, the oxygen consumption stoichiometry was experimentally confirmed. The very low endogenous respiration rates were always taken into account. With the enrichments of Ca. N. uzonensis and of the Ca. N. uzonensis-related strain 5A, a stoichiometry between 1:1.50 and 1:1.53 was obtained, indicating that any contribution of accompanying heterotrophic organisms to respiration by these enrichments was very small (equal to or less than 3% of the total oxygen consumed). Michaelis-Menten plots of total ammonium (NH3 + NH4+), ammonia (NH3), or nitrite oxidation rates versus substrate concentration were obtained by fitting a Michaelis-Menten model to the data using the equation V = (Vmax × [S]) × (Km + [S])-1. Here V is rate, Vmax is the maximum rate (μM h-1), Km is the half saturation constant (μM), and [S] is the substrate concentration (μM). Specific substrate affinity (ao, l g wet cells-1 h-1) was calculated using the equation ao = Vmax × Km-1. Here Vmax is the maximum rate (in g substrate g wet cells-1 h-1) and Km is the half saturation constant (in g substrate). For conversion, the factor of 5.7 g wet weight per g of protein was used40. For N. inopinata only, the measured nitrite oxidation rate was taken into account when calculating the total ammonium (NH3 + NH4+) and ammonia (NH3) oxidation rates. Km(app) values of other nitrifiers shown in Fig. 3 were compiled from the literature3,8–11,41–53. If only total ammonium (NH4 + NH3) information was given by the authors for Km(app), the corresponding ammonia (NH3) values were calculated based on the reported experimental temperatures and pH values54.
Determination of molar growth yield
N. inopinata and N. gargensis were grown at their optimum temperatures of 37 °C and 46 °C, respectively (Extended Data Fig. 3), in 100 ml of AO medium in 250 ml Schott bottles, without agitation in the dark. N. viennensis was grown at 42 °C in 100 ml of FWM medium in 500 ml Schott bottles, without agitation in the dark. For all organisms, ammonium-depleted biomass was used as inoculum to avoid transfer of residual ammonium. The starting concentration of ammonium in all cultures was 500 μM. The ammonium levels in all cultures were measured twice per day, and cultures were re-fed with 500 μM NH4+ once all ammonium had been converted to nitrite or nitrate, respectively. In total, N. gargensis consumed 2 mM NH4+ whereas N. inopinata and N. viennensis consumed 1.5 mM NH4+. The total protein content was determined as described for the substrate-dependent oxygen uptake measurements. It was measured at the beginning of the experiments and immediately after all of the substrate had been consumed (264 h for N. inopinata, 192 h for N. gargensis, and 120 h for N. viennensis). The growth yields in mg protein were normalized to the mol substrate (NH3) consumed, taking into account the amount of protein present at the beginning of each experiment. All experiments were performed in parallel with four biological replicates.
Replication of physiological experiments and statistical analyses
All physiological experiments were conducted with at least three and up to six biological replicates. The numbers of replications are detailed in the subsections for each specific experiment. The supplementary experiments with the newly enriched Ca. N. uzonensis-related strain 5A could only be performed in duplicates because of limiting biomass availability. The experiments were not randomized. No statistical methods were used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment. Statistical significance of differences between N. inopinata, N. gargensis, and N. viennensis for Km(app), Vmax, substrate affinity (ao), and molar growth yield was assessed using pairwise one-way ANOVA tests with correction for multiple comparisons using the Tukey-Kramer method. For Km(app) only, values were log transformed prior to statistical analysis due to the order of magnitude difference in the determined values. The Brown-Forsythe test was used to assess variance in different sample groups for each one-way ANOVA test.
Simulation of competition among N. inopinata, N. gargensis, and soil Nitrosospira spp
The Km(app) and Vmax values (Vmax normalized to protein) determined in this study from physiological measurements were linearized into Lineweaver-Burk plots. The best fit linear regression line was then used to generate consensus Michaelis-Menten best fit curves for N. inopinata and N. gargensis. Normalized, best fit N. inopinata Km(app) and Vmax values were 0.65 μM and 14.5 μM h-1; N. gargensis Km(app) and Vmax values were 5.5 μM and 28.5 μM h-1. For soil Nitrosospira spp., reported Km(app) values were obtained from previous literature18,45,55 (~90 μM NH3 + NH4+), which are well within the range of oligotrophic AOB such as N. oligotropha and Nitrosomonas sp. Is79 (75 to 105 μM NH3 + NH4+). The Vmax values for soil Nitrosospira spp. were inferred from reported specific growth rate (μmax) values (actual value = 0.05 h-1), assuming a linear relationship between the rate of ammonium oxidation and μmax. The inferred value for Vmax for soil Nitrosospira spp. was 59.1 μM h-1. The final best fit Lineweaver-Burk plots were then charted together on a Michaelis-Menten plot.
Electron Microscopy
For scanning electron microscopy, N. inopinata cells were fixed on poly-L lysine coated slides with a filter-sterilized 2.5% glutaraldehyde fixation solution in cacodylate buffer (25 mM sodium cacodylate, 0.7 mM MgCl2, pH 7.0) after letting the cells adhere to the slide by placing it into an active culture for 3 days. Subsequently, fixed cells were washed three times for 10 min in cacodylate buffer and were post-fixed with a 1% OsO4 solution in cacodylate buffer for 40 min. The fixed cells were again washed three times in cacodylate buffer and dehydrated in a 30 to 100% (v/v) ethanol series, washed in acetone, and critical point dried with a CPD 300 unit (Leica). Samples were mounted on stubs, sputter coated with gold using a sputter coater JFC-2300HR (JEOL), and images were obtained with a JSM-IT300 scanning electron microscope (JEOL).
Data availability
Sequence data that support the findings of this study have been deposited in the European Nucleotide Archive (ENA) with the accession codes PRJEB20196 (for the raw sequence data from the N. inopinata pure culture) and PRJEB20386 (for the raw metagenome sequence data from the Ca. N. uzonensis-related AOA enrichment 5A). The pure culture of N. inopinata has been deposited at the Japan Collection of Microorganisms with the accession code JCM 31988. Source data for Extended Data Figures 2b, 3, and 9 are provided with the paper.
Extended Data
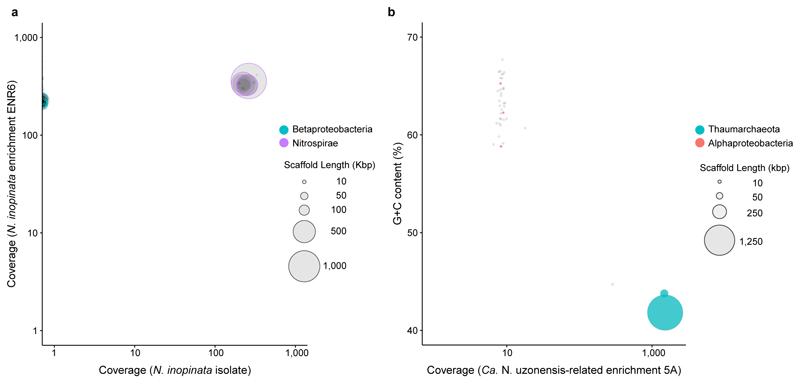
Extended Data Figure 1. Binning of the metagenome scaffolds from nitrifier cultures.
Circles represent scaffolds, scaled by the square root of their length. Only scaffolds ≥5 kbp are shown. Clusters of similarly coloured circles represent potential genome bins. a, Sequence composition-independent binning of the scaffolds from the N. inopinata isolate. The previous enrichment culture ENR6, which contained N. inopinata and only one contaminating betaproteobacterial organism1, was used for comparison in differential coverage binning. The lack of scaffolds from the betaproteobacterium (coverage = 0) shows the absence of this organism in the pure culture of N. inopinata. To further confirm that no trace amounts of the betaproteobacterium were left in the pure culture, the trimmed sequence reads were also mapped to the ENR6 metagenomic assembly that contained the contaminant1. None of 5.8 million reads mapped to the betaproteobacterium, whereas 99.8% of reads mapped to the closed genome of N. inopinata. Purity of the N. inopinata culture was also confirmed by fluorescence in situ hybridization, quantitative PCR targeting the amoA gene of N. inopinata and the soxB gene of the betaproteobacterium1, and the absence of growth in an organic medium. b, Binning of the scaffolds from the Ca. N. uzonensis-related enrichment culture 5A based on coverage and the G+C content of DNA. Aside from the Ca. N. uzonensis-related AOA strain 5A (99.93% 16S rRNA sequence identity), the culture contained an alphaproteobacterium at low abundance. Since the genome coverage of strain 5A was 1,512× and that of the alphaproteobacterium was 8×, the relative abundance of strain 5A in the enrichment culture was 99.5%.
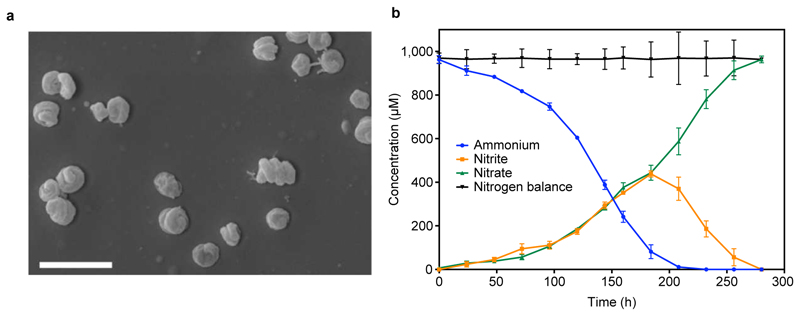
Extended Data Figure 2. Morphology and completely nitrifying activity of the N. inopinata isolate.
a, Scanning electron micrograph of spiral-shaped N. inopinata cells. The cells had a diameter of 0.2 to 0.3 μm and length of 0.7 to 1.7 μm. The scale bar represents 2 µm. b, Near-stoichiometric oxidation of 1 mM ammonium to nitrate with transient accumulation of nitrite. Data points show means, error bars show 1 s.d. of n = 3 biological replicates. If not visible, error bars are smaller than symbols.
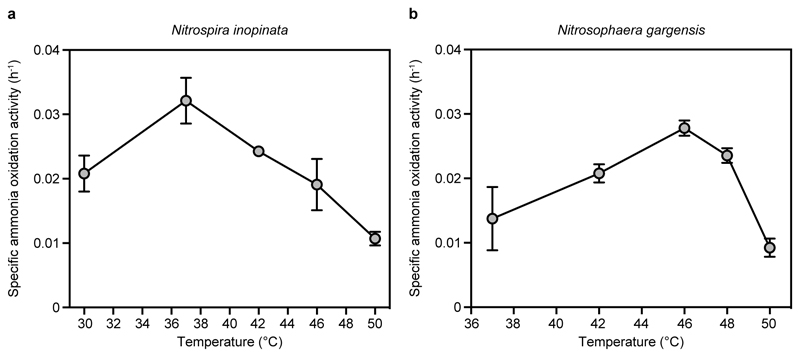
Extended Data Figure 3. Temperature optima of the ammonia-oxidizing activity of N. inopinata and N. gargensis.
a, Ammonia oxidation by N. inopinata over a temperature range from 30 to 50 °C. Since the optimum for activity was at 37 °C, kinetic analyses of N. inopinata were performed at this temperature. b, Ammonia oxidation by N. gargensis over a temperature range from 37 to 50 °C. Since the optimum for activity was at 46 °C, kinetic analyses of N. gargensis were performed at this temperature. Data points in a and b show means, error bars show 1 s.e.m. of n = 3 biological replicates. If not visible, error bars are smaller than symbols.
Extended Data Figure 4. Ammonia oxidation kinetics of N. inopinata.
Apparent half-saturation constants (Km(app)) and maximum oxidation rates (Vmax) for total ammonium were determined by fitting the data to the Michaelis-Menten kinetic equation. The red curve indicates the best fit of the data. Standard errors of the estimates based on non-linear regression are reported. See Methods for details of experiments and calculations. a, b, Total ammonium oxidation rates were calculated from microsensor measurements of NH3 dependent O2 consumption and discrete slopes over many substrate concentrations. Results for two biological replicates are shown here; a third biological replicate is shown in Fig. 1a. c-e, Total ammonium oxidation rates were calculated from microsensor measurements of NH3 dependent O2 consumption in single trace measurements. Results for three biological replicates are shown.
Extended Data Figure 5. Nitrite oxidation kinetics of N. inopinata.
a-c, Nitrite oxidation rates were calculated from microsensor measurements of NO2- dependent O2 consumption and discrete slopes over many substrate concentrations. Results for three biological replicates are shown here; a fourth biological replicate is shown in Fig. 2. Apparent half-saturation constants (Km(app)) and maximum oxidation rates (Vmax) for nitrite were determined by fitting the data to the Michaelis-Menten kinetic equation. The red curve indicates the best fit of the data. Standard errors of the estimates based on non-linear regression are reported. See Methods for details of experiments and calculations. Single trace measurements to analyse nitrite oxidation were not feasible because of the high Km(app) of N. inopinata for NO2- and the low solubility of O2 in water at 37 °C.
Extended Data Figure 6. Ammonia oxidation kinetics of N. gargensis.
Apparent half-saturation constants (Km(app)) and maximum oxidation rates (Vmax) for total ammonium were determined by fitting the data to the Michaelis-Menten kinetic equation. The red curve indicates the best fit of the data. Standard errors of the estimates based on non-linear regression are reported. See Methods for details of experiments and calculations. a, b, Total ammonium oxidation rates were calculated from microsensor measurements of NH3 dependent O2 consumption and discrete slopes over many substrate concentrations. Results for two biological replicates are shown here; a third biological replicate is shown in Fig. 1b. c, Total ammonium oxidation rates were calculated from microsensor measurements of NH3 dependent O2 consumption in a single trace measurement.
Extended Data Figure 7. Ammonia oxidation kinetics of N. viennensis.
Apparent half-saturation constants (Km(app)) and maximum oxidation rates (Vmax) for total ammonium were determined by fitting the data to the Michaelis-Menten kinetic equation. The red curve indicates the best fit of the data. Standard errors of the estimates based on non-linear regression are reported. See Methods for details of experiments and calculations. a, b, Total ammonium oxidation rates were calculated from microsensor measurements of NH3 dependent O2 consumption and discrete slopes over many substrate concentrations. Results for two biological replicates are shown here; a third biological replicate is shown in Fig. 1c. c, Total ammonium oxidation rates were calculated from microsensor measurements of NH3 dependent O2 consumption in a single trace measurement.
Extended Data Figure 8. Ammonia oxidation kinetics of Ca. Nitrosotenuis uzonensis.
Apparent half-saturation constants (Km(app)) and maximum oxidation rates (Vmax) for total ammonium were determined by fitting the data to the Michaelis-Menten kinetic equation. The red curve indicates the best fit of the data. Standard errors of the estimates based on non-linear regression are reported. See Methods for details of experiments and calculations. a, b, Total ammonium oxidation rates were calculated from microsensor measurements of NH3 dependent O2 consumption and discrete slopes over many substrate concentrations. Results for two biological replicates are shown here; a third biological replicate is shown in Fig. 1d. Single trace measurements to analyse ammonium oxidation were hampered by the slow rate of ammonium oxidation by Ca. N. uzonensis.
Extended Data Figure 9. Adaptation of N. inopinata to slow growth under highly oligotrophic conditions.
Data for N. inopinata are depicted in red, for AOA in blue, and for AOB in orange. a, b, Calculated specific affinities (a) and maximum rates of ammonia oxidation (b). Organisms for which parameters were determined in this study are depicted in bold. Remaining values were obtained from the literature3. Values from this study were determined with n = 6 biological replicates for N. inopinata and n = 4 biological replicates each for N. gargensis and N. viennensis. The specific affinity and Vmax of N. inopinata were significantly different from that of N. gargensis (P < 0.0001) and N. viennensis (P = 0.0001). c, Molar growth yield of ammonia-oxidizing microbes expressed as cellular protein formed per mol NH3 oxidized. Organisms for which parameters were determined in this study are depicted in bold. Remaining values were obtained from the literature (see Methods for references). Values from this study were determined with n = 4 biological replicates each for N. inopinata, N. gargensis, and N. viennensis. The mean molar growth yield of N. inopinata was 394.7 μg protein per mol NH3 oxidized (s.d. 20.2, n = 4), that of N. gargensis was 298.4 μg protein per mol NH3 (s.d. 24.2, n = 4), and that of N. viennensis was 304.3 µg protein per mol NH3 (s.d. 17.9, n = 4). The molar growth yield was significantly different between N. inopinata and that of N. gargensis (P = 0.0003) and N. viennensis (P = 0.0004).
Supplementary Material
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Acknowledgements
We thank A. Mueller for assistance with the cultivation of AOA strain 5A, M. Palatinszky for assistance with the cultivation of N. uzonensis, J. Vierheilig for help with molecular analyses and the cultivation of N. inopinata, and D. Gruber, N. Cyran, A. Klocker, and S. A. Eichorst for assistance with sample preparation for electron microscopy. K.D.K., C.J.S., P.H., S.R., and M.W. were supported by the European Research Council Advanced Grant project NITRICARE 294343 (to M.W.). P.P. and H.D. were supported by the Austrian Science Fund (FWF) project P27319-B21, and A.D. and H.D. were supported by FWF project P25231-B21. K.D.K. and L.Y.S. were supported by the Natural Sciences and Engineering Research Council of Canada (RGPIN-2014-03745). M.A. was supported by research grant 15510 from the VILLUM FONDEN.
Footnotes
Author Contributions H.D. and M.W. designed this study and wrote the manuscript with the help of all authors. K.D.K. and C.J.S. performed the kinetic and yield experiments. E.V.L. and A.B. purified N. inopinata. P.H., E.V.L., and A.B. enriched the N. uzonensis-related AOA strain 5A. A.D. and S.R. performed electron microscopy of N. inopinata. P.P. and L.Y.S. helped with data interpretation. M.A. performed purity checks of N. inopinata and AOA strain 5A by Illumina sequencing and bioinformatics analyses.
Author Information Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Daims H, et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Kessel MAHJ, et al. Complete nitrification by a single microorganism. Nature. 2015;528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 4.Prosser JI. In: Nitrification. Ward BB, Arp DJ, Klotz MG, editors. ASM Press; 2011. pp. 347–383. [Google Scholar]
- 5.Stein LY. In: Nitrification. Ward BB, Arp DJ, Klotz MG, editors. ASM Press; 2011. pp. 95–114. [Google Scholar]
- 6.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 7.Leininger S, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- 8.Nowka B, Daims H, Spieck E. Comparison of oxidation kinetics of nitrite-oxidizing bacteria: nitrite availability as a key factor in niche differentiation. Appl Environ Microbiol. 2015;81:745–753. doi: 10.1128/AEM.02734-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schramm A, de Beer D, van den Heuvel JC, Ottengraf S, Amann R. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl Environ Microbiol. 1999;65:3690–3696. doi: 10.1128/aem.65.8.3690-3696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung MY, et al. Enrichment and characterization of an autotrophic ammonia-oxidizing archaeon of mesophilic crenarchaeal group I.1a from an agricultural soil. Appl Environ Microbiol. 2011;77:8635–8647. doi: 10.1128/AEM.05787-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park BJ, et al. Cultivation of autotrophic ammonia-oxidizing archaea from marine sediments in coculture with sulfur-oxidizing bacteria. Appl Environ Microbiol. 2010;76:7575–7587. doi: 10.1128/AEM.01478-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daims H, Lücker S, Wagner M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016;24:699–712. doi: 10.1016/j.tim.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tourna M, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci U S A. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palatinszky M, et al. Cyanate as an energy source for nitrifiers. Nature. 2015;524:105–108. doi: 10.1038/nature14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lebedeva EV, et al. Enrichment and genome sequence of the group I. 1a ammonia-oxidizing archaeon "Ca. Nitrosotenuis uzonensis" representing a clade globally distributed in thermal habitats. PLoS ONE. 2013;8:e80835. doi: 10.1371/journal.pone.0080835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatzenpichler R, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spang A, et al. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: Insights into metabolic versatility and environmental adaptations. Environ Microbiol. 2012;14:3122–3145. doi: 10.1111/j.1462-2920.2012.02893.x. [DOI] [PubMed] [Google Scholar]
- 18.Prosser JI, Nicol GW. Archaeal and bacterial ammonia-oxidisers in soil: the quest for niche specialisation and differentiation. Trends Microbiol. 2012;20:523–531. doi: 10.1016/j.tim.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 19.Schleper C. Ammonia oxidation: different niches for bacteria and archaea? ISME J. 2010;4:1092–1094. doi: 10.1038/ismej.2010.111. [DOI] [PubMed] [Google Scholar]
- 20.Button DK. Biochemical basis for whole-cell uptake kinetics: Specific affinity, oligotrophic capacity, and the meaning of the michaelis constant. Appl Environ Microbiol. 1991;57:2033–2038. doi: 10.1128/aem.57.7.2033-2038.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Könneke M, et al. Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci U S A. 2014;111:8239–8244. doi: 10.1073/pnas.1402028111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa E, Perez J, Kreft JU. Why is metabolic labour divided in nitrification? Trends Microbiol. 2006;14:213–219. doi: 10.1016/j.tim.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Bartelme RP, McLellan SL, Newton RJ. Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing Archaea and comammox Nitrospira. Front Microbiol. 2017;8 doi: 10.3389/fmicb.2017.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palomo A, et al. Metagenomic analysis of rapid gravity sand filter microbial communities suggests novel physiology of Nitrospira spp. ISME J. 2016;10:2569–2581. doi: 10.1038/ismej.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinto AJ, et al. Metagenomic evidence for the presence of comammox Nitrospira-like bacteria in a drinking water system. mSphere. 2015;1:e00054–00015. doi: 10.1128/mSphere.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang YL, et al. Comammox in drinking water systems. Water Res. 2017;116:332–341. doi: 10.1016/j.watres.2017.03.042. [DOI] [PubMed] [Google Scholar]
- 27.Pjevac P, et al. AmoA-targeted polymerase chain reaction primers for the specific detection and quantification of comammox Nitrospira in the environment. bioRxiv. 2017 doi: 10.1101/096891. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark JM, Firestone MK. Kinetic characteristics of ammonium-oxidizer communities in a California oak woodland-annual grassland. Soil Biol Biochem. 1996;28:1307–1317. [Google Scholar]
- 29.Lehtovirta-Morley LE, et al. Isolation of 'Candidatus Nitrosocosmicus franklandus', a novel ureolytic soil archaeal ammonia oxidiser with tolerance to high ammonia concentration. FEMS Microbiol Ecol. 2016;92:fiw057. doi: 10.1093/femsec/fiw057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch H, et al. Growth of nitrite-oxidizing bacteria by aerobic hydrogen oxidation. Science. 2014;345:1052–1054. doi: 10.1126/science.1256985. [DOI] [PubMed] [Google Scholar]
- 31.Daims H, Stoecker K, Wagner M. In: Molecular Microbial Ecology. Osborn AM, Smith CJ, editors. Bios-Garland; 2005. pp. 213–239. [Google Scholar]
- 32.Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol. 2001;67:5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Widdel F, Bak F. In: The Prokaryotes. Balows A, et al., editors. Springer; 1992. pp. 3352–3378. [Google Scholar]
- 34.Stieglmeier M, et al. Nitrososphaera viennensis gen. nov., sp nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int J Syst Evol Microbiol. 2014;64:2738–2752. doi: 10.1099/ijs.0.063172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karst SM, Kirkegaard RH, Albertsen M. mmgenome: a toolbox for reproducible genome extraction from metagenomes. bioRxiv. 2016 doi: 10.1101/059121. Preprint at. [DOI] [Google Scholar]
- 36.Kandeler E, Gerber H. Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol Fertil Soils. 1988;6:68–72. [Google Scholar]
- 37.Hood-Nowotny R, Hinko-Najera Umana N, Inselbacher E, Oswald-Lachouani P, Wanek W. Alternative methods for measuring inorganic, organic, and total dissolved nitrogen in soil. Soil Sci Soc Am J. 2010;74:1018–1027. [Google Scholar]
- 38.Griess-Romijn van Eck E. Physiological and chemical tests for drinking water. NEN 1056 IV-2 Nederlands Normalisatie Instituut; Rijswijk, The Netherlands: 1966. [Google Scholar]
- 39.Miranda KM, Espey MG, Wink DA. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide-Biol Chem. 2001;5:62–71. doi: 10.1006/niox.2000.0319. [DOI] [PubMed] [Google Scholar]
- 40.Button DK. Nutrient uptake by microorganisms according to kinetic parameters from theory as related to cytoarchitecture. Microbiol Mol Biol Rev. 1998;62:636–645. doi: 10.1128/mmbr.62.3.636-645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayatsu M, et al. An acid-tolerant ammonia-oxidizing gamma-proteobacterium from soil. ISME J. 2017;11:1130–1141. doi: 10.1038/ismej.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhagen FJM, Laanbroek HJ. Competition for ammonium between nitrifying and heterotrophic bacteria in dual energy-limited chemostats. Appl Environ Microbiol. 1991;57:3255–3263. doi: 10.1128/aem.57.11.3255-3263.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki I, Dular U, Kwok SC. Ammonia or ammonium as substrate for oxidation by Nitrosomonas europaea cells and extracts. J Bacteriol. 1974;120:556–558. doi: 10.1128/jb.120.1.556-558.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laanbroek HJ, Bodelier PLE, Gerards S. Oxygen consumption kinetics of Nitrosomonas europaea and Nitrobacter hamburgensis grown in mixed continuous cultures at different oxygen concentrations. Arch Microbiol. 1994;161:156–162. [Google Scholar]
- 45.Bollmann A, Schmidt I, Saunders AM, Nicolaisen MH. Influence of starvation on potential ammonia-oxidizing activity and amoA mRNA levels of Nitrosospira briensis. Appl Environ Microbiol. 2005;71:1276–1282. doi: 10.1128/AEM.71.3.1276-1282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stehr G, Böttcher B, Dittberner P, Rath G, Koops HP. The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiology Ecology. 1995;17:177–186. [Google Scholar]
- 47.Sedlacek CJ, et al. Effects of bacterial community members on the proteome of the ammonia-oxidizing bacterium Nitrosomonas sp strain Is79. Appl Environ Microbiol. 2016;82:4776–4788. doi: 10.1128/AEM.01171-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Both GJ, Gerards S, Laanbroek HJ. Kinetics of nitrite oxidation in two Nitrobacter species grown in nitrite-limited chemostats. Arch Microbiol. 1992;157:436–441. [Google Scholar]
- 49.Boon B, Laudelout H. Kinetics of nitrite oxidation by Nitrobacter winogradskyi. Biochem J. 1962;85:440–447. doi: 10.1042/bj0850440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hunik JH, Meijer HJG, Tramper J. Kinetics of Nitrobacter agilis at extreme substrate, product and salt concentrations. Appl Microbiol Biotech. 1993;40:442–448. [Google Scholar]
- 51.Ushiki N, et al. Nitrite oxidation kinetics of two Nitrospira strains: The quest for competition and ecological niche differentiation. J Biosci Bioeng. 2017;123:581–589. doi: 10.1016/j.jbiosc.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 52.Suwa Y, Imamura Y, Suzuki T, Tashiro T, Urushigawa Y. Ammonia-oxidizing bacteria with different sensitivities to (NH4)2SO4 in activated sludges. Water Res. 1994;28:1523–1532. [Google Scholar]
- 53.Ward BB. Kinetic studies on ammonia and methane oxidation by Nitrosococcus oceanus. Arch Microbiol. 1987;147:126–133. [Google Scholar]
- 54.Clegg SL, Whitfield M. A chemical model of seawater including dissolved ammonia and the stoichiometric dissociation constant of ammonia in estuarine water and seawater from -2 to 40°C. Geochim Cosmochim Acta. 1995;59:2403–2421. [Google Scholar]
- 55.Belser LW, Schmidt EL. Growth and oxidation kinetics of three genera of ammonia oxidizing nitrifiers. FEMS Microbiol Lett. 1980;7:213–216. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the European Nucleotide Archive (ENA) with the accession codes PRJEB20196 (for the raw sequence data from the N. inopinata pure culture) and PRJEB20386 (for the raw metagenome sequence data from the Ca. N. uzonensis-related AOA enrichment 5A). The pure culture of N. inopinata has been deposited at the Japan Collection of Microorganisms with the accession code JCM 31988. Source data for Extended Data Figures 2b, 3, and 9 are provided with the paper.