Abstract
Elevated tumor expression of the cell surface GPI-linked CD24 protein signals poor patient prognosis in many tumor types. However, some cancer cells selected to be negative for surface CD24 (surCD24-) still retain aggressive phenotypes in vitro and in vivo. Here we resolve this apparent paradox with the discovery of biologically active, nuclear CD24 (nucCD24) and finding that its levels are unchanged in surCD24- cells. Using the complementary techniques of biochemical cellular fractionation and immunofluorescence, we demonstrate a signal for CD24 in the nucleus in cells from various histological types of cancer. Nuclear-specific expression of CD24 (NLS-CD24) increased anchorage independent growth in vitro and tumor formation in vivo. Immunohistochemistry of patient tumor samples revealed the presence of nucCD24, whose signal intensity correlated positively with the presence of metastatic disease. Analysis of gene expression between cells expressing CD24 and NLS-CD24 revealed a unique nucCD24 transcriptional signature. The median score derived from this signature was able to stratify overall survival in 4 patient datasets from bladder cancer and 5 patient datasets from colorectal cancer. Patients with high scores (more nucCD24-like) had reduced survival. These findings define a novel and functionally important intracellular location of CD24, they explain why surCD24- cells can remain aggressive, and they highlight the need to consider nucCD24 in both fundamental research and therapeutic development.
Keywords: CD24, nucleus, metastasis, cancer, biomarker, prognosis
INTRODUCTION
High CD24 expression in breast, prostate, pancreas, ovary, colorectal and bladder tumors signals poor prognosis (1). Expression of this GPI-linked, canonical cell surface glycoprotein (2) promotes in vitro cell growth as well as in vivo tumor growth, invasion and metastasis (3–12) while depletion reduces these properties (4,7,9,10,13,14). Treatment of tumor bearing mice with CD24 monoclonal antibody leads to reduced tumor burden in mice harboring human bladder (9), pancreatic (4), lung (3,4), ovarian (3), and colon (15) tumors. CD24 knockout mice exposed to chemical carcinogens developed no colorectal tumors (16) and fewer bladder tumors (10). The CD24 knockout mice also had reduced metastasis (10). Together, these findings make CD24 a very attractive therapeutic target.
However, recent evidence casts doubt that antibody-mediated CD24 therapy constitutes the optimal approach in patients. For example, recent work revealed that low CD24 surface expression leads to only a ~50% decrease in metastatic cancer burden while shRNA mediated silencing of CD24 results in a 90% decrease (9). In addition, ourselves (9) and others (17) have shown that cancer cells with little to no surface CD24 (surCD24-) retain significant tumorigenic properties. Together, these data suggest that CD24 exists in additional cellular locations and has significant biological activity. Studies supporting this hypothesis show cytoplasmic CD24 binds G3BP, leading to degradation of mRNAs which drive invasion and metastasis (18), and that cytoplasmic CD24 competitively inhibits ARF binding to NPM, resulting ultimately in decreased levels of p53 (19).
Hence, we sought to define the location of intracellular CD24 and determine if location impacts tumor phenotypes and patient outcomes in order to eventually allow the development of optimal CD24 directed therapy. Here we identify a distinct nuclear population of CD24 (nucCD24) in cancer cells and show that nucCD24 promotes tumorigenic phenotypes both in vitro and in vivo. Analysis of CD24 signal in patient tumor samples revealed a strong association between nucCD24 signal and aggressive disease. This work fundamentally changes our concepts regarding CD24 directed therapy.
MATERIALS AND METHODS
Cell Lines and Culture
Human bladder cancer cell lines UMUC3, UMUC6, UMUC13, MGHU3, SW1710, and UMUC3-Lul2 (9) were cultured at 37°C in 5% CO2 in Modified Eagle’s Medium (MEM) + 10% FBS + 1 mM Sodium Pyruvate. All media and reagents were obtained from Gibco. The above human bladder cancer cell lines and the human cell lines HCC1937, H358, HT29, and DU145 were obtained from the University of Colorado Cancer Center Tissue Culture Shared Resource from 2013–2016. All cell lines in the UCCC Tissue Culture Shared Resource undergo cell line authentication (CLA) using the STR DNA Profiling PowerPlex-16 HS Kit (Promega) in the Molecular Biology Service Center at the Barbara Davis Center, University of Colorado, Anschutz Medical Campus. All experiments in this study were performed within 6 weeks of being thawed from an ampule that was created within 2 weeks of receiving cells from the UCCC Tissue Culture Shared Resource.
Western Blot Analysis, Antibodies and RNA interference
Western blots were carried out as previously described (10) using two anti CD24 antibodies (ML5 and Swa11) as described in Supplementary Methods. The chromatin isolation and subsequent chromatin digestion method used in this study is largely based on protocols previously published (20). The micrococcal nuclease used in this study (Thermo Scientific #88216) was used at 4U in 100 μL of buffer A plus CaCl2 and incubated at 37°C for 5 mins. RNA interference was carried out as previously described (10) using siRNA oligonucleotides as follows: Control CGUACGCGGAAUACUUCGA; CD24 #1 ACAACTGGAACTTCAAGTAAC; CD24 #2 CAACTAATGCCACCACCAA; Xpo1 SMARTpool (Dharmacon M-003030-02-0005). UMUC3-Lul2 cells stably expressing CD24 shRNA were made by lentiviral transduction of a plasmid expressing the CD24 #1 siRNA sequence and subsequent selection by neomycin. More details can be found in Supplementary Methods.
Fluorescent Activated Cell Sorting
To obtain CD24 surface negative cells, UMUC3-Lul2 cells were harvested and placed on ice while incubating with 0.5% BSA/PBS for 20min. To cell suspensions was added either isotype control (IgG-FITC – BD Biosciences #556652) or anti-CD24 (ML5-FITC) antibodies at 1:100 dilution. After 30min incubation on ice cells were washed 1× 0.5% BSA/PBS and resuspended in sorting buffer (1× PBS (Ca/Mg++ free), 1mM EDTA, 25mM HEPES pH 7.0, 1% BSA, DAPI (4ug/mL)). Viable, single cells were assessed on a Moflo XDP 100 and a FITC negative gate was established using IgG-FITC labeled cells. Subsequently, ML5-FITC labeled cells were sorted. After all cells were collected the purity of the sample was assessed by reanalysis of 1,000 events on the Moflo XDP 100. To assess surface levels of CD24 on previously sorted cells and engineered cell lines, cells were prepared as above and CD24 surface signal assessed on a Beckman Coulter Gallios 561. Gating with isotype control labeled cells and CD24 signal quantitation were performed using Kaluza Flow Analysis Software (Beckman Coulter).
In Vitro Cell Growth Assays and Cell Fractionation
Anchorage dependent and independent (soft agar and suspension) cell proliferation was carried out as described in Supplementary Methods. To obtain membrane/cytoplasm and nucleoplasm fractions cells were treated as per manufacturer instructions (NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce #78833)). Equal volumes were loaded for each fraction.
Microscopy
Cells were plated onto glass coverslips (Fisherbrand 18mm # 12-545-100) residing in 12 well dishes. After 24 hrs the cells were washed 1× with PBS followed by the addition of paraformaldehyde (Thermo Scientific #28908) at 4% final concentration in calcium and magnesium free PBS (CMF-PBS) for 12 mins at room temperature (RT). Most experiments also had a final concentration of 0.1% glutaraldehyde (Fluka #49630). Coverslips were washed 1× with CMF-PBS for 5 mins. For permeabilization the coverslips were treated with 0.1% Triton X-100/CMF-PBS for 10 mins at RT before washing 1× with CMF-PBS for 5 mins. For blocking step the coverslips were incubated with 0.5% BSA/CMF-PBS for 30 mins at RT. To this solution was added anti-CD24 antibody (Swa11) at 1:40 dilution and coverslips incubated overnight at 4°C. The next morning the coverslips were washed 3× with CMF-PBS followed by incubation with anti-mouse antibody conjugated with FITC (Cell signaling #4408S) in 0.5% BSA/CMF-PBS at RT for 30 mins. Coverslips were washed 2× CMF-PBS followed by 1× with DAPI (4 μg/mL final) in CMF-PBS for 5 mins at RT. Coverslips were then placed onto glass slides with mounting media (Invitrogen #P36934) and allowed to dry overnight. For methanol fixation of cells on glass coverslips, the coverslips were washed 1× with PBS followed by the addition of cold (−20°C) 100% methanol for 10 mins on ice. Coverslips were then washed 2× with PBS at RT before proceeding to the blocking step as detailed above. Details of the imaging software and techniques are described in Supplementary Methods
In Vivo Tumor Study
UMUC13 and UMUC6 cells were implanted subcutaneously in 6 week old NCr nu/nu mice (NCI-Frederick, Frederick, MD) at 2 sites per mouse, 7 mice for each group (Wt or NLS expressing cells). Starting at 8–14 days implantation the tumors were measured every 3–4 days in two dimensions with a digital caliper and the tumor volume determined with the equation (L × W2)/2. All animals used in this study were treated according to University of Colorado Denver and Institutional Animal Care and Use Committee (IACUC) guidelines (protocol number is B-93413(12)1E).
Immunohistochemistry and Scoring
Immunohistochemistry was performed with a well characterized CD24 (Swa11) antibody (21) on formalin-fixed paraffin-embedded urothelial carcinoma specimens from 105 patients who had undergone cystectomy and lymph node dissection. Further details are provided in Supplementary Methods.
Microarray Analysis
Whole-genome expression data was acquired from UMUC13 bladder cancer cells expressing either Wt-CD24 or NLS-CD24 (n = 3/group) using the following conditions, which conform to MIAME principals (22). Total RNA from each line was assessed using the Affymetrix Clariom S Human Array (Affymetrix, Santa Clara, CA). Expression values were normalized and summarized into transcript clusters using Plier (Guide to Probe Logarithmic Intensity Error (PLIER) Estimation, Affymetrix Technical report, Santa Clara 2005) and log2 transformed for analysis. Linear regression (lm function in R v3.3.1) was used to look for differential expression between WT and NLS groups and p-values were adjusted for multiple comparisons using a False Discovery Rate (FDR). Only candidates with nominal p-value <0.005 were considered. The list of differentially expressed genes and corresponding expression values were assessed by Ingenuity Pathway Analysis software (Qiagen, Hilden, Germany). Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathway Knowledge Base (IPKB), leading to an interpretation to highlight the diseases, biological processes, canonical pathways, and other gene networks that nucCD24 impacts.
Genes differentially expressed between Wt-CD24 and NLS-CD24 were evaluated for their ability to stratify patient outcome in 13 cancer patient datasets. The patient datasets include GSE12276, GSE3141, GSE17538, GSE16560, GSE13507, GSE14333, GSE39582, GSE19915, GSE13507, GSE37892, and GSE41258. Gene expression data for MSKCC was downloaded from supplementary material in publication (23) while gene expression data for Blaveri came from material in publication (24). These datasets consist of gene expression generated by array technology (please see (23–26) for platform specifics) and the signature was created for each dataset separately. To create this signature, the probe sets that correspond to the candidate genes were first identified. For each probe set, a Cox proportional hazards (PH) regression was performed using the probe sets expression value to predict survival. A gene signature score was then calculated by taking a weighted sum of the expression values from the probe sets with the weight corresponding to the estimated regression coefficient from the univariate PH models. Patients were then classified as having either a high or low gene expression signature by the median of the scores. Stratified Kaplan-Meier plots were generated based on high and low score status. To determine if there is a significant difference of survival between high and low gene signatures, a PH regression was performed using high/low classification to predict survival. p-values from the likelihood ratio test are reported.
Statistical Analysis
Values provided are the mean. Error bars on bar and line graphs denote standard error of the mean. Statistical significance was assessed using a two-tailed Student t test with equal variance unless otherwise noted in figure legend. For relationships between CD24 immunohistochemistry staining and phenotype, p-values were calculated using a two-tailed Student t test to compare continuous H-scores across independent samples, and using the Wilcoxon signed-rank test to compare qualitative staining scores across matched samples (Primary Tumor (M+) to Lymph Node Tumor).
RESULTS
Surface CD24 negative cells have residual CD24 protein expression and CD24 driven growth
Human bladder cancer cells (UMUC3-Lul2) expressing CD24 shRNA had little to no metastatic ability in vivo while cells sorted by FACS for no surface CD24 (surCD24-) had only reduced (50%) metastatic ability (9). This suggested CD24 was still driving metastasis in surCD24- cells but that hypothesis remained untested. Here we used FACS to generate a surCD24- population of cells (Supp. Fig. S1A) and confirmed lack of CD24 on the surface using CD24 immunofluorescence (Supp. Fig. S1B). surCD24- cells have increased anchorage independent growth in vitro relative to unsorted cells (shCtrl) and cells lacking CD24 (shCD24) (Fig. 1A). Anchorage dependent assessment demonstrated that surCD24- cells do not simply grow faster than unsorted cells (Supp. Fig. S1C). Western blot analysis of surCD24- cells revealed that low levels of CD24 persist (Supp. Fig. S1D) while FACS analysis confirmed these cells remained surCD24- (Supp. Fig. S1E), demonstrating that our results are not due to reacquisition of surCD24 expression. To determine if intracellular CD24 in surCD24- cells drives in vitro growth we eliminated all CD24 using siRNA. Treatment of surCD24- cells with CD24 siRNA leads to dramatic reduction in CD24 signal, shown here (Fig. 1B) with its characteristic banding pattern owing to the presence of glycans of varying length attached to the protein. This CD24 reduction also correlated with a reduction in anchorage dependent (Fig. 1C) and independent (Fig. 1D) proliferation. These data suggest that the enhanced growth observed in surCD24- cells is driven by intracellular CD24.
Figure 1. FACS sorted cells lacking surface CD24 (surCD24-) have increased tumorigenic properties and residual intracellular CD24.
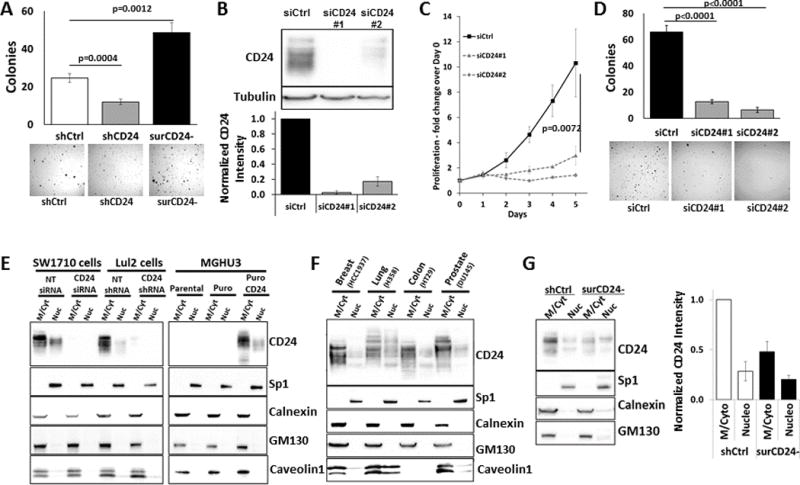
(A) surCD24- UMUC3-Lul2 cells grow better in soft agar anchorage independent assays relative to both unsorted cells and those depleted of CD24. All graphs display the means obtained from combining data of 3 or more independent analyses. Images and blots are representatives from 1 of the independent analyses. Error bars represent standard error. Statistical analysis for all experiments, unless otherwise noted, was a t test and the p-value is presented. (B) Treatment of surCD24- cells with 2 different CD24 siRNA molecules (siCD24#1 and siCD24#2) for 72 hrs leads to elimination of residual CD24 protein. (C) Depletion of total cellular CD24 from surCD24- cells leads to a reduction in anchorage dependent proliferation as assessed by counting cells daily after treating with siRNA. Counting began on the day cells were treated with siRNA (day 0). (D) Depletion of total cellular CD24 from surCD24- cells leads to decreases in soft agar anchorage independent growth. Cells were plated in agar after 48 hrs of siRNA treatment. (E) Subcellular fractionation of bladder cancer cells into membrane/cytoplasm (M/Cyt) and nucleoplasm (Nuc) fractions revealed that CD24 protein exists in the nucleoplasm. This CD24 signal was reduced in cells expressing CD24 siRNA or shRNA. Similarly, this CD24 signal only existed in MGHU3 cells when CD24 was exogenously expressed. (F) CD24 signal was also found in the nucleoplasm fraction from cells of 4 different cancer types. (G) The amount of nucleoplasmic CD24 was the same in unsorted and surCD24- UMUC3-Lul2 cells while the cytoplasmic and membrane bound CD24 decreased dramatically in the same cells.
Cellular fractionation finds CD24 in the nucleoplasm of parental and surCD24- cells
The data above demonstrate a role for intracellular CD24 in cell growth but do not identify in which cellular compartments this CD24 resides. To determine this, we subjected cell lysates to subcellular fractionation. CD24 signal was observed in the cytoplasmic and membrane bound fraction (M/Cyt) as well as the nucleoplasm fraction (Nuc) of SW1710 and UMUC3-Lul2 bladder cancer cells (Fig. 1E). Similarly, CD24 signal was observed in the nucleoplasm fraction of MGHU3 bladder cancer cells stably expressing exogenous CD24. This CD24 signal was absent in SW1710 cells treated with CD24 siRNA, UMUC3-Lul2 cells stably expressing CD24 shRNA, and MGHU3 expressing vector control (Fig. 1E). It is worth noting that the characteristic banding pattern of CD24 signal is different between M/Cyt and Nuc fractions, suggesting that changes in glycosylation of CD24 protein may control the destination of the protein. Next, we confirmed the integrity of the fractions was confirmed by the findings that only the nucleoplasm fraction contained signal for the transcription factor Sp1 and was not contaminated with endoplasmic reticulum (Calnexin), Golgi apparatus (GM130) or lipid rafts (Caveolin1) (Fig. 1E). Importantly, this location of CD24 is not cell line dependent, as CD24 is found in the nucleoplasm fraction of cancer cell lines from different tissues including breast, lung, colon and prostate cells (Fig. 1F). In fact, we found CD24 signal in the nucleoplasm fraction of every cell line we analyzed which expresses CD24 protein.
Given that whole cell lysates from surCD24- cells showed residual CD24 protein (Supp. Fig. S1D), if nucCD24 is contributing to this residual signal it should be observed in the nucleoplasm fractionation of surCD24- cells. This was indeed the case and found to exist at about the same level as unsorted shCtrl cells (Fig. 1G). In contrast, cytoplasmic and membrane bound CD24 decreased dramatically in the same cells. Since surface CD24 is known to reside in lipid rafts, this latter approach has potential caveats since lipid raft proteins can precipitate into the nuclear fraction with use of non-ionic detergents in the lysis buffer (27). We evaluated this and provide several lines of evidence that no lipid raft-based CD24 contamination exists in this case. First, we are assessing the soluble, nucleoplasm material and not the detergent insoluble pellet which is generated from this approach and which is not shown. Second, we show that the lipid raft resident protein Caveolin-1 is not present in this soluble nucleoplasm fractionation (Fig. 1E–F). Third, the nucleoplasm abundance of CD24 is the same in unsorted and surCD24- cells. This would not be observed if cell surface CD24 was contaminating the nucleoplasm fraction.
Immunofluorescence microscopy confirms nucCD24 location
To complement cellular fractionation studies we performed CD24 immunofluorescence. The use of paraformaldehyde (PFA) to fix cells demonstrated high levels of punctate CD24 signal on the plasma membrane (Fig. 2A). Since PFA only fixation is insufficient to fully characterize GPI-linked protein distribution (28), we added glutaraldehyde (GA) to PFA fixation which lead to less aggregation of CD24 signal on the surface of bladder cancer cells (Fig. 2A, Supp. Fig. S2A). Similarly, CD24 signal in methanol fixed cells and GFP-CD24 (Supp. Fig. S2B) signal in unfixed cells also revealed a less punctate, more diffuse distribution of CD24 protein on the cell surface (Fig. 2A). Thus, the true cell surface distribution of CD24 is likely diffuse on the plasma membrane.
Figure 2. Immunofluorescence confirms the presence of nucCD24.
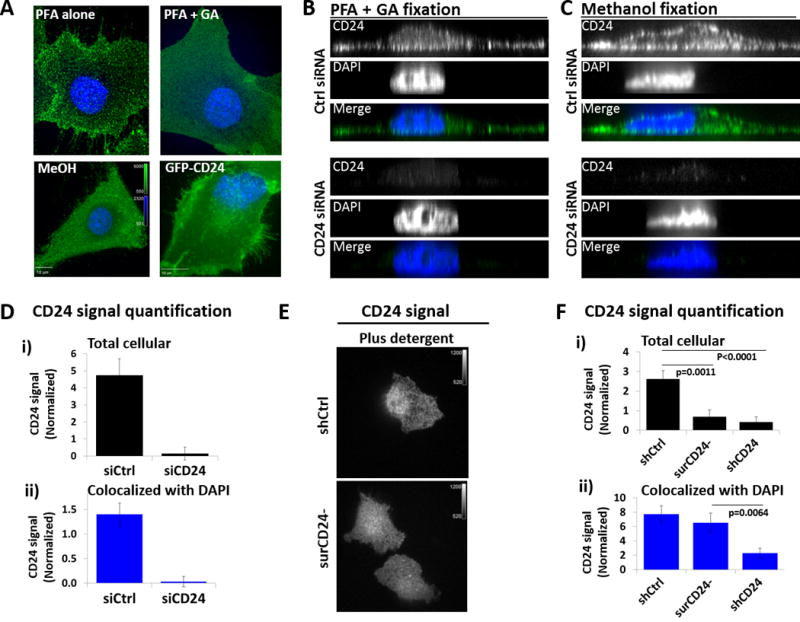
(A) SW1710 bladder cancer cells were fixed with paraformaldehyde (PFA) or PFA+GA (glutaraldehyde). The addition of GA to the fixation method reduces the movement of GPI-anchored proteins and their subsequent antibody-induced aggregation on cell surface. Fixing with methanol (MeOH) also prevents this aggregation. CD24 immunofluorescence signal from PFA+GA fixed cells matches that of unfixed MGHU3 cells (which lack endogenous CD24 expression) expressing GFP-CD24. (B) SW1710 cells were fixed with PFA+GA and probed with anti-CD24 antibody and z-stacks acquired with a confocal microscope. Orthogonal views reveal cytoplasmic and nucCD24 signal, which is lost with CD24 siRNA treatment. (C) SW1710 cells fixed with methanol before probing and imaging as above. (D) i) Total cellular – quantification of CD24 signal in a statistical mask of the whole cell (all planes of a z-stack) confirms the dramatic loss of total cellular CD24 signal with CD24 siRNA treatment. Minimum of 14 cells quantified per treatment. ii) Colocalized with DAPI – quantification of CD24 signal in a statistical mask defined by a minimum DAPI signal confirms the dramatic loss of nucCD24 signal with CD24 siRNA treatment. (E) Immunofluorescence analysis in the presence of cell permeabilization (plus detergent) shows that intracellular CD24 signal is the same between unsorted and surCD24- cells. Channel intensities are identical. (F) i) Total cellular – Using the same approach as in panel D we show that total cellular CD24 signal in unsorted cells is dramatically higher than surCD24- cells and cells expressing CD24 shRNA. ii) Colocalized with DAPI – nucCD24 signal is not statistically different between unsorted and surCD24- cells while there is a significant reduction in nucCD24 signal in CD24 shRNA cells. Minimum of 34 cells quantified per cell type.
PFA/GA fixation revealed CD24 signal in both the cytoplasm and nucleus of bladder cancer cells (Fig. 2B, Supp. Fig. S2C). We also observed these signals in methanol fixed cells (Fig. 2C). This nuclear and cytoplasmic signal was no longer apparent when cells were depleted of CD24 (Fig. 2B–C, Supp. Fig. S2C). Using statistical masks, CD24 signal was quantified within each mask (Supp. Fig. S3A) to reveal that siRNA treatment lead to a >95% decrease in total cellular CD24 signal (Fig. 2Di). Identical results were observed in UMUC3-Lul2 cells stably expressing CD24 shRNA (Supp. Fig. S3B). Quantification of nucCD24 was achieved by generating a segmented statistical mask based on a minimum DAPI signal (Supp. Fig. S3C) and quantifying CD24 signal within that segmented mask. This approach confirmed that CD24 signal exists in the nucleus and that >95% of this signal is lost with CD24 siRNA (Fig. 2Dii) or shRNA (Supp. Fig. S3D). This nucCD24 signal is 15–20% of total cellular CD24, percentages very similar to those observed above in biochemical experiments (Fig. 1E–G).
CD24 immunofluorescence of non-permeabilized surCD24- cells did not detect nucCD24 signal (Supp. Fig. S1B) while permeabilized surCD24- cells had the same amount of nucCD24 signal as unsorted cells (Fig. 2E). We next quantified CD24 signal in control, surCD24-, and shCD24 cells and found total cellular CD24 signal in surCD24- cells was 75% lower than control cells and similar to shCD24 cells (Fig. 2Fi), suggesting the majority of CD24 is at the cell surface. In contrast, nucCD24 (DAPI colocalized) did not decrease in surCD24- cells (Fig. 2Fii), consistent with fractionation (Fig. 1G).
nucCD24 associates with chromatin and promotes an aggressive tumor phenotype
Next we asked if CD24 can associate with the chief component of the nucleus, chromatin. Using a chromatin prep (20) we demonstrated DNA binding specificity by looking for proteins released from the prep as a result of DNA digestion (Fig. 3A). Our chromatin/insoluble protein prep (P1) contained HistoneH3, as expected, as well as CD24 (Fig. 3A). The soluble protein tubulin was not found in the chromatin isolate while the cytoskeletal protein vimentin was, confirming the crude nature of the prep. However, vimentin was not released to the supernatant upon DNA digestion with micrococcal nuclease (MN) while both HistoneH3 and CD24 were released (Fig. 3A) indicating that CD24, like HistoneH3, is bound to chromatin within the nucleus. To determine whether CD24 enters the nucleus passively or actively, we assessed nuclear levels of CD24 following depletion of the nuclear export mediator protein Xpo1 (Crm1). Treatment of cells with Xpo1 siRNA led to an 80% decrease in cellular Xpo1 and a subsequent accumulation of CD24 and RanBP1 in the nucleus (Fig. 3B). Furthermore, inhibition of Xpo1 activity in cells with the drug leptomycin B also led to an accumulation of CD24 and RanBP1 in the nucleus (Fig. 3C, Supp. Fig. S4). These observations suggest that CD24 movement in and out of the nucleus is regulated in an active manner. Since CD24 lacks a known nuclear localization or export signal its movement is likely dependent upon a yet to be identified associating protein.
Figure 3. nucCD24 binds chromatin and its expression enhances growth in vitro and in vivo.
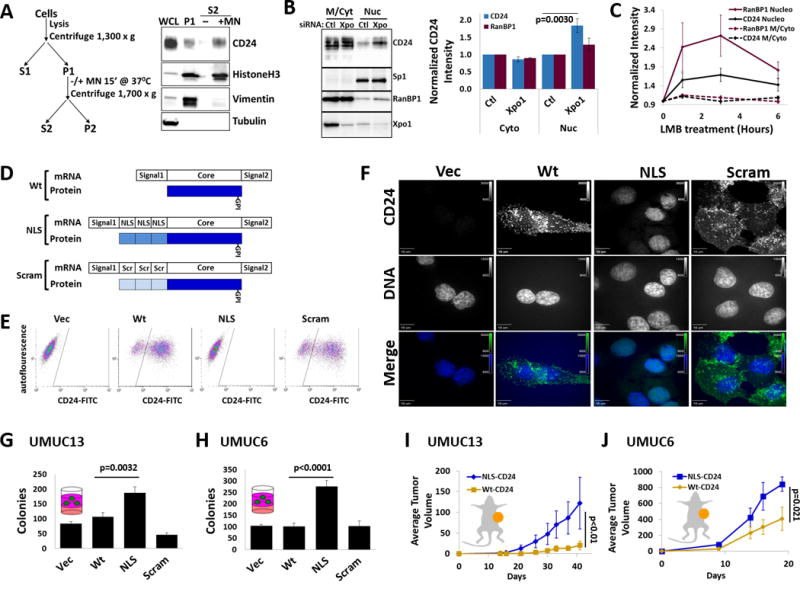
(A) Chromatin isolation strategy and subsequent DNA digestion with micrococcal nuclease (MN). Modified from classic Méndez and Stillman protocol. CD24, HistoneH3 and vimentin were all isolated in the chromatin prep (P1) from UMUC3-Lul2 cells but only CD24 and HistoneH3 are released from the prep (S2) following DNA digestion. (B) Depletion of Xpo1 from UMUC3-Lul2 cells led to an accumulation of CD24 and RanBP1 in the nucleoplasm fraction but no change in Sp1 levels. (C) Inhibition of nuclear export in UMUC3-Lul2 cells by Leptomycin B (LMB) (20nM) also resulted in accumulation of CD24 and RanBP1 in the nucleoplasm fraction. Blot shown in Fig. S4. (D) Schematic of the CD24 cDNA constructs and mature protein products generated in this study to drive CD24 to the nucleus (NLS) or the scrambled (Scram) control. (E) Stable expression of these constructs in MGHU3 cells (which lack endogenous CD24 protein and do not form colonies in soft agar) and subsequent FACS shows that wild-type-CD24 (Wt) and Scram-CD24 (Scram) cells have high CD24 signal on their cell surface while control cells and NLS-CD24 (NLS) cells do not. (F) Immunofluorescence confirms that Wt and Scram-CD24 have identical cellular distribution while NLS-CD24 colocalizes exclusively with DNA (DAPI). Channel intensities are identical across each line for DAPI or CD24 signals. (G) Expression of CD24 constructs in UMUC13 cells, which lack endogenous CD24 protein and which form colonies in soft agar assays, reveals that cells expressing NLS-CD24 form the most colonies. (H) Expression of the CD24 constructs in UMUC6 cells, which express little endogenous CD24 and which form colonies in soft agar assays, reveals that cells expressing NLS-CD24 form the most colonies. (I) UMUC13 cells expressing Wt or NLS-CD24 were injected into the flanks of nude mice and tumor volume analysis revealed that cells expressing NLS-CD24 lead to a tumor growth rate almost 3 times that of cells expressing Wt-CD24. p-value is from two-way RM-ANOVA. (J) UMUC6 cells expressing Wt-CD24 had almost half the tumor growth rate of UMUC6 cells expressing NLS-CD24.
To explore the biological significance of nucCD24 with minimal influence from cytoplasmic CD24, we expressed CD24 exclusively in the nucleus in cells. The fusion of 3 nuclear localization sequences (NLS) to the CD24 N-terminus (Fig. 3D) led to CD24 protein being absent from the cell surface (Fig. 3E) and cytoplasm and present only in the nucleus (Fig. 3F). In contrast, wild type CD24 (Wt) and a 3×-scrambled control CD24 were present at the cell surface and in the entire cell (Fig. 3E–F). Analysis revealed that NLS-CD24 promotes an increase (2–2.7×) in colony formation in agar relative to Wt and Scram-CD24 (Fig. 3G–H). UMUC13 or UMUC6 cells expressing Wt or NLS-CD24 subcutaneously injected into mice showed the latter grew better that the former (Fig. 3I–J). These studies show nuclear CD24 movement is actively regulated and nucCD24 promotes enhanced tumorigenic phenotypes in vitro and in vivo.
nucCD24 is associated with metastasis and poor outcome in bladder cancer patients
To determine the clinical impact of these findings we performed CD24 immunohistochemistry on human bladder urothelial carcinomas (UC) from 105 patients (Fig. 4A). nucCD24 signal was observed in 51% of primary UCs, with that percentage increasing (red arrow) to 68% (p=0.056) for primary UCs which had concomitant lymph node metastasis at time of radical cystectomy (Fig. 4B). CD24 staining on membranes and cytoplasm was observed in over 85% of primary UCs (Fig. 4B), consistent with past findings that CD24 expression is elevated in bladder cancer (9,12) and other cancer types (1). There was a significant (p=0.020) increase (red arrow) in membrane/cytoplasmic staining of lymph node metastatic lesions compared to their matched (n=54) primaries (Fig. 4B). The intensity of CD24 signal as measured by H-score demonstrated a significant increase (p=0.039) in nucCD24 signal in patients whose primary UC had metastasized to the lymph nodes relative to those with non-metastatic (n=51) primary tumors (Fig. 4C). Membrane/cytoplasmic CD24 staining intensity demonstrated a significant (p=0.0055) increase from primary to metastatic lesions in the 54 paired samples (Fig. 4D). Thus, high intensity nucCD24 signal correlates with metastatic disease and nearly all metastatic lesions appear to have significant CD24 expression.
Figure 4. Immunohistochemical analysis of human bladder tumors illustrates that nucCD24 signal correlates with poor patient outcome.
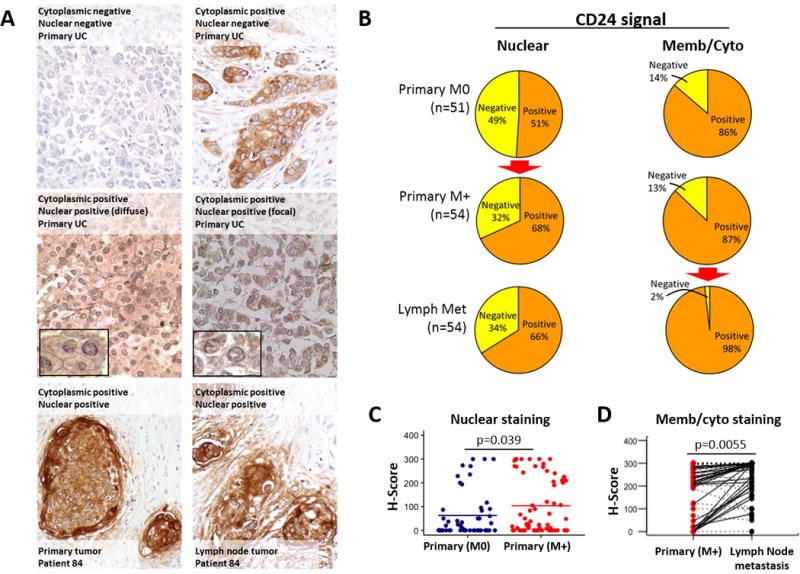
(A) A tissue microarray of bladder cancer tumors and corresponding lymph node metastases from 105 patients was probed for CD24. Staining for CD24 was observed in cytoplasmic/plasma membrane distribution as well as the nucleus. Examples of the various staining patterns are shown here including matched primary and metastatic lesions for patient #84. Blowouts highlight the nuclear staining in two patients. (B) Tissues from each patient was evaluated for plasma membrane/cytoplasmic and nuclear staining. Over half of patients had nucCD24 staining, with this number increasing significantly (red arrow; p=0.056) to 66% in patients who had metastatic disease. Membrane/cytoplasm staining was found in 86% of patients and this number showed a statistically significant increase (red arrow; p=0.006) to 98% (53 of 54 samples) for the metastatic lymph node lesions. (C) CD24 signal intensity was scored as negative (0), weak (1+), moderate (2+), or strong (3+). CD24 staining was analyzed on non-metastatic (Primary (M0)) (n=51) and metastatic (Primary (M+)) (n=54) primary tumors. p-value calculated using a two-tailed Student t test to compare continuous H-scores across independent samples. (D) Membrane/cytoplasmic staining in 54 paired samples. Black lines are solid if there is an increase between Primary Tumor (M+) and lymph met, and are dashed otherwise. p-value calculated using the Wilcoxon signed-rank test to compare qualitative staining scores across matched samples.
Transcriptional signature of nucCD24 is associated with aggressive characteristics in cancer
To begin understanding the mechanisms by which nucCD24 promotes more tumorigenic and metastatic properties we compared gene expression between Wt and NLS-CD24 expressing cells. Microarray analysis revealed that 304 genes (Supp. Data Table 1) were significantly differentially expressed (p<0.005) (Fig. 5A). A transcriptional signature was developed and evaluated for its ability to stratify patient outcome as we have done before (29). We analyzed 4 bladder cancer (23–26) and 5 colorectal cancer (30–34) patient datasets with common microarray technology because of the relevance of CD24 in both bladder cancer (11) and colon cancer (15,35). We analyzed additional patient datasets, based on large sample sizes and common microarray platforms, for 3 other cancer types (36–38) for which we’ve confirmed the existence of nucCD24 (Fig. 1F). Patients were classified as having either a high (above median) or low (below median) gene expression signature score (29) and Kaplan-Meier plots generated. The nucCD24 gene signature was found to have statistically significant correlation with reduced patient survival in cancer patient datasets from 5 different caner types (Fig. 5B, Supp. Fig. S5A–B).
Figure 5. nucCD24 gene expression signature correlates with poor patient outcome.
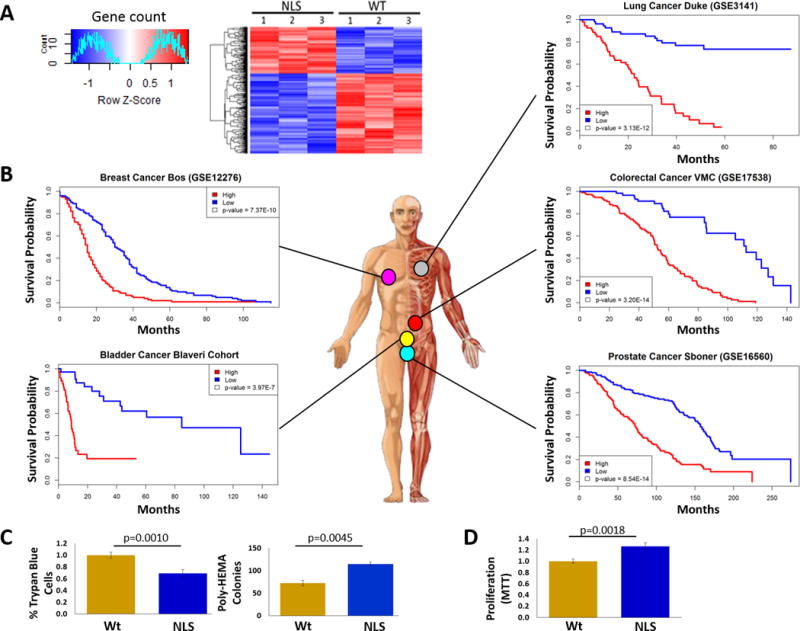
(A) UMUC13 cells expressing Wt-CD24 or NLS-CD24 were subjected to microarray analysis which revealed 304 genes were differentially expressed, shown here as a heat map. (B) These 304 genes were able to stratify patient outcome in 5 different cancer types. p-values from the likelihood ratio test. (C) Inhibition of cell death was observed in UMUC13 cells overexpressing NLS-CD24 relative to Wt-CD24 based on less trypan blue staining and greater colony numbers after cells were grown in poly-HEMA coated plates. (D) Proliferation of UMUC13 cells overexpressing NLS-CD24 is higher relative to Wt-CD24 expressing cells.
Analysis of the 304 genes using Ingenuity Pathway Analysis software predicts that nucCD24 expression decreases (negative activation z-score) “cell line apoptosis” (z = −0.988, p=0.0195) and “cytolysis” (z = −0.152, p=0.0104). This is supported by our finding that expression of NLS-CD24 decreases anchorage independent apoptosis (anoikis) and promotes increased survival (colony numbers) relative to Wt-CD24 (Fig. 5C). Analysis also predicted that “cell activation” (z =2.52, p=0.0281) would be positively impacted (positive activation z-score) by NLS-CD24 expression and this is supported by finding NLS-CD24 increased anchorage independent cell proliferation (Fig. 5D).
DISCUSSION
We previously showed that surCD24- bladder cancer cells had similar in vivo metastatic properties as FACS-unselected cells, yet both cell types lost metastatic ability with total cellular depletion of CD24. Here we demonstrate that surCD24- cells have residual CD24 protein which exists in the nucleus and promotes aggressive tumor properties. This highlights the need to consider nucCD24 in fundamental research and questions the premise that antibody based CD24 directed therapies currently in consideration will be effective monotherapy. These findings also shed some light on a longstanding paradox in the breast cancer field where FACS isolated surCD24- (CD24−/low) cells are considered stem cells and are extremely tumorigenic. We can speculate that this tumorigenicity is driven by nucCD24.
While important to cancer biology and therapeutic development, our data are not the first to show a GPI-linked protein functioning beyond the cell surface. CD59, like CD24, is a GPI-linked, glycosylated protein which exists and functions in blood plasma (39,40). CD24 would also not be the first GPI-linked protein identified to bind chromatin. Prion protein binds nuclear lamina and interacts with chromatin (41,42). At least one group has shown evidence for lipid rafts being present in the nucleus and if so, these specialized microdomains would be excellent environments for the GPI-linked proteins to reside (43) and associate with chromatin. Perhaps this association allows CD24 to help regulate the balance between heterochromatin and euchromatin and subsequently, promote different transcriptional programs.
Finding that nucCD24 is functional in experimental models, and its level prognostic in patients, does not weaken the relevance of surCD24 expression as either a tumor driver or prognostic marker. For example, one can envision a model where the majority of cancer cells in a tumor are expressing high levels of CD24 in all compartments while a few cells express only nucCD24 (i.e. breast cancer surCD24- stem cells). While questioning their effectiveness in the monotherapy setting, our findings should not entirely exclude anti-CD24 monoclonal antibodies (mAb) as potential therapies since the majority of cells assessed in patient tumors have surface CD24 expression. In addition, CD24hiCD19+CD38hi B cells are considered B regulatory cells (Breg or B10) and have been shown to suppress T cell function through secretion of IL-10 (44). Thus, anti-CD24 therapy may promote down-regulation of Breg cells and promote increased T cell activity on tumors. One study supported this notion by showing that anti-CD24 mAb treatment was able to lead to changes in tumor cytokine levels in mice (45). In conclusion, one could imagine combining anti-CD24 mAb with agents targeting transcription factors which regulate CD24 expression such as androgen receptor (10), HIF1 (12), and GON4L (10,12).
Supplementary Material
Acknowledgments
We thank staff of the University of Colorado Cancer Center Flow Cytometry Shared Resource for help with sorting cells and other technical assistance. We also thank Changho Lee, visiting scholar from Soonchunhyang University, Cheonan Hospital, in Korea for help in making mutant constructs of CD24. We thank the University of Colorado Cancer Center Genomics Shared Resource (P30-CA046934) for performing microarray experiments. Lastly, we thank the other members of the Theodorescu research group for advice and technical assistance.
Financial Support: Supported by National Institutes of Health grant CA143971 to D. Theodorescu.
Footnotes
Conflict of Interest: The authors have declared that no conflict of interest exists
References
- 1.Lee J-H, Kim S-H, Lee E-S, Kim Y-S. CD24 overexpression in cancer development and progression: a meta-analysis. Oncol Rep. 2009 Nov;22(5):1149–56. doi: 10.3892/or_00000548. [DOI] [PubMed] [Google Scholar]
- 2.Runz S, Mierke CT, Joumaa S, Behrens J, Fabry B, Altevogt P. CD24 induces localization of beta1 integrin to lipid raft domains. Biochem Biophys Res Commun. 2008 Jan 4;365(1):35–41. doi: 10.1016/j.bbrc.2007.10.139. [DOI] [PubMed] [Google Scholar]
- 3.Bretz N, Noske A, Keller S, Erbe-Hofmann N, Schlange T, Salnikov AV, et al. CD24 promotes tumor cell invasion by suppressing tissue factor pathway inhibitor-2 (TFPI-2) in a c-Src-dependent fashion. Clin Exp Metastasis. 2012 Jan;29(1):27–38. doi: 10.1007/s10585-011-9426-4. [DOI] [PubMed] [Google Scholar]
- 4.Bretz NP, Salnikov AV, Perne C, Keller S, Wang X, Mierke CT, et al. CD24 controls Src/STAT3 activity in human tumors. Cell Mol Life Sci CMLS. 2012 Nov;69(22):3863–79. doi: 10.1007/s00018-012-1055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon MJ, Han J, Seo JH, Song K, Jeong HM, Choi J-S, et al. CD24 Overexpression Is Associated with Poor Prognosis in Luminal A and Triple-Negative Breast Cancer. PloS One. 2015;10(10):e0139112. doi: 10.1371/journal.pone.0139112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lubeseder-Martellato C, Hidalgo-Sastre A, Hartmann C, Alexandrow K, Kamyabi-Moghaddam Z, Sipos B, et al. Membranous CD24 drives the epithelial phenotype of pancreatic cancer. Oncotarget. 2016 May 17; doi: 10.18632/oncotarget.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Z-L, Chen Y-P, Song J-L, Wang Y-Q. Knockdown of CD24 inhibits proliferation, invasion and sensitizes breast cancer MCF-7 cells to tamoxifen in vitro. Eur Rev Med Pharmacol Sci. 2015 Jul;19(13):2394–9. [PubMed] [Google Scholar]
- 8.Mierke CT, Bretz N, Altevogt P. Contractile forces contribute to increased glycosylphosphatidylinositol-anchored receptor CD24-facilitated cancer cell invasion. J Biol Chem. 2011 Oct 7;286(40):34858–71. doi: 10.1074/jbc.M111.245183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overdevest JB, Thomas S, Kristiansen G, Hansel DE, Smith SC, Theodorescu D. CD24 offers a therapeutic target for control of bladder cancer metastasis based on a requirement for lung colonization. Cancer Res. 2011 Jun 1;71(11):3802–11. doi: 10.1158/0008-5472.CAN-11-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Overdevest JB, Knubel KH, Duex JE, Thomas S, Nitz MD, Harding MA, et al. CD24 expression is important in male urothelial tumorigenesis and metastasis in mice and is androgen regulated. Proc Natl Acad Sci U S A. 2012 Dec 18;109(51):E3588–3596. doi: 10.1073/pnas.1113960109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith SC, Oxford G, Wu Z, Nitz MD, Conaway M, Frierson HF, et al. The metastasis-associated gene CD24 is regulated by Ral GTPase and is a mediator of cell proliferation and survival in human cancer. Cancer Res. 2006 Feb 15;66(4):1917–22. doi: 10.1158/0008-5472.CAN-05-3855. [DOI] [PubMed] [Google Scholar]
- 12.Thomas S, Harding MA, Smith SC, Overdevest JB, Nitz MD, Frierson HF, et al. CD24 is an effector of HIF-1-driven primary tumor growth and metastasis. Cancer Res. 2012 Nov 1;72(21):5600–12. doi: 10.1158/0008-5472.CAN-11-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao X-L, Zhao C, Niu M, Chen D. Downregulation of CD24 inhibits invasive growth, facilitates apoptosis and enhances chemosensitivity in gastric cancer AGS cells. Eur Rev Med Pharmacol Sci. 2013 Jul;17(13):1709–15. [PubMed] [Google Scholar]
- 14.Lee K, Ju J, Jang K, Yang W, Yi JY, Noh DY, et al. CD24 regulates cell proliferation and transforming growth factor β-induced epithelial to mesenchymal transition through modulation of integrin β1 stability. Cell Signal. 2012 Nov;24(11):2132–42. doi: 10.1016/j.cellsig.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Sagiv E, Starr A, Rozovski U, Khosravi R, Altevogt P, Wang T, et al. Targeting CD24 for treatment of colorectal and pancreatic cancer by monoclonal antibodies or small interfering RNA. Cancer Res. 2008 Apr 15;68(8):2803–12. doi: 10.1158/0008-5472.CAN-07-6463. [DOI] [PubMed] [Google Scholar]
- 16.Naumov I, Zilberberg A, Shapira S, Avivi D, Kazanov D, Rosin-Arbesfeld R, et al. CD24 knockout prevents colorectal cancer in chemically induced colon carcinogenesis and in APC(Min)/CD24 double knockout transgenic mice. Int J Cancer. 2014 Sep 1;135(5):1048–59. doi: 10.1002/ijc.28762. [DOI] [PubMed] [Google Scholar]
- 17.Bozorgi A, Khazaei M, Khazaei MR. New Findings on Breast Cancer Stem Cells: A Review. J Breast Cancer. 2015 Dec;18(4):303–12. doi: 10.4048/jbc.2015.18.4.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taniuchi K, Nishimori I, Hollingsworth MA. Intracellular CD24 inhibits cell invasion by posttranscriptional regulation of BART through interaction with G3BP. Cancer Res. 2011 Feb 1;71(3):895–905. doi: 10.1158/0008-5472.CAN-10-2743. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Liu R, Ye P, Wong C, Chen G-Y, Zhou P, et al. Intracellular CD24 disrupts the ARF-NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation. Nat Commun. 2015;6:5909. doi: 10.1038/ncomms6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Méndez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000 Nov;20(22):8602–12. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kristiansen G, Machado E, Bretz N, Rupp C, Winzer K-J, König A-K, et al. Molecular and clinical dissection of CD24 antibody specificity by a comprehensive comparative analysis. Lab Investig J Tech Methods Pathol. 2010 Jul;90(7):1102–16. doi: 10.1038/labinvest.2010.70. [DOI] [PubMed] [Google Scholar]
- 22.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001 Dec;29(4):365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol Off J Am Soc Clin Oncol. 2006 Feb 10;24(5):778–89. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 24.Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, et al. Bladder cancer outcome and subtype classification by gene expression. Clin Cancer Res Off J Am Assoc Cancer Res. 2005 Jun 1;11(11):4044–55. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- 25.Kim W-J, Kim E-J, Kim S-K, Kim Y-J, Ha Y-S, Jeong P, et al. Predictive value of progression-related gene classifier in primary non-muscle invasive bladder cancer. Mol Cancer. 2010 Jan 8;9:3. doi: 10.1186/1476-4598-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindgren D, Frigyesi A, Gudjonsson S, Sjödahl G, Hallden C, Chebil G, et al. Combined gene expression and genomic profiling define two intrinsic molecular subtypes of urothelial carcinoma and gene signatures for molecular grading and outcome. Cancer Res. 2010 May 1;70(9):3463–72. doi: 10.1158/0008-5472.CAN-09-4213. [DOI] [PubMed] [Google Scholar]
- 27.Say Y-H, Hooper NM. Contamination of nuclear fractions with plasma membrane lipid rafts. Proteomics. 2007 Apr;7(7):1059–64. doi: 10.1002/pmic.200600849. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka KAK, Suzuki KGN, Shirai YM, Shibutani ST, Miyahara MSH, Tsuboi H, et al. Membrane molecules mobile even after chemical fixation. Nat Methods. 2010 Nov;7(11):865–6. doi: 10.1038/nmeth.f.314. [DOI] [PubMed] [Google Scholar]
- 29.Smith SC, Baras AS, Owens CR, Dancik G, Theodorescu D. Transcriptional signatures of Ral GTPase are associated with aggressive clinicopathologic characteristics in human cancer. Cancer Res. 2012 Jul 15;72(14):3480–91. doi: 10.1158/0008-5472.CAN-11-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birnbaum DJ, Laibe S, Ferrari A, Lagarde A, Fabre AJ, Monges G, et al. Expression Profiles in Stage II Colon Cancer According to APC Gene Status. Transl Oncol. 2012 Apr;5(2):72–6. doi: 10.1593/tlo.11325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freeman TJ, Smith JJ, Chen X, Washington MK, Roland JT, Means AL, et al. Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of β-catenin. Gastroenterology. 2012 Mar;142(3):562–571.e2. doi: 10.1053/j.gastro.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin Cancer Res Off J Am Assoc Cancer Res. 2009 Dec 15;15(24):7642–51. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: a genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A. 2009 Apr 28;106(17):7131–6. doi: 10.1073/pnas.0902232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weichert W, Denkert C, Burkhardt M, Gansukh T, Bellach J, Altevogt P, et al. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin Cancer Res Off J Am Assoc Cancer Res. 2005 Sep 15;11(18):6574–81. doi: 10.1158/1078-0432.CCR-05-0606. [DOI] [PubMed] [Google Scholar]
- 36.Bos PD, Zhang XH-F, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009 Jun 18;459(7249):1005–9. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006 Jan 19;439(7074):353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 38.Sboner A, Demichelis F, Calza S, Pawitan Y, Setlur SR, Hoshida Y, et al. Molecular sampling of prostate cancer: a dilemma for predicting disease progression. BMC Med Genomics. 2010 Mar 16;3:8. doi: 10.1186/1755-8794-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meri S, Lehto T, Sutton CW, Tyynelä J, Baumann M. Structural composition and functional characterization of soluble CD59: heterogeneity of the oligosaccharide and glycophosphoinositol (GPI) anchor revealed by laser-desorption mass spectrometric analysis. Biochem J. 1996 Jun 15;316(Pt 3):923–35. doi: 10.1042/bj3160923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rooney IA, Morgan BP. Characterization of the membrane attack complex inhibitory protein CD59 antigen on human amniotic cells and in amniotic fluid. Immunology. 1992 Aug;76(4):541–7. [PMC free article] [PubMed] [Google Scholar]
- 41.Mangé A, Crozet C, Lehmann S, Béranger F. Scrapie-like prion protein is translocated to the nuclei of infected cells independently of proteasome inhibition and interacts with chromatin. J Cell Sci. 2004 May 1;117(Pt 11):2411–6. doi: 10.1242/jcs.01094. [DOI] [PubMed] [Google Scholar]
- 42.Strom A, Wang G-S, Picketts DJ, Reimer R, Stuke AW, Scott FW. Cellular prion protein localizes to the nucleus of endocrine and neuronal cells and interacts with structural chromatin components. Eur J Cell Biol. 2011 May;90(5):414–9. doi: 10.1016/j.ejcb.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 43.Cascianelli G, Villani M, Tosti M, Marini F, Bartoccini E, Magni MV, et al. Lipid microdomains in cell nucleus. Mol Biol Cell. 2008 Dec;19(12):5289–95. doi: 10.1091/mbc.E08-05-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mauri C, Menon M. The expanding family of regulatory B cells. Int Immunol. 2015 Oct;27(10):479–86. doi: 10.1093/intimm/dxv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salnikov AV, Bretz NP, Perne C, Hazin J, Keller S, Fogel M, et al. Antibody targeting of CD24 efficiently retards growth and influences cytokine milieu in experimental carcinomas. Br J Cancer. 2013 Apr 16;108(7):1449–59. doi: 10.1038/bjc.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


