Abstract
Background
Immunofluorescence (IF) plays a major role in quantifying protein expression in situ and understanding cell function. It is widely applied in assessing disease mechanisms and in drug discovery research. Automation of IF analysis can transform studies using experimental cell models. However, IF analysis of postmortem human tissue relies mostly on manual interaction, often subjected to low-throughput and prone to error, leading to low inter and intra-observer reproducibility. Human postmortem brain samples challenges neuroscientists because of the high level of autofluorescence caused by accumulation of lipofuscin pigment during aging, hindering systematic analyses. We propose a method for automating cell counting and classification in IF microscopy of human postmortem brains. Our algorithm speeds up the quantification task while improving reproducibility.
New method
Dictionary learning and sparse coding allow for constructing improved cell representations using IF images. These models are input for detection and segmentation methods. Classification occurs by means of color distances between cells and a learned set.
Results
Our method successfully detected and classified cells in 49 human brain images. We evaluated our results regarding true positive, false positive, false negative, precision, recall, false positive rate and F1 score metrics. We also measured user-experience and time saved compared to manual countings.
Comparison with existing methods
We compared our results to four open-access IF-based cell-counting tools available in the literature. Our method showed improved accuracy for all data samples.
Conclusion
The proposed method satisfactorily detects and classifies cells from human postmortem brain IF images, with potential to be generalized for applications in other counting tasks.
Keywords: dictionary learning, sparse models, image segmentation, immunofluorescence, postmortem human brain, microscopy
1. Introduction
Target validation and cell selection provide critical insights into mechanisms driving disease, particularly at the early steps in the drug discovery process. Immunofluorescence (IF) microscopy applies antibodies and other affinity reagents with fluorescent proteins on tissue and may unlock information into molecular cascades while preserving the tissue integrity. IF is particularly important in quantifying protein expression and understanding cell function. IF experiments often occur in big batches resulting in high amounts of data, making manual analysis very time-consuming and prone to error including high inter and intra-observer variability and low reproducibility [1].
Several authors developed algorithms to automate IF analysis using unsupervised methods, mostly based on deformable contours and region-based approaches [2, 3, 4, 5, 6, 7, 8] and fewer focused on signal processing [9, 10]. Despite a great deal of publications in cell analyses, most of the methods for detecting and segmenting cells in IF images have been tailored to work with images derived from cell cultures or experimental animals, in which several parameters can be controlled to render IF images with a rather smooth, clean background and high contrast. On the other hand, IF images derived from postmortem human brain present considerable autofluorescence, which gets more pronounced in aged brains, inherent to human brain tissue because of accumulation of lipofuscin pigment [11, 12]. Autofluorescence reduces the signal-to-noise ratio of specific labeling [13], making identification of stained cells harder. Antigen masking caused by rather prolonged formaldehyde fixation compared to cell and experimental animals results in impoverished IF signals [14]. An additional challenge to IF analysis in human tissue is that quantification experiments based on stereological principles require thicker tissue slides (30–60μm, instead of 5–8μm) [15], increasing the amount of neuropil, which also contributes to background fluorescence [16]. This problem can be minimized with the use of confocal microscopes, however the size of the human brain, even for the analysis of small areas, would require a high amount of microscope time and data processing capabilities. Finally, as human brain histological sections usually have a much larger surface in proportion to its thickness, holes, folds, cell fragments and all kinds of clutter are commonplace, making the detection task even more complex. As a result, available cell counting algorithms show a poor performance in analyzing IF images from postmortem human brains, precluding automating cell counting.
In this study, we propose a computer-based segmentation method suitable for handling IF microscopy from our thick postmortem human brain sections, aiming to improve user productivity by reducing the counting time and also reducing inter and intra-observer variability. Our proposal consists in applying dictionary learning and sparse coding for cell modeling. Dictionary learning is a cutting-edge method for learning signal representations that better fit underlying data, thus improving description [17], while sparsity allows better portrayal of salient image properties and produces features more likely to be separable, which is useful for machine learning [17, 18].
First, we present an introduction to the basic sparse coding and dictionary learning framework, together with an in-depth description of our method. Then we show the results obtained with our method tested with real data and data on user experience experiments using our implementation, followed by a comparison between our method with other open-access cell counting methods found in literature. Finally, we discuss our approach when compared to other similar methods.
2. Materials and Methods
2.1. Tissue Acquisition and Preparation
Patients
Our methods examine IF digital images captured from 49 horizontal sections, from 21 human brainstem. The sections come from the Brain Bank of the Brazilian Aging Brain Study Group (BBBABSG) [19, 20] and the Neurodegenerative Disease Brain Bank (NDBB) from the University of California, San Francisco (UCSF). All subjects showed variable degree of Alzheimers disease pathology staged according to the new NIA-AA guidelines [21]. The institutional review boards of both participating institutions deemed this study non-human. All selected cases have a postmortem interval of less than 24h. The brainstems were fixed in 10% buffered formalin from 10 to 20 days.
Tissue Processing and Staining for Generating the Images Used in This Study
Tissue processing and staining protocols have been described previously [15]. In summary, each brainstem block was severed from the brain, fixed in 10% formalin, and embedded in 8% celloidin for subsequent sectioning [22]. Blocks were sectioned horizontally in serial sets, each one containing one 300μm thick and five 60μm thick sections [15]. Selected 60μm thick sections were autoclaved in citrate buffer retrieval solution for 5 min at 121°C, followed by 30 min incubation with 0.1% Sudan black B (Sigma-Aldrich) for blocking autofluorescence and 30 min incubation in protein blocking solution (5% milk in phosphate-buffered saline with 0.05% Tween 20)at room temperature (RT). Sections were immunostained with an antibody against phospho-tau at Serine 202 antibody (CP-13, 1:500, monoclonal mouse, gift of Peter Davies, NY) to detect neurofibrillary tangles and active caspase-6 (aCasp-6, OAAF05316, 1:500, rabbit polyclonal, Aviva System Biology), an apoptosis marker [23, 24].
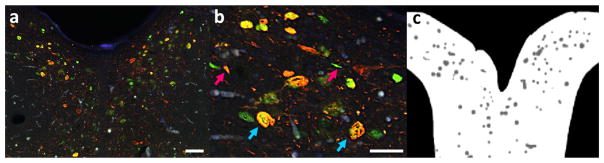
Double immunostaining (CP-13/aCasp-6) was performed overnight at 4°C in PBS, followed by incubation with immunofluorescence species-specific secondary antibodies conjugated to Alexa Fluor 488 (aCasp-6) and 546 (CP-13) for 1h at RT (1:200 in PBS-T; A-11008 and A-11003 respectively, both by Invitrogen, CA). Neuronal cell bodies were labeled with a fluorescent Nissl stain for 30 min at RT (Neurotrace 435/455; 1:50 in PBS-T; Life Technologies, NY, USA). The sections were photographed with a Nikon 6D high-throughput wide-field epifluorescence microscope (Nikon, Tokyo, Japan). Regions of the dorsal raphe nucleus (DRN) were imaged at 10x magnification (Plan Apo 10x/0.45, Nikon, Japan. Pixel resolution was about 0.2μm.), and each image set covering the regions of interest (ROIs) was merged into a single composite using NIS-Elements 4.30 (Nikon, Japan). Figure 1(a) presents one IF image from our dataset, stained using the aforementioned method, and Figure 1(b) illustrates its respective full resolution image.
Figure 1.
IF image and grayscale mask. (a) dorsal raphe nucleus (DRN) image stained for CP-13 and Casp6 (scalebar is 100μm); (b) respective 100% zoom view - the scalebar is 50μm (blue arrows point to real cells while magenta arrows point to debris that can be easily mistaken for cells); (c) respective grayscale mask used for training dictionaries, with white regions delimiting the DRN, while cells appear in gray.
Manual Neuronal Counting and Creation of Ground-Truth Data
In each composite image, DRN boundaries come from traces based on cytoarchitectonic parameters, as proposed in Baker et al. [25] and Rub et al. [26]. Adobe Photoshop CS6’s built-in counting tool (Adobe, CA) was used for manual cell counting. Neurons were recognized by size, shape and the presence of Neurotrace positivity and classified into four groups: red (reactive to CP-13), green (reactive to aCasp-6), yellow (reactive to both CP-13 and aCasp-6) and none (not reactive to CP-13 or aCasp-6). The counting tool embedded in Photoshop tracked the number of markers per group. Counting was conducted on merged images and confirmed on single channel images, for precision. Care was taken to ensure that the counting marks were placed inside the cell boundaries. Users had to be careful during the manual counting to avoid counting cells that were out of focus (i.e. out of the imaging plane), as well as holes and debris. Magenta arrows in Figure 1(b) point to debris that are easily mistaken for cells by inexperienced observers and most computational tools. Blue arrows point to real cells. All counting results were stored in Photoshop file format.
Grayscale masks delimiting the DRN, cell bodies and background were manually created on top of cell counting files. These masks were used for training our segmentation method and constraining processing to the DRN region thus saving computation time, as described in-depth in the following sections. Figure 1(c) shows a example of a manual mask, where the white region delimits the DRN, and all cells are painted in gray.
2.2. Computerized Cell Counting
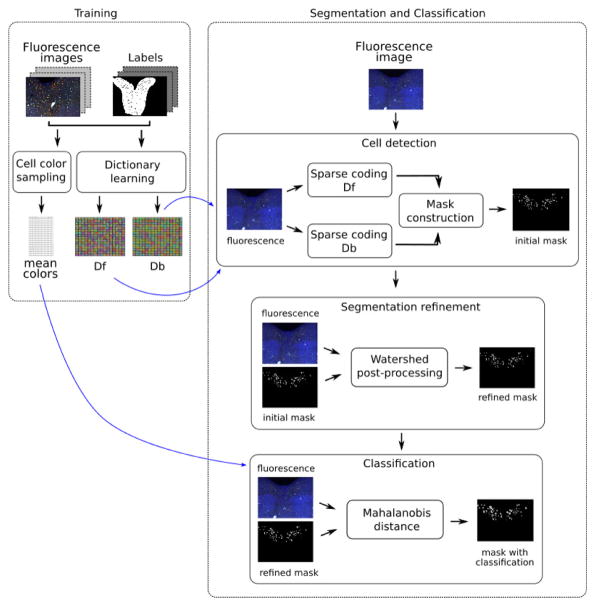
We propose a semi-automatic method for assisting cell counting in IF images. Our method guides the users during the counting process by highlighting the detected cells, while allowing them to select the real cells and ignore debris and other mislabeled structures. Our method is based on dictionary learning and sparse coding for classification and is composed of two major stages: training, where dictionaries that best represent data are learned and color samples are computed; and segmentation-classification, where cells are detected using the trained dictionaries and classified into red (CP-13), green (aCasp-6) and yellow (CP-13 and aCasp-6 overlap) using color samples as references. Figure 2 shows the outline of our method.
Figure 2.
Proposed cell counting methodology.
2.2.1. The Dictionary Learning and Sparse Coding Framework
Traditional signal processing uses analytic bases, also known as dictionaries, such as Fourier [27], Wavelets [28] and Curvelets [29], to represent data. Those are popular for having closed mathematical formulations with well-known properties and allowing fast computations. Analytical bases are, nonetheless, one-fits-all methods that have the downside of poorly representing many types of signals [30]. Recently, a set of methods for learning dictionaries from the underlying data emerged and proved successful in fine tuning representations in a data-set specific way [31]; these methods are known as dictionary learning.
A dictionary contains a set of atoms (elementary signals or vectors) that can be used to decompose more complex signals. In mathematical terms, a dictionary D is the matrix D = [d1|d2| … |dL] ∈ ℝN×L where the columns d represent the atoms. A signal x ∈ ℝN can be represented as a linear combination of atoms in D:
| (1) |
If the atoms in D form a basis, signals can be uniquely represented by the equation 1. However, if L > N the dictionary is said to be overcomplete and the system in 1 is overdetermined. In that case, optimization methods are used to compute a by minimizing an objective function of the general form [30]:
| (2) |
where C(.) is a cost function that enforces particular encoding characteristics. When working with overcomplete dictionaries we want the cost function to enforce sparsity, meaning that the encoding will have few non-zero coefficients. Moreover, a good choice of C(.) will seek the most informative vectors. Thus, the problem is usually stated as:
| (3) |
where ||.||0 is the l0 pseudo-norm, where a norm lp is defined as:
| (4) |
l0 is basically the number of non-zero coefficients in a. It promotes sparsity by penalizing low value coefficients while tolerating large ones. Solving 3 is known as sparse coding, meaning that a signal is represented by a few number of atoms (k) in D. Ideally, k is much smaller than N. However, problem 3 is NP-hard [32] and is often replaced by a relaxed objective function that uses the l1 norm [33]:
| (5) |
Here, the λ parameter balances the trade-off between reconstruction error and sparsity. The problem in 5 is convex and can be solved very efficiently. Moreover, the l1 norm is proved to promote sparsity and is more stable than l0 [33].
Sparse dictionary learning deals with the task of training such dictionaries ensuring that the final atoms enforce sparsity. Currently, there are two main methods for training sparse dictionaries, the MOD (methods of optimal directions)[34] and K-SVD (k-singular value decomposition)[31] algorithms. K-SVD became popular for being more efficient than MOD. In K-SVD, a dictionary of K atoms is learned by using the SVD (singular value decomposition) process to update an atom at a time. The objective function can be written as:
| (6) |
where ||.||F if the Frobenius norm, Y = [y1|…|yi] is a coefficient matrix and D = [d1|…|di] a dictionary. Ek is the residual matrix, are rows of Y, and dk, are the values being updated in the k-th iteration. These values are computed by minimizing 6 by finding a low-rank approximation of Ek. The second term in 6 measures the number of non-zero coefficients in Y and sets a limit T thus enforcing sparsity. Notice that this method allows more flexibility to represent the data because it lacks constraints such as that the atoms (basis vectors) are orthogonal.
Dictionary learning and sparse coding have been successfully used in many signal and image processing task, such as denoising [35] and in painting [36]. More recently, it has also been successfully used in supervised and unsupervised classification tasks [37, 38].
The rationale behind using dictionaries for image classification is learning as many dictionaries as classes considered in the problem, each trained with data drawn from a particular class. These dictionaries are used to encode the data that needs to be classified, which in turn is reconstructed and compared to the original data-set. The smallest reconstruction error is obtained when the dictionary belonging to the correct class is used thus, we are able to predict the correct classes.
2.2.2. Training
Two dictionaries, one for foreground (Df) and one for background (Db), are learned in this stage. The learning task is guided by the grayscale masks built from the manual labeled ground-truth (GT) images (Figure 1(c)) that indicate the ROI regions (data outside the ROIs are not processed). All training images are converted to the LAB colorspace and normalize to the [0, 1] interval. The LAB colorspace is a color representation scheme designed to mimic the human visual color and brightness perception. The L (luminance) channel values relates linearly to the human perception of brightness, while A and B carry the color information. Moreover, the Euclidean distance between colors in LAB corresponds to color difference perceived by human vision[42]. An 11-by-11 pixel sliding window is placed on top of every pixel of interest, on each channel, and used to extract the patches which form a training matrix. Df and Da are learned using the on-line dictionary learning algorithm described in Mairal et al. [39]. Our final dictionaries are composed of 256 vectors, each.
A set of reference cell color samples, which are used during classification, is also computed along with the dictionary learning training. Here, 5×5 pixel windows are centralized on top of each cell in the mask and the mean LAB values are computed and stored.
2.2.3. Segmentation and Classification
Cell Detection
The use of dictionary learning and sparse coding in image classification relies on the fact that a signal is better represented by a dictionary learned on data drawn from its own class, which yields the decomposition with the smallest reconstruction error.
Our segmentation approach consists of encoding image patches using the background and foreground dictionaries and measuring the reconstruction error. Pixels whose patches are better represented by Df are marked as foreground.
More specifically, all images are converted to the LAB color space and normalized. An 11-by-11 pixels sliding window is placed on top of every pixel of interest, on each channel, and used to extract the patches x. The computation is constrained to the ROI, in our case the DRN, indicated by the manually labeled masks. A patch x can be written as x = [xL|xA|xB]T where xL, xA, xB are vectors containing all pixels from the L,A and B channels. The encoding tasks can be written as:
| (7) |
| (8) |
Equations 7 and 8 are solved using the Lasso [40] algorithm. The results are two versions of patch x encoded by both dictionaries. The reconstruction error ℛ is measured by:
| (9) |
| (10) |
where N is the number of atoms in D. A pixel is included in the segmentation mask if ℛf < ℛb.
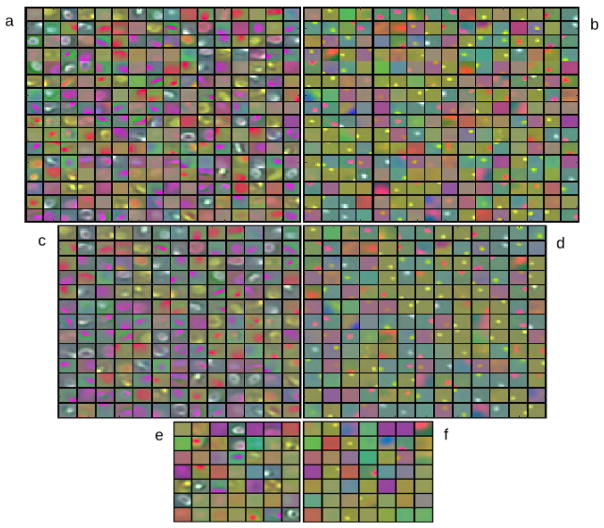
One should note that atoms representing features common to both classes appear in both dictionaries and tend to have high coefficient values, thus reducing the error value. These atoms can be detected by computing where most correlated atoms will have high inner product values [38]. Exclusion of these highly correlated atoms can sensibly improve discriminatory power [38]. Thus, Df and Db are thresholded to remove atoms whose inner products are equal or larger than 0.99. In our experience, ℛf and ℛb are highly separable when computed on pixels from cell regions, thus being a good method for detecting cells. On the other hand, ℛf and ℛb are more prone to ambiguity, having closer values when computed over background pixels. This means that DL segmentation alone can yield false positives and needs some kind of post-processing. Figure 3 shows examples of the dictionaries before and after threshold and discarded atoms. Visual inspection, especially on the discarded dictionaries, reveals that atoms very similar in both dictionaries are removed, together with repeated atoms. It is also interesting to note how the foreground dictionary captures mostly cell-like features while the background dictionary captures wide homogeneous spaces and debris-like features.
Figure 3.
Trained dictionaries in LAB colorspace. (a) and (b) are examples of foreground and background dictionaries. (c) and (d) are the same dictionaries after thresholding most correlated atoms. (e) and (f) are the discarded atoms.
Segmentation Refinement and Post-processing
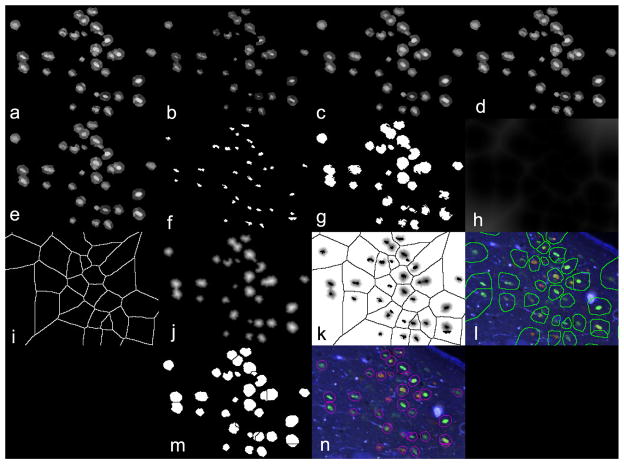
The initial segmentation may not separate cells that are touching. So, we used a watershed-based approach to refine its result. Watershed segmentation models images as a set of catchment basins (dark pixels) and ridges (bright pixels), where the latter is used to guide the actual segmentation task. Watershed is a computationally efficient but prone to over segmentation when directly applied to the original images. Thus, our strategy is to use the original IFs together with the DL segmentation mask to locate background and cell markers and impose those on the initial mask so that the watershed transform consistently segments the boundaries between cells. We provide a detailed explanation of our refinement methods in the following paragraphs. Figure 4 summarizes our strategy.
Figure 4.
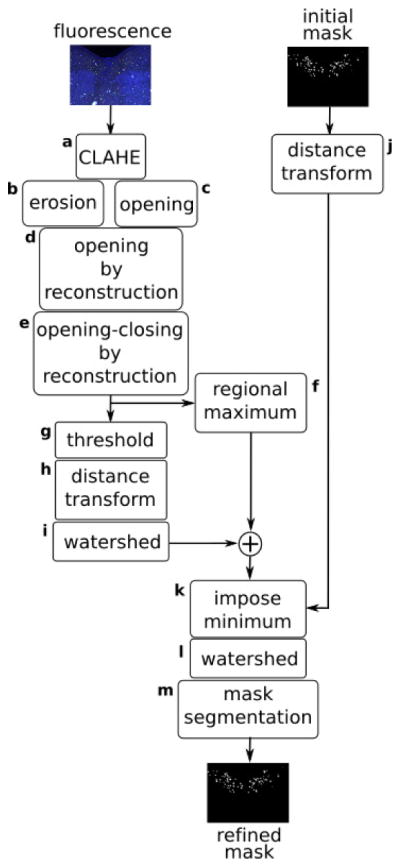
Watershed-based segmentation refinement strategy
The first step (Figure 4:a) is contrast enhancement of fluorescence images using local histogram equalization (CLAHE [41]). CLAHE helps to locally improve the contrast of badly defined boundaries and enhances faded cell bodies that are mixing with background. Next, the newly enhanced images are eroded and, in parallel, are opened using a disk-shaped structuring element of 5 pixels radius which yields images sets Io and Ie. The eroded set is left with high intensity peaks while the opened set has a smoothed version of the images. In step d (Figure 4:d), Ie are used as markers and Io are used as masks for morphological opening-by-reconstruction [42]. Intuitively, this operation forces the peaks in Ie to spread, having Io as a template. It results in a smoothed image where the cells are bright and background is suppressed. These images are further refined in the opening-closing step that operates on the image complement (Figure 4:e). The resulting image has its regional maximums computed [42] (Figure 4:f) resulting in Imax and, in parallel, it is thresholded using Otsu’s method [43] (Figure 4:g). Next, the distance transform is computed on the thresholded image (Figure 4:h) and the watershed transform is used to find its ridges (Figure 4:i) resulting in image Imin. Meanwhile, the distance transform is also computed on the segmentation mask resulting in Imask. Step k (Figure 4:k) composes a map of regional maximums and minimums from Imax and Imin and uses it to impose local minimums on Imask. These minimums work as markers for watershed computation. Next, the watershed transform is computed on Imask and the resulting ridge lines are used to separate cells on the segmentation mask. Figure 5 shows step-by-step refinement results. Figure 5:a shows a segmented image after CLAHE enhancement. Figure 5:b shows the erosion result and 5:c opening result. Figure 5:d and 5:e show opening by reconstruction and opening-closing by reconstruction results respectively. Figure 5:f shows the regional maximums and 5:g the thresholded image. Figures 5:h and 5:h show the distance transform and its respective watershed. Figure 5:j show the distance transform computer over the segmentation mask, as described in Figure 4:j. Figure 5:k shows the imposed minimums, which work as seeds for the final watershed transform. Figure 5:l shows the watershed overlayed on the original images and 5:m the refined segmentation mask. Finally, the final segmentation is displayed in 5:n.
Figure 5.
Step-by-step watershed refinement results.
The DL-based segmentation may recognize artifacts such as debris, as real cells. Especially when they have the same color of actual cells. False positives may also occur during watershed refinement, where smaller parts of the segmentation mask may be broken into independent structures. Thus, we employ a set of computationally efficient filters to reduce the number of false positives and improve counting results. First, a color-wise filter removes structures whose color is closer to the background than to cells. A color difference map (ΔE) is computed between background pixels and the original image and thresholded. ΔE works as a distance map between a reference (here, the background) color and the rest of the image. Regions where the colors are similar (or closer) to the reference will be dark (small distance) while regions where colors are different will be bright (large distance - computing the ΔE is explained in details in the next section). Thresholding it removes most of the undesired structures. The segmentation mask obtained in the previous steps is used as a guide for drawing background samples. We set the threshold value to the 75th percentile value as ΔE does not follow a normal distribution. Second, an area-based threshold is performed to remove structures that have a smaller number of pixels than a particular set number. Here, we set the threshold to the 5th percentile of the area distribution. Finally, we perform a shape-wise filtering based on the solidity coefficient [?], which measures an object’s convexity. Solidity is computed as the ratio S = A/Ac where A in the number of pixels in the structure and Ac is the number of pixels in its convex hull, yielding a value between 0 and 1. The more convex the object is, the smaller is its solidity. In our images, cells are approximately round, thus convex structures are very likely to be debris that should be removed. We set our solidity threshold to the 10th percentile. It is noteworthy that more aggressive filtering would greatly reduce the number of false positives at the expense of missing more real cells.
Classification
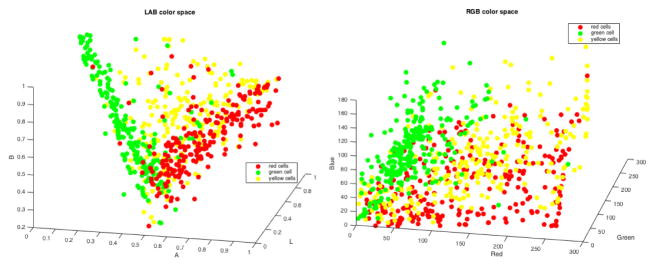
Finally, our method classifies cells in red (stained for CP-13), green (stained for aCasp-6) and yellow (stained for both markers). Inspection of our fluorescence images reveals bleed-through between channels, meaning that cell classification based on the color channels alone is inaccurate, as there is no pure color. Even cells considered red or green during manual labeling are composed of a mix of signals. In fact, the RGB scatter plot in Figure 6 show how mixed the cells are in the color space.
Figure 6.
Cell colors scatter plots in LAB (left) and RGB (right) color space.
In order to circumvent this problem, instead of using thresholding, our approach consists in finding a cell mean color location in the LAB color space, which improves separation as shown in Figure 6, and computing its distance from red, green and yellow regions. The color samples extracted during the training step are used as LAB region references here.
Notice that the segmentation process lacks perfect boundaries results, often leaving some background around the cells. These extra pixels allow suitable visualizations but impair the classification, as their presence change the mean color values. We remove the offending pixels from each segmented cell individually, by computing the mean color difference (ΔE) map between background pixels and the original image, and thresholding it. First, we compute difference values Ldiff, Adiff, Bdiff from the pixels within each cell, as:
where LB, AB and BB are the background LAB channels, Lc, Ac, Bc are the IF image LAB channels and μ(.) the mean. ΔE is computed by:
| (11) |
Since background is the reference here, pixels whose color value is closer to it are dark in ΔE (smaller distance) while cell pixels, whose color are further away, are brighter (larger distance). This map is hard thresholded, yielding cells that are mostly free of background. In our experiments we empirically set the threshold to 0.2.
Classification is performed by computing the Mahalanobis distance [44] between each cell mean LAB values (μcell = [L|A|B]) and sample sets from red (μ1 =[L|A|B]), green (μ2 = [L|A|B]) and yellow (μ3 = [L|A|B]) cells (computed in training). The cell is assigned to the class that has the smallest distance:
where ℳ(.) is the Mahalanobis distance. The cell class is defined as is advantageous to use a distance metric, since it does not require that all classes are present in the image, unlike clustering methods that always expect a fixed number of classes. However, Mahalanobis does require that all classes are minimally represented in the training set, otherwise it won’t be able to account for them during classification. Figure 7(e) shows an example of classified fluorescence image.
Figure 7.
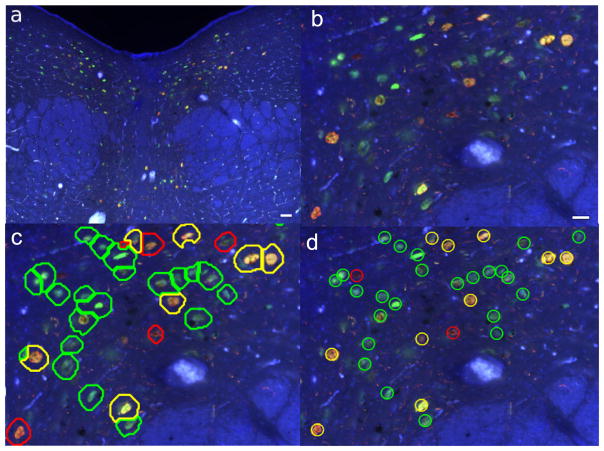
Example of a result obtained using our method. (a) test image (scalebar is 100μm); (b) zoom in view of (scalebar is 40 μm) (a); (c) final segmentation together with classification (the boundary color indicates predicted cell class); (d) ground truth.
2.3. Experimental Design and Comparative Strategies
We tested our segmentation strategy using a common leave-one-out scheme [45], where for each iteration one image was left out of the training phase, which was performed with the remaining data. This image was then used in the test phase. Foreground and background dictionaries, together with the cell color samples, were trained during each training phase and used for testing. This process was repeated until all images were segmented. Post-processing filter thresholds were the same for all images throughout the leave-one-out test. Segmentation results were saved as binary masks and cell classification results were saved as grayscale masks, where each class is color-coded as a specific intensity value, during each testing phase. Segmentation quality was assessed in terms of true positive (TP), false positive (FP), false negative (FN), precision, recall, false positive rate (FPR) and F1 score, where:
The scores were computed between the binary mask and respective ground truth (GT). Finally, cell classification quality was computed as the percentage of correct classification for green, red and yellow classes.
GT data was extracted from the Photoshop files (section 2) using a Javascript routine made for reading the cell markers coordinates and cell class information. We notice that the manual counting procedure can yield inadequate centered markers, thus a TP was identified as a segmented cell whose centroid was 19 pixels away from a marker in the GT data. Information regarding true negatives (TNs) was not available for our dataset, so we employed the strategy similar to Xu et al.[46] for computing an estimate of the number of TNs: we used a sliding window, as used in segmentation, to count the number of patches that could be extracted from the DRN without touching the cells, having the manual labeled masks as reference. That processes gave the approximate number of TNs.
2.3.1. Comparison with other methods
We compared our pipeline to four different cell segmentation systems found in the literature. These methods were chosen for being distributed as free software and having been used in other segmentation studies. None of the selected methods classified the cells, so we only compared the cell detection performance. For all methods, we saved the binary masks resulting from their segmentations and used them for comparison with our GT data, as described in the previous paragraph. TP, FP, FN, precision, recall, FPR, and F1 scores were computed for all methods. We also plotted the Precision-recall and ROC (receiver operating characteristic) graphs to compare performance between all methods and the precision and recall box-plots for all methods to show the results dispersions. It is important to note that our segmentation method outputs a discrete binary classification instead of a class probability score, and this is also true for other methods compared in this manuscript. Thus, traditional ROC curves were discarded in detriment of other graphs. A discrete binary classifier yields only a (TPR, FPR) point in ROC space, so we used a discrete plot instead of a curve to show compare results, as described in Fawcett et al. [47?]. Moreover, the tested methods were unsupervised, so there was no need to perform cross-validation on them.
Automatic cell counting (AC)[48]: Here, the author proposes a basic cell segmentation scheme, where a fluorescence image is split in 3 channels (R,G,B), which are segmented by threshold followed by blob detection. Their method was implemented as ImageJ macros. We modified their macros to save the segmentations as binary masks, which were compared to our GT data as described above.
CellProfiler (CP)[49]: CellProfiler is a general purpose tool for cell image analysis. It works with pipelines that can be designed to drive the image processing according to the users needs. We assembled a CellProfiler pipeline suitable for processing fluorescence images and counting cells based on the studies in Padmanabhan et al. [50] and Soliman et al. [51]. Briefly, in our pipeline the input were the original color images. They were (a) resized to 15% of the size (to reduce memory footprint); (b) converted to grayscale; (c) the identify primary objects filter was used to detect cells in the green channel; (d) the identify primary objects filter was used to detect cells in the red channel; and (e) all detected cell coordinates were exported to a spreadsheet. The coordinates were then transformed to reflect the original image size. Here, TPs were identified as a cell whose (x, y) coordinate was 19 pixels away from a marker in the GT data.
-
Method proposed by Ushizima et al. (DU)[52]: the super-pixel Voronoi diagram (SPVD) method [52] was selected the best performing algorithm during the Overlapping Cervical Cytology Image Segmentation Challenge [52, 53]. We tested a modified version provided by the authors, tuned to work with fluorescence instead of bright field. It was implemented as a ImageJ macro.
This algorithm directly processes color images, yielding a binary mask for green and another for red channels. We combined all masks in a single binary mask that was compared to GT as previously described.
CellC (CC)[54]: CellC was developed for counting bacterial cells in microscopy images. Although it is capable of processing fluorescence images, it does not directly work with color information and the different fluorescence channels must be entered as grayscale images. For this test we split our images into red, green and blue channels and segmented the red and green channels separately. The results were combined in a single binary mask that was compared to GT as previously described.
2.3.2. User experience quantification
Our final test aimed at assessing how much actual user time would be saved by using our method instead of manual counting. In our setup, six users without any previous experience in cell counting received a basic training for performing the counting task, following the same protocol applied for labeling GT data: The DRN ROI should first be traced using cyto-architectonic parameters based on Baker et al. for LC [25] and Rub et al. [26]. Adobe Photoshop CS6’s built-in counting tool was used for manual cell counting. Neurons were classified into 3 classes: Red, Green, and Yellow (overlap) and separated into three different groups inside Photoshop using the counting tool. Counting was conducted on the RGB images and confirmed on single channel images, for precision. Each file was saved in Photoshop file format.
After training, three users (referred to as user #1, user #2 and user #3) counted the original fluorescence images independently of each other. Each file was uninterruptedly counted and the necessary time for performing the task was measured with a stopwatch. Two other users (referred to as user #4, user #5 and use #6)) were presented with the same fluorescence images, but this time the detected cells were highlighted by thin boundaries. Again each file was uninterruptedly counted and the necessary time for performing the task was measured with a stopwatch. We assessed the resulting statistical significance using Student’s pair-wise t-test.
3. Results
Our segmentation method was implemented in Matlab (MathWorks Inc), and the dictionary learning and sparse coding functions come from the SPAMS Toolbox [39, 55]. Our experiments were performed in a Linux workstation with an Intel(R) Xeon(R) 6-core CPU 2.40GHz E5-2620 and 16GB of RAM.
Figure 7 shows the segmentation result for an image from our dataset: (a) is the original IF image and (b) a zoomed in region. (c) shows the final segmentation results. Note that here we also show the classification results. Boundary colors indicate the class attributed to each cell. (d) is the ground truth from the manual counting, placed here for comparison. Is is possible to see that most cells were correctly segmented and classified, although some mistakes are visible.
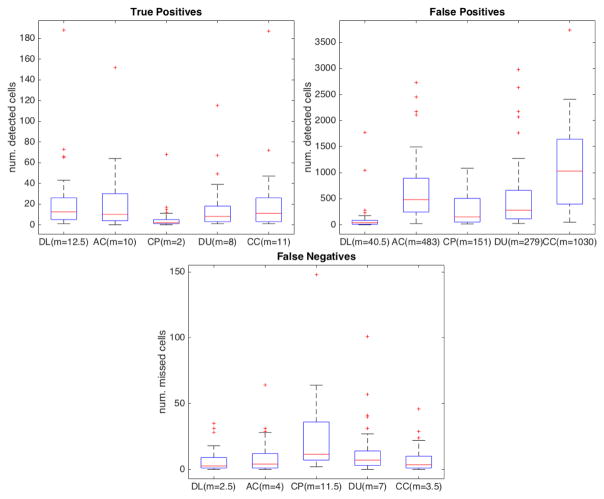
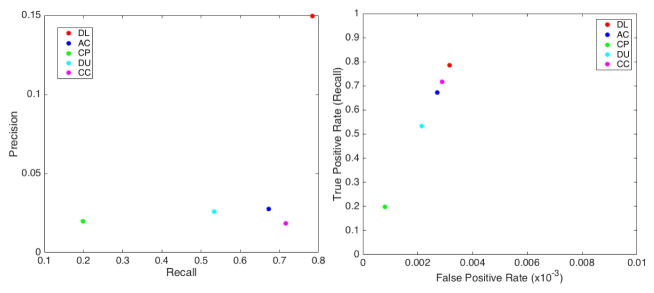
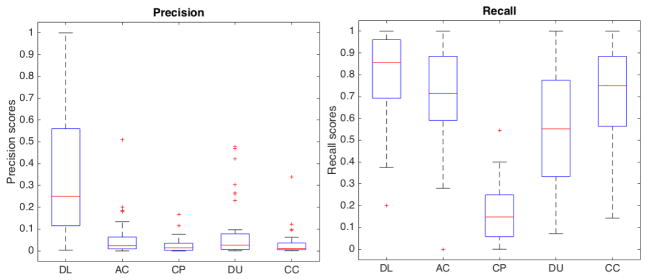
For segmentation efficiency, we obtained a TP rate (recall) of approximately 0.78 (in [0, 1] interval, where 1 is best) while AC obtained 0.67, CP obtained 0.2, DU obtained 0.53 and CC, the tested method with highest TP, obtained 0.71. We obtained a modest precision score of 0.15 (in [0, 1] interval, where 1 is best), which was, nonetheless, one order of magnitude higher than 0.018 obtained by CC, 0.026 obtained by DU, 0.02 obtained by CP and 0.028 obtained by AC. The precision scores were lowered by the number of FPs in all methods. Here, even though we have a set of post-processing filters to removes FPs, we avoided string filtering in order to keep FNs low. FPs also caused low F1 scores, since it measures the balance between precision and recall. Our method obtained a F1 score of 0.25 (in [0, 1] interval, where 1 is best) while CC obtained 0.027, DU obtained 0.028, CP obtained 0.008 and AC obtained 0.038. On the other hand, we obtained a very low FNR of 0.22 (in [0, 1] interval, where 0 is best) while CC obtained 0.28, DU obtained 0.46, CP obtained 0.8 and AC obtained 0.32. As a practical example, the mean recall for algorithms from the literature was 0.53 while ours was 0.785, yielding a raw mean difference of about 0.25. It suggest that our method was able to detect 25% more real cells than the other algorithms. Regarding cell numbers, we detected 277 more cells (our ground truth contained 1107 cells). Moreover, the algorithms from the literature obtained mean false negative rate of 0.47 while our method obtained 0.215, coincidentally yielding about 0.25 raw mean difference. It shows that our method yields 25% less false negatives among the detected cells (277 less undetected cells than other methods). Table 1 summarizes the scores obtained in our tests and Figure 8 shows true positive (TP) (a), false positive (FP) (b) and false negative (FN) (c) rates of our method (DL) in comparison to the automatic cell counting (AC), CellProfiler (CP), Ushizima et al. (DU) and CellC (CC) methods. As illustrated in these box-plots, our method outperformed the other cell counters, having the highest mean TP rate while keeping the number of FNs very low. The box-plots also reveal that our method had the lowest variability throughout the data set in regard to FP and FN rates. We used the precision-recall and ROC plots in Figure 9 to aid in comparing our method to the others in literature. Since our method is based on discrete binary classification, we use discrete plots instead of curves. The same is true for the considered methods in literature, which output a binary mask instead of confidence values. In Figure 10 we have the precision and recall box-plots that show result scores dispersions for all considered methods. Finally, our average cell color correct classification rate was 0.71 (± 0.21).
Table 1.
Precision, recall, false negative rate (FNR - the percentage of non detected cell), and F1 score for all tested methods: Automatic Cell Counting (AC), CellProfiler (CP), Ushizima et al. (DU), CellC (CC), and Dictionary Learning (DL).
| Methods | Precision | Recall | FNR | F1 score |
|---|---|---|---|---|
| AC | 0.028 | 0.672 | 0.328 | 0.040 |
| CP | 0.020 | 0.199 | 0.801 | 0.008 |
| DU | 0.026 | 0.534 | 0.466 | 0.029 |
| CC | 0.018 | 0.716 | 0.284 | 0.028 |
| DL | 0.150 | 0.785 | 0.215 | 0.251 |
Figure 8.
True positive, false positive and false negative scores obtained by our method (DL) in comparison to other cell counters (AC, CP, DU and CC). Red bar indicates the median (m) for each method (also in parenthesis). Red stars indicate files whose results were outliers.
Figure 9.
Precision-recall (left) and ROC (right) plots on the detection accuracy of our method (DL) compared to AC, CP, DU and CC
Figure 10.
Precision (left) and recall (right) box-plots of our method (DL) compared to AC, CP, DU and CC
Table 1 shows that we had a low precision rate, which was caused by the number of FPs. However, our method outperformed other cell counting methods in literature by far. This fact can also be note when analyzing the precision-recall and ROC graphs in Figure 9.
In a different experiment, we measured user time differences when counting the original images and pre-segmented images. We were interested in quantifying the improvement (if any) in user experience when using our cell detection tool for analyzing our IF images. Figure 1(a) exemplify an image used in this experiment. We were careful to keep the background of the segmented images in order to avoid confusing the users. All users counted a total of 49 images, each. Users #1, #2 and #3 counted the original images, while users #4, #5 and #6 counted the images with detected cells highlighted. Mean count time for original images was 318.02s (5.3 minutes) with 318s standard deviation. Mean count time for segmented images was 122.2s (2.03 minutes – 38% of the original counting time) with 99.75s standard deviation. Computer-aided users were, on average, 2.6 times faster when using our methods. The mean time difference was statistically significant (pair-wise t-test yielded p-value ¡ 0.0001). Figure 11 shows the box-plot of the time distribution per user.
Figure 11.
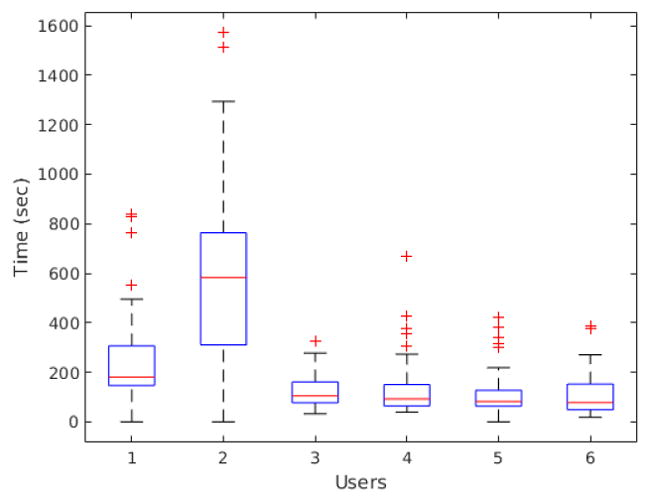
Counting time distribution per user in seconds. User #1, #2 and #3 counted the original images, while user #4, #5 and #6 counted the images with detected cells highlighted
Finally, we measured the walltime (i.e. the total time a computer spent running a process) for the dictionary learning segmentation task together with watershed and filters post-processing. Our method, on average, ran for (segmentation + watershed + post-processing time) 2.7 minutes per image (2.19 minutes), from which 2.47 minutes were used for running the segmentation and 14s (0.23 minutes) for post-processing. The computational time required by our method is way below the average 5.3 minutes required by manual counting. Also, while the user can modify the post-processing filters parameters, the DL-based segmentation, which is the most expensive part, does not require user interaction. Meaning that the main segmentation could easily run in batches and later be fine-tuned by the user.
4. Discussion
This paper proposes a semi-automatic method for segmenting cells in IF images derived from postmortem human brain tissue, followed by their classification according to their markers. The method is appropriated for double-labeled IF and accounts for cells labeled by both markers. We used dictionary learning and sparse coding for performing the cell detection, which is a versatile technique that bypasses intricate data pre-processing and can be trained with small sample sets (when compared to other state-of-the-art machine learning methods, such as deep learning [56]).
We evaluated our results using manually counted images as GT (the gold-standard) and compared our results to four open-access cell counter systems available in literature. We obtained a high recall rate of approximately 0.78, meaning that our system was able to detect most of the cells in the image. However, we obtained a modest precision rate of approximately 0.15, meaning that among the detected cells there were many incorrect detections. This was caused by a high number of FPs, which in turn were caused by excessive amounts of debris and artifacts in our image backgrounds. We included a set of post-processing filters to reduce FPs. However, the same parameter were used for all images during the leave-v-out test and we chose conservative values to avoid too aggressive filtering. In a practical setup the user can easily change these parameters to better suite each images, thus considerably improving the final segmentations. Our method, nonetheless, outperformed all other tested cell counters. In Table 1 we can see that our precision score was one order of magnitude higher that the other tested methods. FPR was also an important problem for the other methods, as can be seen in Figure 8. They were actually much less sensitive and specific when tested with our images. We decided to keep a low amount of FNs in expense of having a high rate of FPs as those could be easily ignored by the user. However, we are aware that excessive amounts of FPs are detrimental, requiring a lot of effort to be removed. Conversely, our system yielded the lowest FNR when compared with the other considered methods, as can be seen in graphs in Figure 8. Our results show that cell counting was on average 2.6 times faster when the users were guided by our method compared to manual counting, meaning that the semi-automated counting proposed here required only 38% of the time needed by manual counting to complete the same task. Finally, instead of criticizing other methods, our comparisons show how challenging handling human brain IF images can be and why we needed a more versatile method tailored to this kind of imagery.
In fact, open-access published algorithms for IF imaging analysis have been rarely tested in postmortem human brain tissue. Moreover, the majority of published algorithms are based on unsupervised, region-based and deformable contour methods. For instance, Lin et al. [57] proposed a method based on watershed for segmenting cells in IF images and created a distance transform that considers both geometric information and image gradients. Their method was tested with rat brain IF images. Elter et al. [58] developed a graph-based approach for segmentation cells in IF images where the cells are modeled as oriented graphs. This model is very computationally efficient, running in linear time, but it requires the background to be smooth, thus not suitable for human brains. Dzyubachyk et al. [2] proposed a multiple levelset method for jointly segmenting and tracking cell in IF images. This study obtained good results on images of HeLa cell cultures. However, their images were homogeneous with smooth backgrounds. Fish et al. [3] took on a slightly different goal: segmenting immunoreactive puncta in fluorescence images of synapses using multi-scale morphology filtering. However, the authors propose a methodology that requires considerable manual interaction, rather than an algorithm per se. Their methodology was tested with synthetic and monkey brain images, yielding good quality masks. Srinavasa et al. [4] focused on segmenting specifically punctate patterns (i.e. bright dots distributed over a dark background) in IF images. The authors presented an active contours formulation that includes region-based and curvature-based forces. They tested their method with IF images of HeLa cell cultures, achieving good agreement with manual labeled GT. Bergeest et al. [5] also proposed the use of active contours for segmenting IF images and presented a new energy functional formulation. Their method was tested in several cell culture images and resulted in high colocalization (Dice coefficient) between ground truth masks and segmentation. Ouertani et al. [6] worked on segmenting and counting Leishmaniasis parasites in serum IF images. They proposed a segmentation pipeline that uses k-means clustering and Otsu thresholding for removing the background from their IF. They obtained mild results, having trouble with images with non-homogeneous gray intensities. Kong et al. [59], unlike most of the studies in this field, presented a 3D segmentation approach using gradient vector flow fields where the cells are located in the point where the vector converge. Their method was tested with human glioblastoma cell culture IF images and obtained high agreement with their manual GT. Finally, in a very recent article, Afshar et al. [8] proposed a distributed-memory algorithm for segmentation where each boundary pixel is modeled as a particle system that evolves in time. Authors showed good results in synthetic images and in cell culture IF images.
Despite the large variety, most computational techniques applied for this problem are unsupervised, meaning that they do not need to be trained nor require labeled data. Unsupervised methods seem attractive at first glance but have been highly outperformed by supervised learning in recent years [60], since the former is a one-fits-all solution while the latter is tuned to better represent underlying data. For this reason, we chose to use a supervised learning method in our segmentation. Dictionary learning has been used in general purpose image classification with good results [38, 61] in recent years. It is easy to train with modest amounts of samples and does not require extensive pre-processing. In addition to that, we designed our method tailored to human brain tissue images. In contrast to cell cultures and experimental animals that show clean background, postmortem human brain tissue, especially from older individuals naturally presents a high amount of background autofluorescence emitted by lipofuscin pigments that increases the noise. Treatment with Sudan Black reduces the problem, without eliminating it though [11, 14, 12, 16]. Also, unpredictable periagonal conditions may increase the background. All these factors sum up to create a challenging image, full of cell debris and low contrast. These debris were often detected as cells by all other counting methods we tested, including ours. However, the use of supervised learning allowed us to reduce the number of FPs, since the DL algorithm learns most representative image features.
In future studies we also plan to include the unstained cells in our counting, as they are important for quantifying the total number of neurons in our IF images. A natural evolution of our method would be the use of deep learning techniques, which have been presenting highly encouraging results for cell detection [62, 63] and histological diagnosis [64], in recent publications. One of our biggest challenges in incorporating deep learning in our studies would be the lack of enough samples for training deep learning neural networks. These models are large and complex, comprising a large number of parameters that need to be estimated, requiring an equally large number of training sets [65]. On the other hand, we could try to circumvent this problem by experimenting with data augmentation methods [66] and transfer learning [67].
In summary, we presented an alternative approach for automated cell counting in IF images that showed a better performance than other available methods. Our code will be available to other groups that may benefit from our method since it is trainable and suitable to work with a broad range of images.
Highlights.
Our method detects and classifies cells in human brain fluorescence microscopy
Dictionary learning and sparse coding learn to better represent cells in our images
Segmented cells are automatically classified to speed up the cell counting process
Our method outperforms several open-access methods in literature
Efficient cell detection can be used in several fluorescence analysis tasks
Acknowledgments
Fund was provided by the National Institute of Health (NIH) grants R01AG040311 and K24AG053435 to Lea T. Grinberg, and by institutional grants, P50AG023501 and P01AG019724, Hospital Israelita Albert Einstein, Sao Paulo, Brazil. This work was partially supported by the Director, Office of Science, Advanced Scientific Computing Research, of the U.S. Department of Energy. Ushizimas Early Career Research project is under Contract No. DE-AC02-05CH11231. This research used resources of the National Energy Research Scientific Computing Center, which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. We thank FAPESP, LIM-22 Faculdade de Medicina Universidade de São Paulo and staff for their invaluable contributions to research. We also thank E. Wes Bethel for supporting the investigation and development of scientific analysis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Herculano-Houzel S, von Bartheld CS, Miller DJ, Kaas JH. How to count cells: the advantages and disadvantages of the isotropic fractionator compared with stereology. Cell and tissue research. 2015;360(1):29–42. doi: 10.1007/s00441-015-2127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dzyubachyk O, Niessen W, Meijering E. Advanced level-set based multiple-cell segmentation and tracking in time-lapse fluorescence microscopy images. ISBI. doi: 10.1109/TMI.2009.2038693. [DOI] [PubMed] [Google Scholar]
- 3.Fish KN, Sweet RA, Deo AJ, Lewis DA. An automated segmentation methodology for quantifying immunoreactive puncta number and fluorescence intensity in tissue sections. Brain research. 2008;1240:62–72. doi: 10.1016/j.brainres.2008.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Srinivasa G, Fickus MC, Guo Y, Linstedt AD, Kovacevic J. Active mask segmentation of fluorescence microscope images. IEEE transactions on image processing : a publication of the IEEE Signal Processing Society. 2009;18(8):1817–1829. doi: 10.1109/TIP.2009.2021081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergeest JP, Rohr K. Efficient globally optimal segmentation of cells in fluorescence microscopy images using level sets and convex energy functionals. Medical image analysis. 2012;16(7):1436–1444. doi: 10.1016/j.media.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 6.Ouertani F, Amiri H, Bettaib J, Yazidi R, Salah AB. Adaptive automatic segmentation of leishmaniasis parasite in indirect immunofluorescence images. Conference proceedings : …Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual Conference; 2014; 2014. pp. 4731–4734. [DOI] [PubMed] [Google Scholar]
- 7.Lee S, Salama P, Dunn KW, Delp EJ. Boundary fitting based segmentation of fluorescence microscopy images. SPIE Imaging and Multimedia Analytics in a Web and Mobile World [Google Scholar]
- 8.Afshar Y, Sbalzarini IF. A parallel distributed-memory particle method enables acquisition-rate segmentation of large fluorescence microscopy images. PloS one. 2016;11(4):e0152528. doi: 10.1371/journal.pone.0152528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price JH, Hunter EA, Gough DA. Accuracy of least squares designed spatial fir filters for segmentation of images of fluorescence stained cell nuclei. Cytome-try. 1996;25(4):303–316. doi: 10.1002/(SICI)1097-0320(19961201)25:4<303::AID-CYTO1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 10.Mueller JL, Harmany ZT, Mito JK, Kennedy SA, Kim Y, Dodd L, Geradts J, Kirsch DG, Willett RM, Brown JQ, Ramanujam N. Quantitative segmentation of fluorescence microscopy images of heterogeneous tissue: Application to the detection of residual disease in tumor margins. PloS one. 2013;8(6):e66198. doi: 10.1371/journal.pone.0066198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duong H, Han M. A multispectral led array for the reduction of background autofluorescence in brain tissue. Journal of neuroscience methods. 2013;220(1):46–54. doi: 10.1016/j.jneumeth.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowson JH. The evaluation of autofluorescence emission spectra derived from neuronal lipopigment. Journal of microscopy. 1982;128(Pt 3):261–270. doi: 10.1111/j.1365-2818.1982.tb04628.x. [DOI] [PubMed] [Google Scholar]
- 13.Diez-Fraile A, Hecke NV, Gurin CJ, DHerde K. Optimizing multiple immunostaining of neural tissue. applications of immunocytochemistry. InTech Publishing; [Google Scholar]
- 14.Davis AS, Richter A, Becker S, Moyer JE, Sandouk A, Skinner J, Taubenberger JK. Characterizing and diminishing autofluorescence in formalin-fixed paraffin-embedded human respiratory tissue. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2014;62(6):405–423. doi: 10.1369/0022155414531549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theofilas P, Polichiso L, Wang X, Lima LC, Alho AT, Leite RE, Suemoto CK, Pasqualucci CA, Jacob-Filho W, Heinsen H, Group BABS, Grinberg LT. A novel approach for integrative studies on neurodegenerative diseases in human brains. Journal of neuroscience methods. 2014;226:171–183. doi: 10.1016/j.jneumeth.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clancy B, Cauller LJ. Reduction of background autofluorescence in brain sections following immersion in sodium borohydride. Journal of neuroscience methods. 1998;83(2):97–102. doi: 10.1016/s0165-0270(98)00066-1. [DOI] [PubMed] [Google Scholar]
- 17.Schnass K. A personal introduction to theoretical dictionary learning. Internationale Mathematische Nachrichten. :228. [Google Scholar]
- 18.Guo Y, Liu Y, Oerlemans A, Lao S, Wu S, Lew MS. Deep learning for visual understanding: A review. Neurocomputing. :187. [Google Scholar]
- 19.Grinberg LT, Ferretti REL, Farfel JM, Leite R, Pasqualucci CA, Rosemberg S, Nitrini R, Saldiva PHN, Filho WJ. Brain bank of the brazilian aging brain study group - a milestone reached ad more than 1600 collected brains. Cell Tissue Banking. 8(151) doi: 10.1007/s10561-006-9022-z. [DOI] [PubMed] [Google Scholar]
- 20.Ferretti REL, Damin AE, Brucki SMD, Morillo LS, Perroco TR, Campora F, Moreira EG, Balbino ES, Lima MCA, Battela C, Ruiz L, Grinberg LT, Farfel JM, Leite RE, Suemoto CK, Pasqualucci CA, Rosemberg SR, Saldiva PHN, Jacob-Filho1 W, Nitrini R. Post-mortem diagnosis of dementia by informant interview. Dementia and Neuropsychologia. 4(2) doi: 10.1590/S1980-57642010DN40200011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montine TJ, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Dickson DW, Duyckaerts C, Frosch MP, Masliah E, Mirra SS, Nelson PT, Schneider JA, Thal DR, Trojanowski JQ, Vinters HV, Hyman BT on Aging, A. Association. National institute on aging-alzheimer’s association guidelines for the neuropathologic assessment of alzheimer’s disease: a practical approach. Acta Neuropathologica. 2012;123(1):1–11. doi: 10.1007/s00401-011-0910-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinsen H, Arzberger T, Schmit C. Celloidin mounting (embedding without infiltration) - a new, simple and reliable method for producing serial sections of high thickness through complete human brains and its application to stereological and immunohistochemical investigations. Journal of chemical neuroanatomy. 20(49) doi: 10.1016/s0891-0618(00)00067-3. [DOI] [PubMed] [Google Scholar]
- 23.Kadowaki M, Karim MR. Cytosolic lc3 ratio as a quantitative index of macroautophagy. Methods in enzymology. 2009;452:199–213. doi: 10.1016/S0076-6879(08)03613-6. [DOI] [PubMed] [Google Scholar]
- 24.Rosenfeldt MT, Nixon C, Liu E, Mah LY, Ryan KM. Analysis of macroautophagy by immunohistochemistry. Autophagy. 2012;8(6):963–969. doi: 10.4161/auto.20186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker KG, Tork I, Hornung JP, Halasz P. The human locus coeruleus complex: an immunohistochemical and three dimensional reconstruction study. Experimental brain research. 1989;77(2):257–270. doi: 10.1007/BF00274983. [DOI] [PubMed] [Google Scholar]
- 26.Rub U, Tredici KD, Schultz C, Thal DR, Braak E, Braak H. The evolution of alzheimer’s disease-related cytoskeletal pathology in the human raphe nuclei. Neuropathology and applied neurobiology. 2000;26(6):553–567. doi: 10.1046/j.0305-1846.2000.00291.x. [DOI] [PubMed] [Google Scholar]
- 27.Oppenheim AV. Discrete-time Signal Processing. Prentice Hall; 1989. [Google Scholar]
- 28.Mallat S. A wavelet tour of signal processing. Academic Press; New York: 1999. [Google Scholar]
- 29.Candes E, Donoho D. Curvelets a surprisingly effective nonadaptive representation for objects with edges. Curves and Surface Fitting; Saint-Malo: [Google Scholar]
- 30.Rubinstein R, Bruckstein AM, Elad M. Dictionaries for sparse representation modeling. Proceedings of IEEE. 98(6) [Google Scholar]
- 31.Aharon M, Elad M, Bruckstein AM. The k-svd: an algorithm for designing of overcomplete dictionaries for sparse representation. IEEE Transactions on Signal Processing. 54(11) [Google Scholar]
- 32.Davis G, Mallat S, Avellaneda M. Adaptive greedy approximations. Constructive Approximations. :13. [Google Scholar]
- 33.Lee H, Raina A, Ng AY. Efficient sparse coding algorithms. Advances in Neural Information Processing Systems. :17. [Google Scholar]
- 34.Engan K, Aese SO, Husoy JH. Method for optimal directions for frame design. IEEE International Conference on Acoustics, Speech, Signal Processing; p. 5. [Google Scholar]
- 35.Elad M, Aharon M. Image denoising via sparse and redundant representations over learned dictionaries. IEEE Transactions on Image Processing. :54. doi: 10.1109/tip.2006.881969. [DOI] [PubMed] [Google Scholar]
- 36.Mairal J, Sapiro G, Elad M. Learning multiscale sparse representations for image and video restoration. SIAM Multiscale Modeling and Simulation. 7(1) [Google Scholar]
- 37.Raina R, Battle A, Lee H, Packer B, Ng AY. Self-taught learning: transfer learning from unlabeled data. ICML [Google Scholar]
- 38.Ramirez I, Sprechmann P, Sapiro G. Classification and clustering via dictionary learning with structured incoherence and shared features. CVPR [Google Scholar]
- 39.Mairal J, Bach F, Ponce J, Sapiro G. Online dictionary learning for sparse coding. ICML [Google Scholar]
- 40.Tibshirani R. Regression shrinkage, selection via the lasso. Journal of the Royal Statistical Society, Series B. 58(1) [Google Scholar]
- 41.Pizer SH, Amburn EP, Austin JD, Cromartie R, Greer AG, Romeny BTH, Zimmerman JB. Adaptive histogram equalization, its variations. Computer Vision, Graphics, and Image Processing. 39(3) [Google Scholar]
- 42.Gonzalez RCC, Richard RE, Woods E. Digital Image Processing. 3. Pearson; 2007. [Google Scholar]
- 43.Otsu N. A threshold selection method from gray-level histograms. IEEE Transactions on System, Man and Cybernetics. 9(1) [Google Scholar]
- 44.Mahalanobis PC. On the generalised distance in statistics. Proceedings of the National Institute of Sciences of India. 2(1) [Google Scholar]
- 45.Duda RO, Hart PE, Stork DG. Pattern Classification. Wiley; 2001. [Google Scholar]
- 46.Xu J, Xiang L, Liu Q, Gilmore H, Wu J, Tang J, Madabhushi A. Stacked sparse autoencoder (ssae) for nuclei detection on breast cancer histopathology images. IEEE Transactions on Medical Imaging. 35(1) doi: 10.1109/TMI.2015.2458702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fawcett T. Technical Report HPL-20034. Roc graphs: notes and practical considerations for data mining researchers. [Google Scholar]
- 48.Grishagin IV. Automatic cell counting with imagej. Analytical Biochemistry. 2015;473:63–65. doi: 10.1016/j.ab.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. Cellprofiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Padmanabhan K, Eddy WF, Crowley JC. A novel algorithm for optimal image thresholding of biological data. Journal of neuroscience methods. 2010;193(2):380–384. doi: 10.1016/j.jneumeth.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 51.Soliman K. Cellprofiler: Novel automated image segmentation procedure for super-resolution microscopy. Biological Procedures Online. 2015;17 doi: 10.1186/s12575-015-0023-9. 11–015–0023–9 eCollection 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ushizima DM, Bianchi AGC, Carneiro CM. Segmentation of subcellular compartiments combining superpixel representation with voronoi diagrams. International Symposium on Biomedical Imaging. [Google Scholar]
- 53.Ushizima DM, Alessandra AH, Gomes H, Carneiro CM. Automated pap smear cell analysis: Optimizing the cervix cytological examination. ICMLA [Google Scholar]
- 54.Selinummi J, Seppala J, Yli-Harja O, Puhakka JA. Software for quantification of labeled bacteria from digital microscope images by automated image analysis. BioTechniques. 2005;39(6):859–863. doi: 10.2144/000112018. [DOI] [PubMed] [Google Scholar]
- 55.Mairal J, Bach F, Ponce J, Sapiro G. Online learning for matrix factorization and sparse coding. Journal of Machine Learning Research. :11. [Google Scholar]
- 56.Mao R, Zhu HH, Zhang L, Chen A. A new method to assist small data set neural network learning. International Conference on Intelligent Systems Design and Applications. [Google Scholar]
- 57.Lin G, Adiga U, Olson K, Guzowski JF, Barnes CA, Roysam B. A hybrid 3d watershed algorithm incorporating gradient cues and object models for automatic segmentation of nuclei in confocal image stacks. Cytometry. Part A : the journal of the International Society for Analytical Cytology. 2003;56(1):23–36. doi: 10.1002/cyto.a.10079. [DOI] [PubMed] [Google Scholar]
- 58.Elter M, Daum V, Wittenberg T. Maximum-intensity-linking for segmentation of fluorescence-stained cells. MIAAB [Google Scholar]
- 59.Kong J, Wang F, Teodoro G, Liang Y, Zhu Y, Tucker-Burden C, Brat DJ. Automated cell segmentation with 3d fluorescence microscopy images. Proceedings / IEEE International Symposium on Biomedical Imaging: from nano to macro. IEEE International Symposium on Biomedical Imaging; 2015; 2015. pp. 1212–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.LeCunn Y, Bengio Y, Hinton G. Deep learning. Nature. :521. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 61.Bull G, Gao J, Antolovich M. Image segmentation using dictionary learning and compressed random features. Digital lmage Computing: Techniques and Applications [Google Scholar]
- 62.Ciresan DC, Giusti A, Gambardella LM, Schmidhuber J. Mitosis detection in breast cancer histology images with deep neural networks. Medical image computing and computer-assisted intervention : MICCAI …International Conference on Medical Image Computing and Computer-Assisted Intervention. 2013;16(Pt 2):411–418. doi: 10.1007/978-3-642-40763-5_51. [DOI] [PubMed] [Google Scholar]
- 63.Chen T, Chefdhotel C. Deep learning based automatic immune cell detection for immunohistochemistry images. MICCAI [Google Scholar]
- 64.Litjens G, Sanchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I, van de Kaa CH, Bult P, van Ginneken B, van der Laak J. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Scientific reports. 2016;6:26286. doi: 10.1038/srep26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. 2. 2009. [Google Scholar]
- 66.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Neural Information Processing Systems [Google Scholar]
- 67.Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep convolutional neural networks for computer-aided detection: Cnn architectures, dataset characteristics and transfer learning. IEEE Transactions on Medical Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]