Abstract
The bone marrow microenvironment influences malignant hematopoiesis but how it promotes leukemogenesis has not been elucidated. Additionally, the role of the bone marrow stroma in regulating clinical responses to DNA methyltransferase inhibitors (DNMTi) is also poorly understood. In this study, we conducted a DNA methylome analysis of bone marrow-derived stromal cells from myelodysplastic syndrome (MDS) patients and observed widespread aberrant cytosine hypermethylation occurring preferentially outside CpG islands. Stroma derived from 5-azacytidine-treated patients lacked aberrant methylation and DNMTi treatment of primary MDS stroma enhanced its ability to support erythroid differentiation. An integrative expression analysis revealed that the WNT pathway antagonist FRZB was aberrantly hypermethylated and underexpressed in MDS stroma. This result was confirmed in an independent set of sorted, primary MDS-derived mesenchymal cells. We documented a WNT/β-catenin activation signature in CD34+ cells from advanced cases of MDS where it associated with adverse prognosis. Constitutive activation of β-catenin in hematopoietic cells yielded lethal myeloid disease in a NUP98-HOXD13 mouse model of MDS, confirming its role in disease progression. Our results define novel epigenetic changes in the bone marrow microenvironment which lead to β-catenin activation and disease progression of MDS.
Keywords: DNA methylation, MDS, Stroma, WNT, FRZB
Introduction
Myelodysplastic syndromes (MDS) comprise a heterogeneous group of acquired clonal bone marrow (BM) disorders characterized by varying degrees of cytopenias, morphological and functional abnormalities of hematopoietic cells and the risk of transformation into acute myeloid leukemia (AML) (1). Studies have traditionally focused on hematopoietic cells in an effort to understand hematologic disease development with the goal of pursuing therapeutic solutions. The hematopoietic cells in MDS have been shown to contain numerous genetic and epigenetic aberrations (2) and these studies have helped elucidate the pathobiology of MDS. However, there is growing evidence that microenvironmental defects also contribute to ineffective hematopoiesis and, hence, progression of the disease(3,4).
The bone marrow microenvironment consists of a mixture of different cell types – mostly represented by stromal or mesenchymal cells, macrophages, fibroblasts, adipocytes, endothelial cells, osteoblasts and glial cells(5–7). The microenvironment is critically important in supporting the growth of hematopoietic stem and progenitor cells and is a source of growth factors that drive the self-renewal and differentiation of the hematopoietic cells. Alterations in the marrow niche can, therefore, result in hematopoietic disorders such as MDS. In fact, a recent study showed that genetic deletion of Dicer in marrow mesenchymal progenitor cells led to myelodysplasia and development of leukemia in vivo (8). Another study demonstrated that activation of beta-catenin in the murine osteoblastic niche led to MDS/AML, further supporting the role of stromal dysfunction in the genesis of these diseases (9). Yet, studies of human MDS marrows have generally not revealed gene mutations or cytogenetic alterations as seen in hematopoietic cells (10,11), and the molecular basis of stromal dysfunction in human MDS is currently not known. Since epigenetic alterations are important in regulating gene transcription and are present in the hematopoietic cells in MDS, it is conceivable that such alterations also occur in the stromal cells. Further, even though the DNA methyltransferase inhibitor, 5-Azacytidine (Aza), is approved for therapy of MDS, its hypomethylating actions, presumed to affect the hematopoietic cells, fail to show a strong correlation with therapeutic responses, leading us to hypothesize that methylation patterns in stroma may be relevant.
To evaluate the epigenome of the MDS marrow microenvironment, we used the HELP (HpaII tiny fragment Enrichment by Ligation-mediated PCR) assay to study cytosine methylation patterns in primary stromal cells from patients with MDS. The HELP assay relies on differential digestion by a pair of isoschizomer enzymes, HpaII and MspI, which differ on the basis of their methylation sensitivity. The HpaII and MspI genomic representations can be co-hybridized to a custom microarray and their ratio can be used to indicate the methylation of particular CCGG sites at these loci. The HELP assay has been successfully used to reveal novel epigenomic alterations in leukemias, MDS and other cancers (12–14). Here we used this assay to demonstrate that MDS stromal cells contain aberrant hypermethylation that affects the Wnt/β-catenin pathway. We further showed that β-catenin activation cooperates and results in a lethal MDS or myeloid leukemia in an MDS mouse model. We showed that treatment with 5-Aza leads to abrogation of hypermethylation in stromal cells and, in fact, enhanced the ability of MDS stromal cells to support hematopoiesis (of healthy donor cells) in vitro. Taken together, these results support the concept that aberrant epigenetic marks in MDS stroma contribute to the disease progression and can be targeted for therapeutic interventions with DNMT inhibitors.
Methods
Cell lines, MDS Stromal Samples and Nucleic Acid Extraction
The HS 27a human stroma cell line was derived from a healthy marrow donor and immortalized by transduction with a human papilloma virus E6/E7 construct(15). The KG1a cell line was obtained and authenticated from ATCC by STR profiling in 2015 (Manasses, VA). Cell lines were low passage and were regularly checked for mycoplasma contamination monthly by Plasmotest (Invivogen). BM specimens were obtained from patients diagnosed with MDS and from controls after obtaining informed consent, as approved by the Fred Hutchinson Cancer Research Center and Albert Einstein College of Medicine Institutional Review Boards (Supp Table 1). The patient sample collection were conducted in accordance with declaration of Helsinki. BM mononuclear cells (MNC) were isolated using ficoll-paque gradient separation. Bone marrow cells, 25–30 × 106 cells per 75-ml cell flasks were cultured in nonhematopoietic expansion medium (Miltenyi Biotech Inc. Auburn, CA) at 37°C in 5% CO2, with weekly medium replacement until adherent cells reached 70% confluence. Adherent cells were analyzed by flow cytometry. CD45+ hematopoietic cells were discarded immunomagnetically (Miltenyi). The resulting CD45-ve, low passage, BM-stromal cells were used for DNA and RNA extraction.
DNA Methylation Analysis by HELP assay
The HELP assay was carried out as described previously(14,16) to determine methylome of 50,000 CpGs corresponding to 14,000 genes. Detailed description provided in supplementary methods (GEO (GSE60233)). Pathway Analysis was performed using the IPA software (Redwood City, CA) (17). The list of hypermethylated genes was examined for enrichment of conserved gene-associated regions using the Molecular Signatures Database (MSigDB) (18). Transcription factor binding sites in the differentially methylated regions was determined by the HOMER algorithm (19)
Quantitative DNA Methylation Analysis by Mass Array Epityping
Validation of HELP microarray findings was carried out by MALDI-TOF mass spectrometry using EpiTYPER™ by MassArray (Sequenom, CA) on bisulfite-converted DNA as described previously(20,21).
HELP-tagging Analysis for HS27 Stromal Cells
HS27a stromal cells were grown to 80% confluency in RPMI1640 and then cocultured with KG1a cells (CD45+) at a ratio of 1:3 for 48hrs. Adherent cells were washed, trypsinized and depleted of residual KG1a cells by CD45 MicroBeads (Miltenyi Biotech Inc. Auburn, CA). DNA was isolated from HS27a cells and used for high resolution HELP tagging assay as previously performed (22,23) for methylation status of 1.8 million CpGs.
Gene Expression Analysis
Gene expression data were obtained using Affymetrix Human Genome U133A 2.0 or Plus2 GeneChips; mRNA isolation, labeling, hybridization, and quality control were carried out as described before.(24)
Hematopoietic Progenitor Cell Assays and Flow Cytometry
Hematopoietic progenitor colony formation was determined by clonogenic assays in methylcellulose, as in our previous studies (25,26). BM-stroma cells from controls and MDS patients were expanded and treated with either 0.5µM 5-Aza in alpha-minimal essential medium (MEM) and 10% FBS daily for 5 days. After treatment (~ 40% confluency), 1ml of Methocult H4434 containing 5000 healthy CD34+ cells was layered on top. At day 10, colony formation was assessed, and FACS analysis conducted using CD34 - PE, CD45 - PE-Cy7, CD14 - PB, CD11b - APC, Glycophorin A - PE-Cy5, CD71 – FITC antibodies.
Immunohistochemistry for FRZB
MDS patient stromal cells were cultures onto 4-chambered slides with two chambers having received no 5-AZA treatment and two chambers receiving 5-AZA (0.5uM for five days). Immunohistochemistry for FRZB was performed with FRP-3(H-170) rabbit monoclonal antibody (Cat# sc-13941, Santa Cruz Biotechnology, San Diego, CA) and matched isotype control diluted to 1:100, for 30 min.
Mice
The doxycycline-inducible constitutively active β-catenin mice (KH2-Col1A1-tetO-CTNN1S33Y/Rosa-rtTA; S33Y) were generated by the Hochedlinger laboratory (details in supplementary methods) (27). These mice were crossed to transgenic mice expressing a NUP98-HOXD13 fusion gene in hematopoietic tissues, resulting in a transgenic NUP98-HOXD13 mouse with doxycycline-inducible constitutively active β-catenin. All experimental mice were heterozygous in both Col1a1 (S33Y under tetOP) and Rosa26 (rtTA) loci and for the NUP98-HOXD13 transgene. Transgenic primary NUP98-HOXD13 mice aged 14–18 months were used for analysis and were verified to display clinical hallmarks of MDS and cytopenias. Details on FACS analysis and bone marrow transplantation are provided in supplementary methods.
Analysis of WNT signature in MDS cohort
WNT target genes obtained from a comprehensive database (http://web.stanford.edu/group/nusselab/cgi-bin/wnt/target_genes) that were expressed in MDS derived and healthy CD34+ cells were analyzed in a large cohort of gene expression profiles (28) and used to calculate a composite score.
Primary MDS mesenchymal cell isolation and RNA-seq analysis
Control bone marrow was obtained from donors for allogeneic transplantation (median age: 45 (35–61), after approval by the IRB of Erasmus Medical Center(29). Mesenchymal cells from human MDS patients were FACS sorted using the FACSAria III systems (BDBioscience) with the following antibodies using optimized dilutions: CD45-PE-Cy7 (1:200), CD235a-BV421-A (1:100), CD271-PE (1:100), CD105-APC (1:50), CD31-APC-CY7 (1:50). Sorted cells were kept in TRIzol (Ambion). Smarter Ultra Low RNA kit for Illumina Sequencing (Clonetech) was used for cDNA synthesis according to the manufacturer’s protocol. Sample preparation, sequencing, demultiplexing and alignment were performed as previously described (30) with modifications specific to the application of Smarter kit. Details on analysis are provided in supplementary methods.
RESULTS
Primary stromal cells in MDS are characterized by aberrant hypermethylation
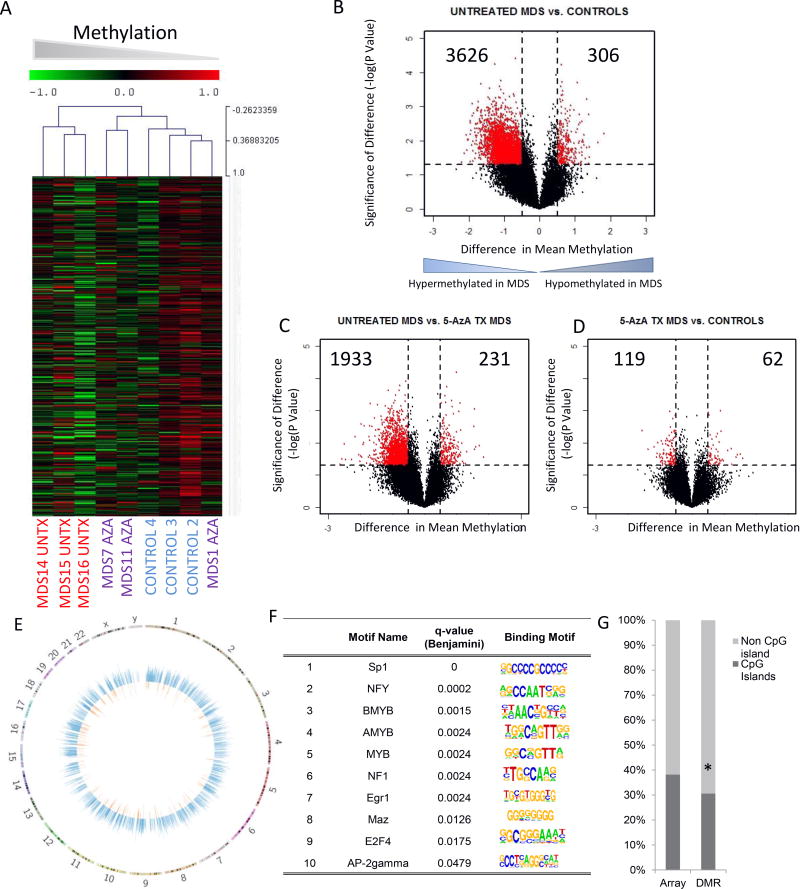
Primary cultures of stromal cells were established from MDS bone marrow samples and controls (Supplemental Table 1 with clinical characteristics). The MDS samples included patients who had been treated (MDS Tx) and never been treated (MDS UnTX) with the DNMT inhibitor 5-Azacytidine (5-Aza). Controls were age matched and had blood counts in the normal range. CD45 negative non-hematopoietic cells from the cultures were immunomagnetically sorted and used for DNA/RNA extraction after low passage numbers (up to 3 passages). Genome wide cytosine methylation was analyzed by the HELP assay, that uses differential methylation-specific digestion by HpaII and MspI followed by amplification, two color labeling and hybridization to quantitatively determine individual promoter methylation of 50,000 CpGs loci covering 14,000 promoters (13,31) Unsupervised clustering based on cytosine methylation profiles demonstrated that untreated MDS stromal cells were epigenetically distinct from healthy controls, (Fig 1A), while MDS stromal cells from 5-Aza treated patients clustered closer to healthy controls. Next, to determine the qualitative epigenetic differences between these groups, we performed a supervised analysis of the respective DNA methylation profiles. A volcano plot comparing the differences between mean methylation of individual loci between MDS stromal cells and controls plotted against the significance (log (p value) based on T Test) of the difference was used to represent these data shown in Fig 1B. We observed that MDS stromal cells were characterized by aberrant hypermethylation when compared to controls (3626 hypermethylated vs 306 hypomethylated loci in untreated MDS stromal cells). Comparison of 5-Aza treated samples demonstrated a lesser degree of methylation and an epigenomic pattern similar to that in healthy controls (Figs 1C,D).
Fig 1. Widespread Epigenetic alterations are seen in MDS stroma.
Unsupervised clustering of primary MDS stromal cells from untreated patients (MDS UnTx), MDS stromal cells from patients treated with 5-Azacytidine (MDS Aza) and healthy controls shows that MDS UnTx stroma has a distinct DNA methylation profile (Hierarchical clustering, Wards) (A). Volcano plot shows that most of differentially methylated genes in stroma from untreated patients are hyper-methylated (B). Comparison of 5-Aza treated stroma with Untx MDS stroma and healthy controls shows that 5-Aza treated samples do not have increased numbers of hypermethylated loci (C, D). Circos plots show that aberrant hypermethylation (blue) occurs throughout the genome and is more frequent than aberrant hypomethylation (orange). (E). Transcription factor binding sites that are enriched at differentially methylated regions (DMRs) are shown with motifs (F). DMRs in UnTx MDS stroma were predominantly present in non CpG island locations (87.9%) and were significantly different from the distribution of HpaII loci in the whole HELP array (61.8%) (Test of Proportions, P Value<0.001) (G).
Even though aberrant methylation in MDS stromal samples occurred genome wide (Fig 1E), there was a significant enrichment at chromosomal regions chr12q15, chr5q32, chr4q21, chr7q31, chr3q13, chr2p12 and chr8q24 when compared with the genomic distribution of all HpaII fragments from the HELP array (P value <0.05; MDSig Program). Further, to determine whether these hypermethylated loci shared any common DNA elements, we performed a search for transcription factor-binding sites enriched in these regions. We observed a significant over-representation of binding sites for Sp1, NFY, MYB, and other transcription factors in differentially methylated regions in MDS stroma (P value <0.05; HOMER Program) (Fig 1F). Finally, we sought to determine if CpG islands were predominantly affected by differential methylation in MDS stromal cells. We observed that a large proportion of aberrantly methylated loci were located outside of CpG islands (Test of Proportions, P value <0.01, Fig 1G), consistent with recent data demonstrating epigenetic changes at less CpG dense regions of the genome (32).
Important functional pathways are epigenetically dysregulated in MDS Stroma
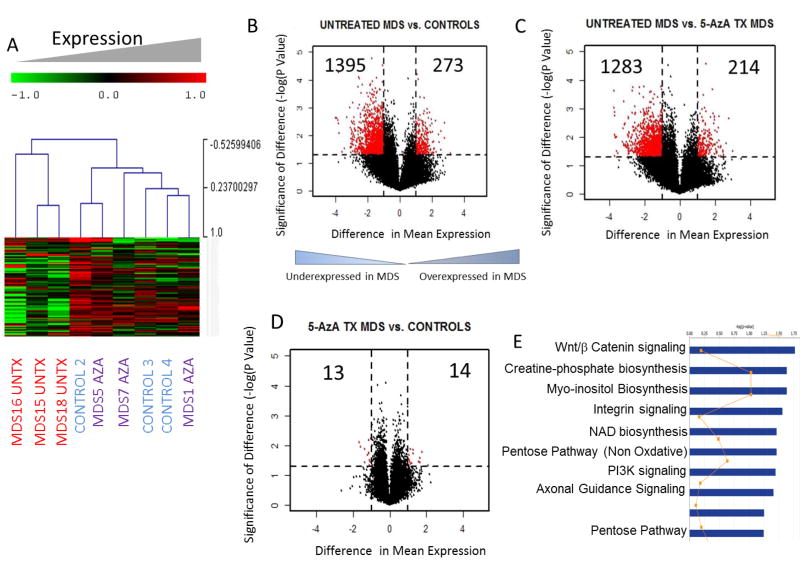
Gene expression analysis on MDS stromal cells also showed transcriptomic differences when compared to controls (Fig 2A). Most transcriptomic changes were seen in the untreated MDS stromal samples and consisted of aberrantly underexpressed genes (Fig 2B). Due to cell limitations due to low passage numbers we did not get adequate RNA from samples MDS14 and MDS11. Samples MDS18 and MDS5 were thus used for gene expression analysis and clustered similarly to untreated and 5-Aza treated status. There were very few differences between 5-Aza treated stromal samples and healthy controls (Fig 2C,D). Thus, these data demonstrated both methylomic and transcriptomic changes in primary MDS stromal samples. Integrative analysis revealed that differentially expressed genes that were also accompanied by aberrant methylation belonged to important functional pathways, such as those controlling cell morphology, signaling and transport (Supp Table 2). Determination of epigenetically regulated signaling pathways included those controlling WNT/Beta catenin signaling, Integrin signaling and other metabolic pathways and included WNT antagonists FRZB (SFRP3) and SFRP1 and cellular receptors belonging to the integrin and ephrin families (Fig 2E, Supp Table 3).
Fig 2. Widespread transcriptomic alterations are seen in MDS stroma.
Unsupervised hierarchical clustering of 3 primary untreated MDS stromal cells (MDS UnTx), MDS stromal cells form patients treated with 5-Azacytidine (MDS Aza) and healthy controls shows that MDS stroma has distinct gene expression profiles (A). Volcano plot shows that the majority of differentially expressed genes in untreated MDS stroma are underexpressed (B). Comparison of 5-Aza treated with Untx MDS and healthy controls shows that 5-Aza treated samples are similar to controls and do not have increased numbers of aberrantly expressed genes (C, D). Ingenuity functional pathway analysis of signaling pathways that are differentially expressed and differentially methylated between Untx MDS and control samples or UnTx and Aza treated samples (E).
FRZB is repressed and the Wnt/Beta catenin pathway activated in highly purified mesenchymal cells from human MDS patients
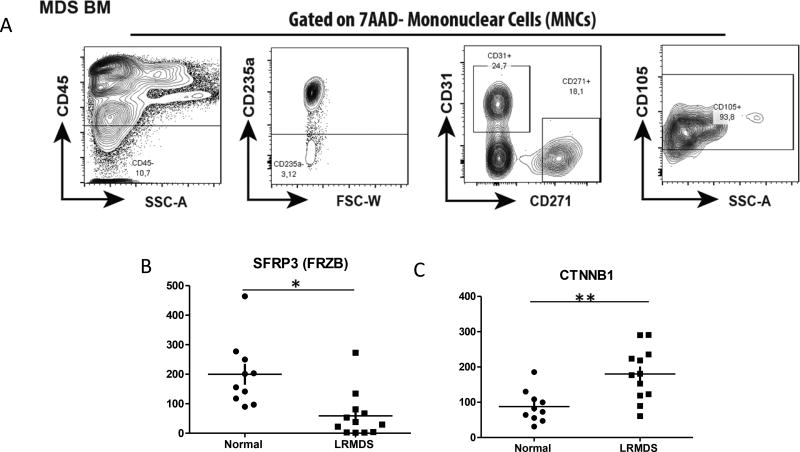
Next, we sought to confirm the relevance of these epigenetic and transcriptional aberrancies in ex vivo expanded stromal cells in primary highly purified unexpanded mesenchymal cells. We interrogated a recently established transcriptome database of prospectively isolated, highly FACS purified CD45−CD235−CD31−7AAD−CD271+CD105+ mesenchymal cells from a cohort of MDS patients (29) (Fig 3A)(Supp Table 4). The mesenchymal nature of CD271+ cells was confirmed by their colony forming unit-fibroblast (CFU-F) capacity and differential expression of mesenchymal, osteolineage and HSPC-regulatory genes. Massive parallel RNA sequencing was performed on these purified mesenchymal cells in comparison to their normal counterparts obtained from allogeneic bone marrow donors (29).
Figure 3. Transcriptional analysis of SFRP3 and Wnt target genes in primary mesenchymal cells in human MDS.
Highly purified CD45−CD235−7AAD−CD31−CD271+CD105+ mesenchymal cells were FACS-sorted (A) and subjected to massive parallel RNA sequencing. Reduced FRZB (SFRP3) expression was seen in MDS samples when compared to Healthy controls (B)(TTest, P Value<0.05). Increased expression of CTNNB1 in seen in MDS MSCs(C,D). * p< 0.05, ** p<0.001; MDS n=12, normal controls n=10.
FRZB (SFRP3) was significantly underexpressed in mesenchymal cells from MDS (n=12) in comparison to normal controls (n=10)(Fig 3B, TTest, P =0.02). This was further corroborated by significantly increased expression of the gene encoding β-catenin (CTNNB1) (Fig 3C)(TTest, P value <0.001). Together, the data confirm that reduced expression of SFRP3 in expanded stromal cells is of relevance to mesenchymal biology in human disease.
WNT pathway antagonists are epigenetically silenced in MDS stroma and can lead to activation of β-catenin in co-cultured HSCs
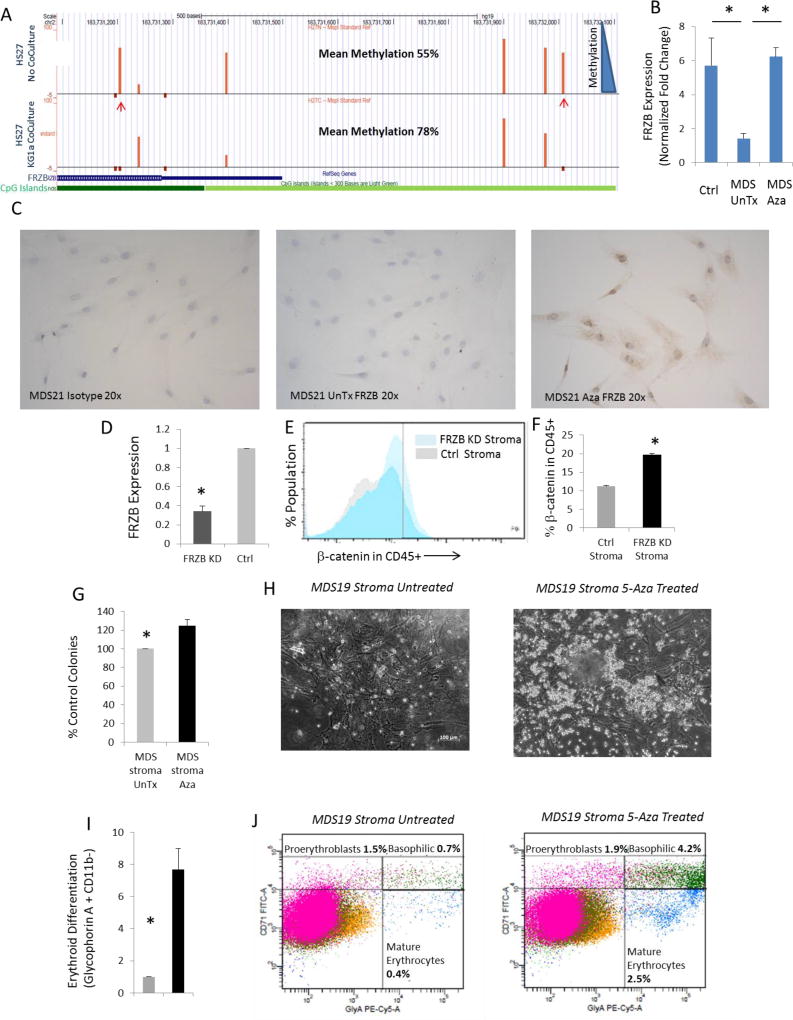
Next, to further validate the hypermethylation and underexpression of FRZB, we evaluated whether the epigenetic changes observed in primary cells could be replicated in vitro. Human stromal HS-27 cells were co-cultured with the leukemic cell line KG1a, and cytosine methylation changes were analyzed at a high resolution by next-gen sequencing-based HELP-Tagging assay. Co-culture led to hypermethylation of various CpGs in the FRZB and SFRP1 promoters in stromal cells (Fig 4A, Supp Fig 1). The differentially methylated CpGs were located in canionical CpG islands as well as 2KB flanking regions (CpG shores) in the promoter region. Other differentially methylated sites were also validated by sequencing and quantitative massarray epityper analysis (Supp Fig 2). Underexpression of the WNT antagonist FRZB was also validated by qRTPCR analysis in primary expanded MDS stromal samples (Fig 4B). FRZB protein levels were shown to be epigenetically downregulated in an independent set of 3 MDS stromal cultures (MDS 21–23) and increased after in vitro treatment with 5-Aza (Fig 4C, Supp Fig 3). Having shown underexpression of secreted WNT antagonist FRZB in MDS stroma, we next wanted to determine whether this can lead to activation of WNT/β-catenin in surrounding HSCs. We used specific siRNAs to knock down FRZB in primary human MSCs (Fig 4D) and co-cultured them with primary marrow derived CD34+ cells in methylcellulose. Intracellular flowcytometry demonstrated that co-culture with FRZB knockdown stromal cells led to activation of nuclear β-catenin in CD34+, CD45+ hematopoietic cells (Fig 4E,F).
Fig 4. WNT antagonist FRZB is hypermethylated and underexpressed in MDS stroma and treatment of MDS stroma with 5-Aza increases hematopoietic activity.
DNA methylation analysis by HELP-tagging assay shows hypermethylation of selected loci (marked by arrows) in the FRZB promoter in the HS27 stromal cells that are co-cultured with KG1a cells. Dark green denotes CpG islands, while light green denoted CpG shores (A). qRT-PCR shows decreased expression of FRZB in untreated MDS samples (n=4) when compared to control stroma (n=4) or Aza treated MDS (n=2) (T test, P value<0.05) (B). Immunohistochemistry shows increased expression of FRZB in MDS stroma treated with 5-Aza (0.5uM for 5 days) (C). siRNA mediated knockdown of FRZB was achieved in primary MSCs (D). Co-culture with FRZB knockdown MSCs led to increased nuclear β-catenin in CD45+ cells (Representative image shown in E; TTest, P Value <0.01, N=2, F). Healthy CD34+ cells were grown with MDS stromal cells (MDS19 and MDS20) in methylcellulose media. MDS stromal cells (MDS19 and MDS20) that were pretreated with 5-Aza for 5 days led to greater colony formation from healthy CD34 cells, (T test, P value <0.001) (G). Dysplastic colonies seen after co-culture of healthy CD34 cells with MDS stroma (H, left panel). 5-Aza pre-treatment leads to increased size of colonies (H, right panel). FACS analysis of co-cultured cells shows increase in Glycophorin A positive cells in 5-Aza treated MDS stromal co-cultures (n=2, T test, P value<0.5) (I). Increased percentages of all stages of erythroid cells are seen in 5-Aza pretreated stromal co-cultures (J).
Treatment with 5-Azacytidine improves the ability of MDS stroma to support erythropoiesis
Having shown that MDS stroma contains hypermethylated loci that are not seen in 5-Aza treated samples, we wanted to test the efficacy of Aza treatment on the stroma. Primary MDS stromal samples (MDS19, 20) were pretreated with 5-Aza for 5 days and then co-cultured with healthy human CD34+ cells. Co-culture with MDS stromal cells from 2 patients (mock-treated, not exposed to 5-Aza) led to dysplastic colony formation (Fig 4G,H). When primary MDS stroma was pre-treated with 5-Aza the numbers of hematopoietic colonies increased, and colonies were of larger size than in mock treated controls (Fig 4G,H). Next, co-cultured hematopoietic cells were collected and examined for differentiation markers by FACS. There was a significant increase in erythroid differentiation after co-culture with Aza treated MDS stroma, as evident from Glycophorin A positivity (Fig 4I,J). Increased erythroid differentiation was seen in all stages as evident from increased percentages of proerythroblasts, basophilic and mature erythrocytes (Fig 4J). Furthermore, we grew low passage stromal cells from another MDS patient treated them with 5-Aza after FRZB knockdown. We observed that 5-Aza consistently led to increased erythroid differentiation in co-cultured healthy CD34+ cells. The erythroid differentiation caused by 5-Aza was partially inhibited in the presence of FRZB knockdown (Supp Fig 4), thus demonstrating a potential role of FRZB expression in 5-Aza mediated effects on MDS stroma.
Activation of β-catenin leads to disease progression in vivo
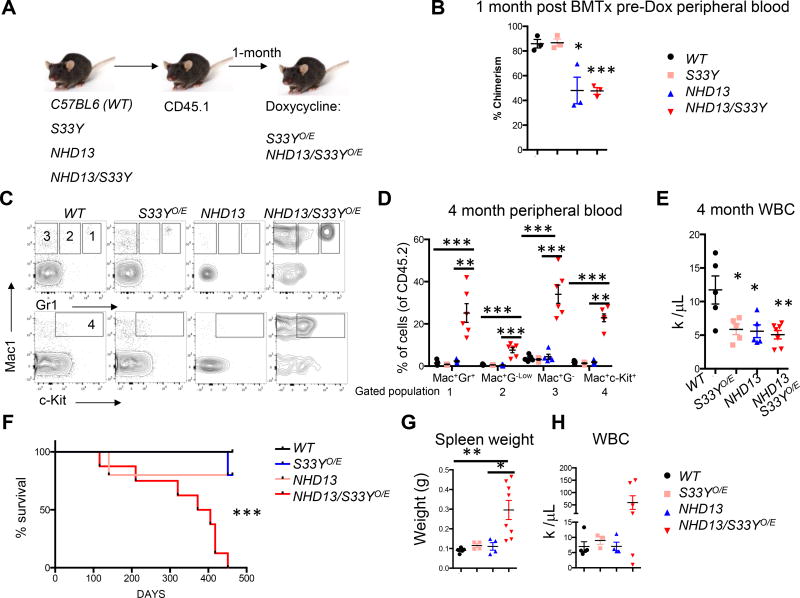
Downregulation of the Wnt pathway antagonists FRZB and SFRP1 suggest that β-catenin activation may contribute to MDS pathogenesis. In order to test the role of activated β-catenin in the context of MDS, we utilized an established murine model of MDS, the NUP98-HOXD13 transgenic model (NHD13). This model recapitulates many of the salient features of MDS including pancytopenias accompanied by hypercellular or normocellular bone marrow at 4–7 months(33–36). Also, 12–17% of the marrow contains dysplastic erythroid, myeloid and rare megakaryocytic cell types(33). Similar to patients with MDS, a significant cohort of the primary mice can progress and develop an aggressive AML. However, if the bone marrow of the NHD13 mice is transplanted, the recipient animals succumb to a fully penetrant form of MDS that rarely and only after 1 year progresses to AML(35). Although the NHD13 transplanted bone marrow cells engraft poorly, they still retain the clinical features of MDS (~10–20% chimerism, data not shown)(35). To test if β-catenin can alter MDS disease, the NHD13 transgene was crossed into a tetracycline inducible and constitutively activated human β-catenin (KH2-Col1A1-tetO-CTNN1S33Y/Rosa-rtTA; S33Y) overexpression mouse model described previously(27).
We performed bone marrow transplants of C57BL6 (WT), S33Y, NHD13 or compound NHD13/S33Y cells into lethally irradiated CD45.1+ recipients (Fig 5A). After equivalent engraftment was verified in the peripheral blood (Fig 5B), mice were treated with doxycycline to constitutively activate β-catenin (S33YO/E and NHD13/S33YO/E) and were then followed for disease progression. At four months, flow cytometric analysis revealed that the NHD13/S33Y mice had an increase in mature and immature myeloid cells among the donor cells (Fig 5C-D). Additionally, the S33YO/E, NHD13 and NHD13/S33YO/E animals all had reduced white blood cell counts compared to the control mice (Fig 5E). Surprisingly, the NHD13/S33YO/E succumbed to a lethal myeloid disease with a median latency of 388 days while other mice were followed until 451 days and then sacrificed to assess their phenotype (Fig 5F). The NHD13/S33YO/E mice mainly died of a myeloid leukemia (5 out 8) with enlarged spleens and increased white blood cell counts while the other mice died of an MDS-like phenotype (3 out 8), (Fig 5G, H). These data suggest that activation of β-catenin drives a lethal and aggressive myeloid disease with an increased likelihood of transformation.
Figure 5. Constitutive activation of β-catenin in an MDS model accelerates myeloid disease progression.
(A) Experimental scheme demonstrating donor bone marrow C57BL6; WT, S33YO/E, NHD13 and NHD13/S33YO/E mice. These cells were transplanted, allowed to engraft into congenic CD45.1 recipients for 1 month and then fed doxycycline feed (B) CD45.1 recipient mice were assessed for donor cell chimerism in the peripheral blood before doxycycline (Dox) addition (n=3 per group, TTest, *P Value<0.05, **P Value<0.01, ***P Value<0.001). (C) Representative Mac1, Gr1, and c-Kit flow cytometric staining of peripheral blood from 4 months post engraftment. Gate numbering represents Mac+Gr1+ (1), Mac+Gr1Low (2), Mac+Gr1− (3), Mac+c-Kit+ (4). (D) Quantification of flow cytometric analysis and gating as described in (C), (C57BL6; WT n=5, S33YO/E; n=5, NHD13 n=5, and NHD13/S33YO/E n=6, TTest, *P Value<0.05, **P Value<0.01, ***P Value<0.001). (E) White blood cell (WBC) counts in peripheral blood 4 months post-transplant, (C57BL6; WT n=5, S33YO/E; n=5, NHD13 n=5, and NHD13/S33YO/E n=7, TTest, *P Value<0.05, **P Value<0.01, ***P Value<0.001) (F) Kaplan-Meier curve of bone marrow transplantation described in (A), C57BL6; WT n=5, S33YO/E; n=5, NHD13 n=5, and NHD13/S33YO/E n=8, TTest, *P Value<0.05, **P Value<0.01, ***P Value<0.001). (G) Spleen weights from moribund mice or mice sacrificed at 463 day endpoint analysis, C57BL6; WT n=5, S33YO/E; n=4, NHD13 n=4, and NHD13/S33YO/E n=8, TTest, *P Value<0.05, **P Value<0.01, ***P Value<0.001) (H) WBC from moribund mice or mice sacrificed at 463 day endpoint analysis, C57BL6; WT n=5, S33YO/E; n=3, NHD13 n=4, and NHD13/S33YO/E n=6, TTest, *P Value<0.05, **P Value<0.01, ***P Value<0.001). All statistical analyses were compared to control mice or between NHD13 and NHD13/S33YO/E.
WNT activation signature is present in MDS and is marker of adverse prognosis
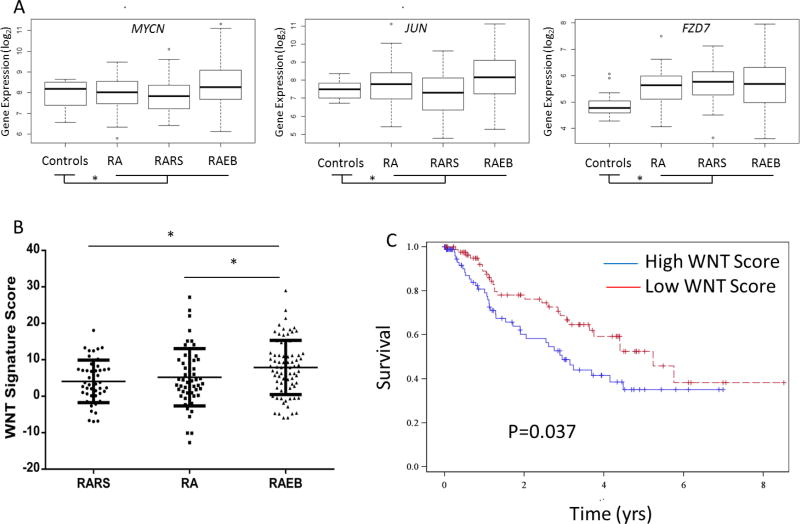
Next, we wanted to determine whether WNT pathway activation was seen in independent cohort of MDS samples. The expression patterns of known WNT pathway targets, (Supp Table 5) was determined in transcriptomic data from a cohort of 183 MDS marrow CD34+ cells and 17 healthy controls. Numerous WNT targets including MYC, JUN, FZD7 and others (Fig 6A, Supp Fig 5) were found to be overexpressed in MDS samples (18/39 overexpressed, Supp Fig 5) when compared to healthy controls and demonstrated a trend towards elevated mean expression in higher risk preleukemic RAEB (Refractory anemia with excess of blasts) subtypes. Since WNT/β-catenin pathway controls numerous downstream genes, we next developed a composite signature based on degree of activation of all expressed downstream targets (Supp Table 5) and correlated it with clinical subtypes of MDS. We observed that the higher risk cases of MDS with higher blast counts (RAEB) and higher propensity of transformation to AML had the highest levels of WNT pathway activation when compared to lower risk RA (refractory anemia) and RARS (refractory anemia with ringed sideroblasts) subtypes (Fig 6B, TTest, P Value<0.05). Correlation with overall survival also revealed that a higher WNT activation signature correlated with shorter overall survival with a median of 2.95 yrs in patients with high levels of WNT activation vs 5.24 yrs in patients with low WNT expression (Fig 6C, Log Rank P =0.037). These data taken together (Fig 7A,B) with in vivo murine data demonstrate that stroma mediated WNT activation is a pathogenic and prognostic event in MDS progression.
Figure 6. WNT activation is seen in higher risk subtypes of human MDS and is associated with worse overall survival.
Expression of WNT target genes were evaluated in 183 MDS marrow derived CD34+ samples when compared to healthy controls and representative candidates are shown (A). WNT score based on degree of activation of target genes is significantly elevated in higher risk RAEB subtypes when compared to lower risk RA and RARS subtypes (TTest, * P Value<0.05)(B). MDS patients with high WNT score (above median) have a worse overall survival (median survival = 2.95 years) when compared to those with low WNT score (median survival = 5.24 years)(Log rank P Value =0.037).
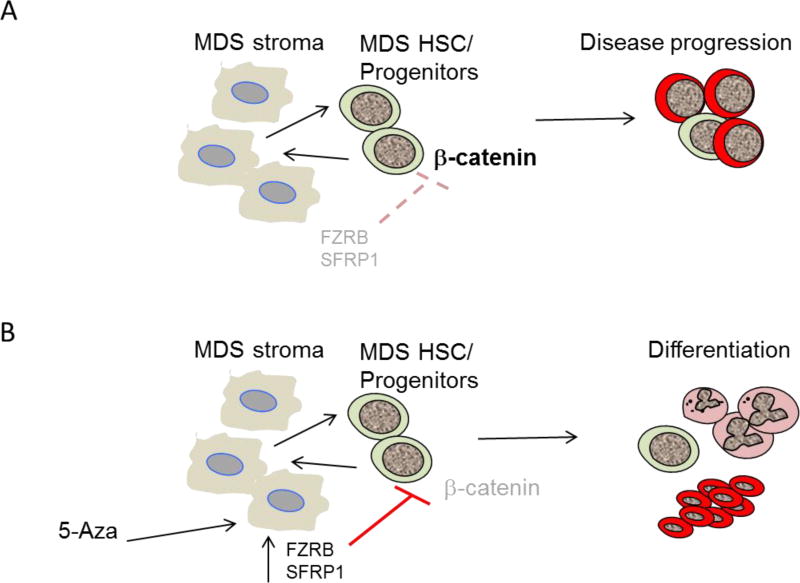
Figure 7. Proposed model of stroma mediated activation of WNT/β-Catenin signaling in MDS.
Aberrant methylation and underexpression of WNT/β-Catenin antagonists FRZB and SFRP1 is seen in MDS stroma. Activation of β-Catenin leads to disease progression in vivo and a WNT/β-catenin activation signature correlates with advanced disease in human samples (A). 5-Azacytidine treated MDS stroma samples have higher FRZB levels and in vitro treatment can lead to increased erythroid differentiation (B). (LSC, Leukemia stem cells).
Discussion
We show that marrow stroma in patients with MDS is aberrantly hypermethylated and that these marks are abrogated in stroma derived from 5-Aza treated patients. These findings demonstrate that the marrow microenvironment is also affected by epigenetic alterations and contributes to the MDS pathophysiology. These data raise the possibility that the marrow niche may also be targeted by epigenetically active drugs (DNMT inhibitors).
Even though numerous studies have highlighted the importance of the marrow microenvironment in the pathogenesis of bone marrow failure and malignant disorders, it is not understood how the stroma is reprogrammed to perpetuate the diseased phenotypes. Previous studies have evaluated stromal cells in MDS for cytogenetic alterations and have generally failed to show a high incidence of these abnormalities (21,37–39). A recent study showed cytogenetic abnormalities in 16% of mesenchymal stem cells in MDS, but none of them showed any mutations (10). Nevertheless, the stroma is presumed to participate in the pathogenesis of ineffective hematopoiesis in MDS and has been shown previously to have altered gene expression patterns (40). Our findings of epigenetic alterations that affect important regulatory pathways provide a molecular basis to explain microenvironmental reprogramming in MDS. A different recent study reported that deletion of Dicer in the bone marrow niche in mice induced MDS/AML with ineffective hematopoiesis and dysmorphic hematopoietic cells (8). Dicer is involved in microRNA processing, providing further evidence that epigenetic dysregulation in the microenvironment can lead to hematopoietic alterations. Another recent report showed that an activating mutation of β-catenin in the osteoblastic niche can lead to an MDS and AML like phenotypes in vivo (9). Activation of β-catenin in the osteoblastic niche was present in a large proportion of MDS patients and led to upregulation of Jagged in osteoblasts, which resulted in altered hematopoiesis via activation of Notch signaling in hematopoietic stem cells (9). Building on these findings, our study provides a mechanism for β-catenin activation in the niche. We demonstrate reduced expression of FRZB (SFRP3) via epigenetic silencing and consequent activation of the Wnt/ B-catenin pathway, not only in ex vivo expanded cells but also in primary mesenchymal cells directly isolated from patient marrows. These findings are, to our knowledge, the first describing molecular congruence between ex vivo expanded stromal cells and their in situ counterparts and provide novel insights into the lineage hierarchy of the stromal system in human bone marrow. The data support the view that a small population of mesenchymal colony forming stem and progenitor cells (CFU-F, comprising <2% of CD271+ cells in MDS; Chen et al, manuscript submitted) is epigenetically altered in human MDS and maintains the CD271+ reticular network in the bone marrow.
Additionally, we demonstrate that β-catenin activation within hematopoietic cells can result in an aggressive myeloid disease and transformation using a model of MDS, providing a complementary mechanism for disease progression (Fig 7). Our study is in line with other reports that suggest that β-catenin can cooperate with HOXA9/MEIS1 in GMPs and with BCR-ABL in CML models (41,42). Another study demonstrated that haploinsufficiency of del(5q) genes, Egr1 and Apc, cooperate with Tp53 loss to induce acute myeloid leukemia in mice(43). Our study shows that β-catenin activation in the NHD mouse model of MDS model can cause transformation of hematopoietic cells. It has also been demonstrated that hematopoietic cells are sensitive to differential levels of cell intrinsic WNT signaling, with varying effects of expression levels on HSCs, myeloid precursors, and T lineage precursors during hematopoiesis(44). Previous studies have also suggested that constitutive β-catenin activation in normal hematopoietic stem and progenitor cells resulted in a block in differentiation and rapid lethality of the mice (45,46). In contrast to this study, our model failed to alter hematopoietic differentiation or cause lethality, unless we combined our model with NHD13. One potential explanation for these disparate findings is that our model utilizes a different promoter to activate β-catenin (Col1A1/Rosa versus the endogenous promoter). Therefore, we suggest that this mouse model allows for the study of β-catenin activation without the toxicity that was previously observed. The NHD13 model used in our studies demonstrates dysplasia and cytopenias coupled with progression to accelerated disease, thus serving as a representative model of human MDS(36,47,48) . Taken together, we provide a novel mechanism for epigenetic suppression of the WNT pathway antagonists FRZB and SFRP1 in the niche, which can be epigenetically reversed by 5-Aza treatment.
5-Aza and Decitabine are inhibitors of DNMT, approved for treatment of MDS. These drugs lead to hematopoietic improvements and 5-Aza has been show to prolong survival. Even though the mechanism of action is presumed to be reversal of aberrant DNA methylation in hematopoietic cells, studies so far have not been able to correlate aberrant hypermethylation in pretreatment samples with response. All of these studies have examined hematopoietic cells - our data raise the possibility that stromal methylation may also have to be considered as a factor in the therapeutic efficacy of these drugs. In addition to altering transcription of various important genes in the stromal compartment, hypomethylation caused by 5-Aza may also affect transcription factor (TF) binding dynamics. We observed that binding sites of many TFs with roles in hematopoiesis such as Myb and NF1 were enriched in differentially methylated regions in MDS stroma. Thus, taken together, our results demonstrate that MDS stromal cells have widespread epigenetic alterations that modify the disease pathophysiology and can be targeted by DNMT inhibitor treatment.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by National Institutes of Health (R01s HL116336, DK103961), Leukemia and Lymphoma Society and Department of Defense (A Verma). M.G. Kharas was supported by the US National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases Career Development Award and NIDDK NIH R01-DK101989-01A1, Louis V Gerstner Young Investigator Award and the American Society of Hematology Junior Scholar Award, Kimmel Scholar Award and V-Scholar Award. M.H.G.P. Raaijmakers was supported by grants from the Dutch Cancer Society (KWF Kankerbestrijding) (EMCR 2010-4733), the Netherlands Organization of Scientific Research (NWO 90700422) and the Netherlands Genomics Initiative (40-41009-98-11062). A. Pellagatti and J. Boultwood were supported by Bloodwise UK. T.D. Bhagat is supported by a fellowship grant from NYSTEM.
Footnotes
Conflicts of Interest: None
References
- 1.Aul C, Bowen DT, Yoshida Y. Pathogenesis, etiology and epidemiology of myelodysplastic syndromes. Haematologica. 1998;83:71–86. [PubMed] [Google Scholar]
- 2.Zhou L, Opalinska J, Sohal D, Yu Y, Mo Y, Bhagat T, et al. Aberrant epigenetic and genetic marks are seen in myelodysplastic leukocytes and reveal Dock4 as a candidate pathogenic gene on chromosome 7q. J Biol Chem. 2011;286:25211–23. doi: 10.1074/jbc.M111.235028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medyouf H, Mossner M, Jann JC, Nolte F, Raffel S, Herrmann C, et al. Myelodysplastic cells in patients reprogram mesenchymal stromal cells to establish a transplantable stem cell niche disease unit. Cell Stem Cell. 2014;14:824–37. doi: 10.1016/j.stem.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Geyh S, Oz S, Cadeddu RP, Frobel J, Bruckner B, Kundgen A, et al. Insufficient stromal support in MDS results from molecular and functional deficits of mesenchymal stromal cells. Leukemia. 2013;27:1841–51. doi: 10.1038/leu.2013.193. [DOI] [PubMed] [Google Scholar]
- 5.Naveiras O, Nardi V, Wenzel PL, Hauschka PV, Fahey F, Daley GQ. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460:259–63. doi: 10.1038/nature08099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell stem cell. 2010;6:251–64. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamazaki S, Ema H, Karlsson G, Yamaguchi T, Miyoshi H, Shioda S, et al. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell. 2011;147:1146–58. doi: 10.1016/j.cell.2011.09.053. [DOI] [PubMed] [Google Scholar]
- 8.Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–7. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kode A, Manavalan JS, Mosialou I, Bhagat G, Rathinam CV, Luo N, et al. Leukaemogenesis induced by an activating beta-catenin mutation in osteoblasts. Nature. 2014;506:240–4. doi: 10.1038/nature12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blau O, Baldus CD, Hofmann WK, Thiel G, Nolte F, Burmeister T, et al. Mesenchymal stromal cells of myelodysplastic syndrome and acute myeloid leukemia patients have distinct genetic abnormalities compared with leukemic blasts. Blood. 2011;118:5583–92. doi: 10.1182/blood-2011-03-343467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopez-Villar O, Garcia JL, Sanchez-Guijo FM, Robledo C, Villaron EM, Hernandez-Campo P, et al. Both expanded and uncultured mesenchymal stem cells from MDS patients are genomically abnormal, showing a specific genetic profile for the 5q-syndrome. Leukemia. 2009;23:664–72. doi: 10.1038/leu.2008.361. [DOI] [PubMed] [Google Scholar]
- 12.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046–55. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson RF, Reimers M, Khulan B, Gissot M, Richmond TA, Chen Q, et al. An analytical pipeline for genomic representations used for cytosine methylation studies. Bioinformatics. 2008;24:1161–7. doi: 10.1093/bioinformatics/btn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roecklein BA, Torok-Storb B. Functionally distinct human marrow stromal cell lines immortalized by transduction with the human papilloma virus E6/E7 genes. Blood. 1995;85:997–1005. [PubMed] [Google Scholar]
- 16.Zhou L, McMahon C, Bhagat T, Alencar C, Yu Y, Fazzari M, et al. Reduced SMAD7 leads to overactivation of TGF-beta signaling in MDS that can be reversed by a specific inhibitor of TGF-beta receptor I kinase. Cancer Res. 2011;71:955–63. doi: 10.1158/0008-5472.CAN-10-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alvarez H, Opalinska J, Zhou L, Sohal D, Fazzari MJ, Yu Y, et al. Widespread hypomethylation occurs early and synergizes with gene amplification during esophageal carcinogenesis. PLoS Genet. 2011;7:e1001356. doi: 10.1371/journal.pgen.1001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figueroa ME, Reimers M, Thompson RF, Ye K, Li Y, Selzer RR, et al. An integrative genomic and epigenomic approach for the study of transcriptional regulation. PLoS ONE. 2008;3:e1882. doi: 10.1371/journal.pone.0001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueroa ME, Skrabanek L, Li Y, Jiemjit A, Fandy TE, Paietta E, et al. MDS and secondary AML display unique patterns and abundance of aberrant DNA methylation. Blood. 2009;114:3448–58. doi: 10.1182/blood-2009-01-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu W, Bhagat TD, Yang X, Song JH, Cheng Y, Agarwal R, et al. Hypomethylation of noncoding DNA regions and overexpression of the long noncoding RNA, AFAP1-AS1, in Barrett’s esophagus and esophageal adenocarcinoma. Gastroenterology. 2013;144:956–66. doi: 10.1053/j.gastro.2013.01.019. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhattacharyya S, Yu Y, Suzuki M, Campbell N, Mazdo J, Vasanthakumar A, et al. Genome-wide hydroxymethylation tested using the HELP-GT assay shows redistribution in cancer. Nucleic Acids Res. 2013;41:e157. doi: 10.1093/nar/gkt601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starczynowski DT, Vercauteren S, Telenius A, Sung S, Tohyama K, Brooks-Wilson A, et al. High-resolution whole genome tiling path array CGH analysis of CD34+ cells from patients with low-risk myelodysplastic syndromes reveals cryptic copy number alterations and predicts overall and leukemia-free survival. Blood. 2008;112:3412–24. doi: 10.1182/blood-2007-11-122028. [DOI] [PubMed] [Google Scholar]
- 25.Verma A, Deb DK, Sassano A, Kambhampati S, Wickrema A, Uddin S, et al. Cutting edge: activation of the p38 mitogen-activated protein kinase signaling pathway mediates cytokine-induced hemopoietic suppression in aplastic anemia. J Immunol. 2002;168:5984–8. doi: 10.4049/jimmunol.168.12.5984. [DOI] [PubMed] [Google Scholar]
- 26.Verma A, Deb DK, Sassano A, Uddin S, Varga J, Wickrema A, et al. Activation of the p38 mitogen-activated protein kinase mediates the suppressive effects of type I interferons and transforming growth factor-beta on normal hematopoiesis. J Biol Chem. 2002;277:7726–35. doi: 10.1074/jbc.M106640200. [DOI] [PubMed] [Google Scholar]
- 27.Hirata A, Utikal J, Yamashita S, Aoki H, Watanabe A, Yamamoto T, et al. Dose-dependent roles for canonical Wnt signalling in de novo crypt formation and cell cycle properties of the colonic epithelium. Development. 2013;140:66–75. doi: 10.1242/dev.084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pellagatti A, Cazzola M, Giagounidis A, Perry J, Malcovati L, Della Porta MG, et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia. 2010;24:756–64. doi: 10.1038/leu.2010.31. [DOI] [PubMed] [Google Scholar]
- 29.Chen S, Zambetti NA, Bindels EM, Kenswill K, Mylona AM, Adisty NM, et al. Massive parallel RNA sequencing of highly purified mesenchymal elements in low-risk MDS reveals tissue-context-dependent activation of inflammatory programs. Leukemia. 2016 doi: 10.1038/leu.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groschel S, Sanders MA, Hoogenboezem R, de Wit E, Bouwman BA, Erpelinck C, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–81. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Oda M, Glass JL, Thompson RF, Mo Y, Olivier EN, Figueroa ME, et al. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raza-Egilmez SZ, Jani-Sait SN, Grossi M, Higgins MJ, Shows TB, Aplan PD. NUP98-HOXD13 gene fusion in therapy-related acute myelogenous leukemia. Cancer research. 1998;58:4269–73. [PubMed] [Google Scholar]
- 34.Lin Y-W, Slape C, Zhang Z, Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood. 2005;106:287–95. doi: 10.1182/blood-2004-12-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung YJ, Choi CW, Slape C, Fry T, Aplan PD. Transplantation of a myelodysplastic syndrome by a long-term repopulating hematopoietic cell. Proc Natl Acad Sci U S A. 2008;105:14088–93. doi: 10.1073/pnas.0804507105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu H, Menendez S, Schlegelberger B, Bae N, Aplan PD, Gohring G, et al. Loss of p53 accelerates the complications of myelodysplastic syndrome in a NUP98-HOXD13-driven mouse model. Blood. 2012;120:3089–97. doi: 10.1182/blood-2012-01-405332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blau O, Hofmann WK, Baldus CD, Thiel G, Serbent V, Schumann E, et al. Chromosomal aberrations in bone marrow mesenchymal stroma cells from patients with myelodysplastic syndrome and acute myeloblastic leukemia. Exp Hematol. 2007;35:221–9. doi: 10.1016/j.exphem.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 38.Klaus M, Stavroulaki E, Kastrinaki MC, Fragioudaki P, Giannikou K, Psyllaki M, et al. Reserves, functional, immunoregulatory, and cytogenetic properties of bone marrow mesenchymal stem cells in patients with myelodysplastic syndromes. Stem cells and development. 2010;19:1043–54. doi: 10.1089/scd.2009.0286. [DOI] [PubMed] [Google Scholar]
- 39.Soenen-Cornu V, Tourino C, Bonnet ML, Guillier M, Flamant S, Kotb R, et al. Mesenchymal cells generated from patients with myelodysplastic syndromes are devoid of chromosomal clonal markers and support short- and long-term hematopoiesis in vitro. Oncogene. 2005;24:2441–8. doi: 10.1038/sj.onc.1208405. [DOI] [PubMed] [Google Scholar]
- 40.Roela RA, Carraro DM, Brentani HP, Kaiano JH, Simao DF, Guarnieiro R, et al. Gene stage-specific expression in the microenvironment of pediatric myelodysplastic syndromes. Leuk Res. 2007;31:579–89. doi: 10.1016/j.leukres.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–67. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327:1650–3. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoddart A, Fernald AA, Wang J, Davis EM, Karrison T, Anastasi J, et al. Haploinsufficiency of del(5q) genes, Egr1 and Apc, cooperate with Tp53 loss to induce acute myeloid leukemia in mice. Blood. 2014;123:1069–78. doi: 10.1182/blood-2013-07-517953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luis TC, Naber BA, Roozen PP, Brugman MH, de Haas EF, Ghazvini M, et al. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell. 2011;9:345–56. doi: 10.1016/j.stem.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 45.Kirstetter P, Anderson K, Porse BT, Jacobsen SE, Nerlov C. Activation of the canonical Wnt pathway leads to loss of hematopoietic stem cell repopulation and multilineage differentiation block. Nature immunology. 2006;7:1048–56. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- 46.Scheller M, Huelsken J, Rosenbauer F, Taketo MM, Birchmeier W, Tenen DG, et al. Hematopoietic stem cell and multilineage defects generated by constitutive beta-catenin activation. Nature immunology. 2006;7:1037–47. doi: 10.1038/ni1387. [DOI] [PubMed] [Google Scholar]
- 47.Slape CI, Saw J, Jowett JB, Aplan PD, Strasser A, Jane SM, et al. Inhibition of apoptosis by BCL2 prevents leukemic transformation of a murine myelodysplastic syndrome. Blood. 2012;120:2475–83. doi: 10.1182/blood-2012-05-430736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beachy SH, Aplan PD. Mouse models of myelodysplastic syndromes. Hematol Oncol Clin North Am. 2010;24:361–75. doi: 10.1016/j.hoc.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.