Abstract
Greater than 50% of estrogen receptor (ER)-positive breast cancers co-express the progesterone receptor (PR), which can directly and globally modify ER action to attenuate tumor growth. However, whether this attenuation is mediated only through PR-ER interaction remains unknown. To address this question, we assessed tumor growth in ER/PR-positive PDX models of breast cancer where both natural and synthetic progestins were found to antagonize the mitogenic effects of estrogens. Probing the genome-wide mechanisms by which this occurs, we documented that chronic progestin treatment blunted ER-mediated gene expression up to 2-fold at the level of mRNA transcripts. Unexpectedly, <25% of all ER DNA binding events were affected by the same treatment. The PR cistrome displayed a bimodal distribution. In one group, >50% of PR binding sites were co-occupied by ER, with a propensity for both receptors to coordinately gain or lose binding in the presence of progesterone. In the second group, PR but not ER was associated with a large fraction of RNA polymerase III (Pol III)-transcribed tRNA genes, independent of hormone treatment. Notably, we discovered that PR physically associated with the Pol III holoenzyme. Select pre-tRNA and mature tRNA that colocalized with PR and POLR3A at their promoters were relatively decreased in estrogen+progestin treated tumors. Our results illuminate how PR may indirectly impede ER action by reducing the bioavailability of translational molecules needed for tumor growth.
Keywords: progesterone receptors, progesterone, estrogen, estrogen receptors, breast cancer, RNA polymerase III
Introduction
Progesterone receptors (PR) have been routinely measured in breast cancers since the 1970s and have been traditionally thought to signify functional estrogen receptor alpha (ER) and a positive response to endocrine therapy (1). However, PR itself exerts a wide array of autonomous activity in breast cancer cells including liganded and unliganded transcriptional activity, rapid activation through kinases, metabolic alterations, and increased cancer stem cell activity (reviewed in 2, 3–5). PR exerts paradoxical effects on breast cancer growth depending on the experimental conditions. In 2D cultures, progestins generally inhibit cell growth following a transient push through the cell cycle (6, 7). Conversely, in 3D culture progestins increase clonogenicity and mammosphere formation, and in some models increase tumor growth independent of estrogen (8–11). In addition, the two natural isoforms of PR differ in their activity: PR-B exerts more proliferative signals whereas the truncated PR-A isoform is trans-repressive on PR and other steroid receptors (12, 13). The dual nature of PR has also confounded clinical use of progestins. In some studies for advanced breast cancer treatment, high doses of the synthetic progestin medroxyprogesterone acetate (MPA) were as effective as tamoxifen (reviewed in 14). However, in postmenopausal hormone replacement trials synthetic progestins in combination with estrogens increased breast cancer incidence, and their use rapidly diminished (15, 16). There is presently a renewed interest in utilizing the natural hormone progesterone (P4) or new synthetic PR-specific ligands for breast cancer management. However, distinguishing which tumors will benefit from positively or negatively targeting PR may depend on its convergence with ER signaling, an equally complex subject.
PR is a direct estrogen-induced gene in most target tissues, and can synergize with or antagonize ER to influence downstream biological processes. In promoter interference assays PR represses ER transcriptional activity predominantly through PR-A (12, 17). Several groups have reported that PR physically associates with ER, which may facilitate receptor crosstalk (18–21). In fact, Mohammed et al used chromatin immunoprecipitation followed by sequencing (ChIP-seq) to discover that progestins can alter global DNA binding events of ER in breast cancer cell lines (21). In particular, in the presence of progestins a net gain of ER binding occurs at novel transcriptionally active loci, many of which coincide with nearby PR binding events, suggestive of a cooperative and functional ER-PR complex. In this study, progesterone abrogated estrogen-induced tumor growth and was additive with tamoxifen, prospectively due in part to movement of ER from mitogenic loci to PR-controlled apoptotic and cell death genes (21). In a similar study, Singhal et al reported that estrogens or progestins alone regulate many of the same genes in the same direction in breast cancer cell lines or patient tumor explants, but that combined treatment led to decreased expression of oncogenic gene programs (22). In this study, ER genomic binding in the presence of estrogens and progestins resembled that of PR alone. Furthermore, a selective PR antagonist was sufficient to abrogate estrogen-mediated tumor growth and showed functional synergy with tamoxifen (22). While these studies illustrate the potential for co-targeting PR with ER in select breast cancers, context and ligand choice remain unresolved, and comparable studies in heterogeneous preclinical breast tumor models are needed.
We have utilized highly ER+PR+ breast cancer patient-derived xenografts (PDX) whose estrogen dependent growth is inhibited by natural and synthetic progestins to probe the underlying mechanisms by which this occurs. Our data indicate that sustained progestin treatment alters up to half of the estrogen-dependent transcriptome, albeit with minimal alteration of the ER cistrome. Interestingly, PR DNA binding in PDX tumors was present in a substantial fraction of RNA Polymerase III (Pol III)-transcribed tRNA genes. Moreover, PR was physically associated with the Pol III complex, and levels of select tRNAs were decreased in progestin treated tumors. These data indicate that in chronically treated solid tumors, in addition to direct interference of ER transcription, progestins may regulate overall tumor growth by modifying Pol III-mediated transcription and downstream translation. Pinpointing the tumors in which this occurs could provide appropriate contexts for prospective clinical use of progestins.
Materials and Methods
Tumor propagation
Development and transplantation of PDX tumors including UCD4 was as previously described (23). UCD65 was derived from a lymph node metastasis of a 41 year-old woman and was ER+PR+, Her2 unamplified. For the present experiments, tumors were partitioned into female NOD/SCID/ILIIrg−/− (NSG) mice supplemented with subcutaneous silastic pellets containing placebo (cellulose), 17β-estradiol (E2), E2+P4, E2+MPA, or MPA only (Sigma-Aldrich) as described previously (23, 24). Tamoxifen treatment was administered by intraperitoneal injection of 1 mg tamoxifen dissolved in peanut oil (vehicle) 3× weekly for 3 weeks. Tumors were measured weekly with a digital caliper and volume was estimated by the formula lw2/2. Tumors were profiled by RNA sequencing (RNA-seq, described below) and Illumina CytoSNP Arrays; UCD4 contains the D538G ER mutation while UCD65 has amplification at the ER locus 6.25.q1. All animal experiments were performed under a protocol approved by the University of Colorado Institutional Animal Care and Use Committee.
Immunohistochemistry
Immunohistochemistry (IHC) was performed as previously described (23). Antibodies used were as follows: AR (AR441, 1:500, DAKO), ERα (SP1, 1:100, Thermo-Fisher), and PR (1294, 1:500, DAKO). Images were captured using the Aperio Digital Pathology system (Leica Biosystems), assembled in Adobe Photoshop CC, and percent positive immunoreactivity determined for triplicate samples using Imagescope software (Leica).
Gene expression profiling
RNA was prepared from tumor fragments stored in Qiazol (Qiagen) and microarray expression profiling was performed on triplicate samples of UCD4 tumors treated with placebo, E2, E2+P4, or E2+MPA using Affymetrix Human Gene 1.1 ST Array Strips representing 28,875 genes as previously described (23). Microarray data were analyzed using Partek Genomics Suite® v6.6 and MetaCore™. Array data have been deposited in the Gene Expression Omnibus (GEO) database (GSE93109). For directional mRNA-seq, total RNA from triplicate PDX tumors treated with E2 or E2+MPA and the TruSeq Stranded Total Library kit were used to prepare libraries which were then sequenced using the Illumina HiSeq2500 System. Single-end 50nt reads were aligned to the human genome version GRCh37.64 using TopHat v2.0.4 and differential gene expression was analyzed using the Cufflinks Suite. Reads were normalized to FPKM values and student’s t-test performed to determine statistical significance (P<0.05).
ChIP-seq
For ChIP-seq experiments, biological triplicate tumors grown under continuous E2 or E2+P4 were analyzed. Flash frozen tumor specimens were pulverized, homogenized, then crosslinked with formaldehyde prior to sonication; chromatin was sheared using an S220 Focused Ultrasonicator (Covaris). ChIP was performed using ChIP-IT Express (Active Motif) using antibodies (5 ug per sample) to PR (Santa Cruz Biotechnology, sc-7208), ERα (Santa Cruz Biotechnology, sc-543), or a corresponding IgG negative control of the same species. Input DNA was also used as a control. For ChIP-seq, ER and PR ChIPseq reads were processed with cutadapt (https://code.google.com/p/cutadapt/; (25)) to remove 3’ adaptor sequences and 3’ bases with QUAL < 13. Trimmed reads were aligned to the human genome (grch37/hg19 build) using bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml; (26). Peaks were called on aligned reads using HOMER-IDR (https://github.com/karmel/homer-idr), an implementation of the Irreproducible Discovery Rate (IDR) framework (https://sites.google.com/site/anshulkundaje/projects/idr; (27)) using the HOMER peak caller (http://homer.salk.edu/homer/index.html). Peak sets were compared using bedtools (https://github.com/arq5x/bedtools2; (28)), and visualized using deeptools (https://github.com/fidelram/deepTools; (29)). Motif enrichment analyses were performed using MEME suite (http://meme-suite.org/; (30)). Raw sequence data and peak calls have been deposited in the Gene Expression Omnibus (GEO) database (GSE93109).
Rapid immunoprecipitation and mass spectrometry of endogenous proteins (RIME)
UCD4 and UCD65 tumors grown under E2+MPA conditions were analyzed by Active Motif’s RIME service. Detailed methods of analysis of MS/MS samples are provided in Supplementary Methods.
Immunoprecipitation (IP) and immunoblotting
For IP, 100–200 mg of flash frozen tumor was pulverized and homogenized in lysis buffer (50mM Tris pH 7.4, 140 nM NaCL, 2mM EGTA, 1.0% Tween-20). IP was performed overnight by combining 750 ug precleared lysate with 5 ug of antibody prebound to protein G Dynabeads (Thermo Fisher). Antibodies for IP were to PR (Santa Cruz Biotechnology, sc-7208, sc-166169; DAKO, 1294), and ERα (sc-543). Each IP was coupled with a corresponding IgG negative control of the same species. Beads were washed with wash buffer (50mM Tris pH 7.4, 140 nM NaCL, 2mM EGTA, 0.1% Tween 20), then boiled at 100°C for 10 min in 30 uL of 1× SDS loading buffer.
Antibodies for immunoblots were PR (sc-7208; DAKO, 1294), ERα (sc-56836), POLR3A (Abcam, 12825), and POLR3B (sc-515362). For imaging, the Odyssey Infrared Imaging System (Li-Cor Biosciences) was used, with secondary antibodies IRDye800CW Goat-Anti-Mouse-IgG and IRDye680LT Goat-Anti-Rabbit-IgG (Li-Cor Biosciences).
ChIP and qPCR
ChIP was performed as described above using antibodies to PR (sc-7208) and POLR3A (Abcam, 12825). qPCR was performed on ChIP DNAs or RNA prepared from tumor tissue as described above and normalized to β-actin using the Verso cDNA kit and ABsolute Blue Sybr Green (Thermo-Fisher Scientific). Primers for ChIP and qPCR are provided in Supplementary Table 1.
Northern blot
For Northern blots, 1 ug of total tumor RNA was run on a 14% acrylamide/urea gel, transferred to charged nylon membrane, UV crosslinked, and probed using biotinylated DNA oligonucleotide probes targeting specific tRNA families (sequences provided in Supplementary Table 2). Oligo probes were biotinylated using the PHOTOPROBE® Biotin Labeling Kit (Vector Labs). Hybridizations were performed overnight at 42°C. The Ambion NorthernMax system (Life Technologies) was used for washes; blocking and detection were performed with Odyssey Blocking Buffer and IRDye800CW Streptavidin (Li-Cor Biosciences). Membranes were stripped between each subsequent hybridization. Mature tRNA expression was analyzed using Image Studio (Li-Cor Biosciences) and normalized to 5.8S rRNA staining.
Statistics
Statistics were performed using Graphpad Prism 7.0. Two-tailed Student’s t-tests, or one-way ANOVA followed by Tukey post hoc multiple comparison tests were used as indicated. P<0.05 were considered significant.
Results
Progestins antagonize estrogen-induced growth of breast cancer PDX
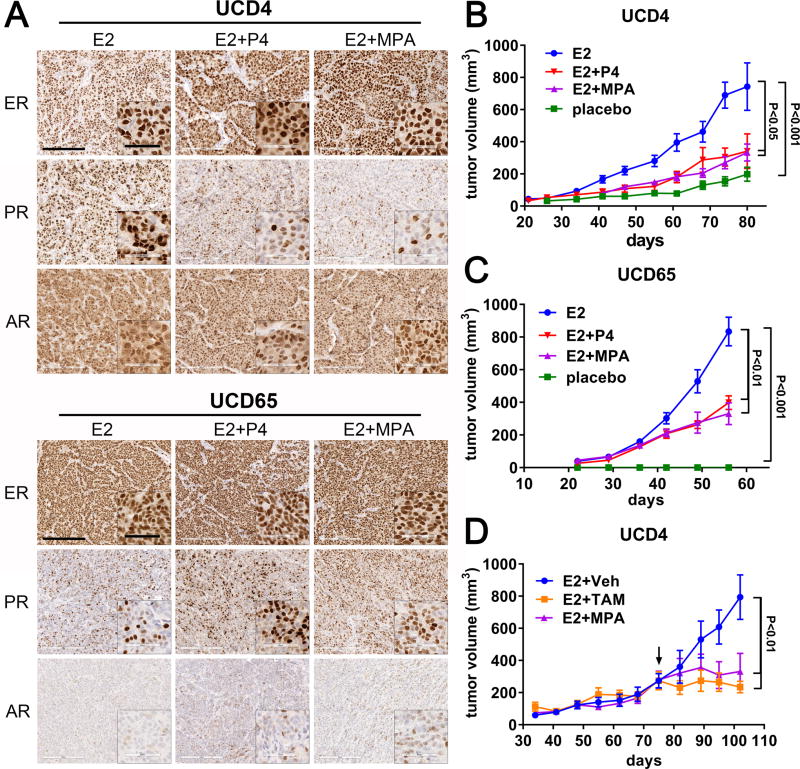
Our group has generated a collection of ER+ breast cancer PDX in which to study steroid hormone and receptor interactions (23, 31). Two PDX, UCD4 and UCD65, express robust ER and PR stably over multiple passages. We assessed levels of ER, PR, and androgen receptor (AR) by IHC in UCD4 and UCD65 tumors grown in mice supplemented with E2 alone, E2 plus P4, or E2 plus the synthetic progestin MPA (Fig. 1A; quantification in Supplementary Fig. S1A). UCD4 tumors given E2 had high ER and PR (>80% immunoreactive cells) and AR (>50%) expression; PR was partially downregulated with progestin treatment (45–50% PR+ cells). UCD65 tumors showed consistent high expression of ER (>95% of cells), PR positivity in ~42% of cells which increased with progestins (to 52–72% PR+ cells), and lower AR levels (<10% positive cells). UCD4 and UCD65 each expressed both natural PR isoforms, with PR-A:PR-B ratios of 1.4 and 1.1, respectively (Supplementary Fig. S1B).
Figure 1.
Progestins inhibit the estrogen-dependent growth of ER+PR+ breast cancer PDX. A, Tumors UCD4 and UCD65 were grown in mice supplemented with continuous E2, E2+P4, or E2+MPA. Sections were stained by IHC for ER, PR, and AR under each of the conditions as indicated. Scale bars, 200 um. Scale bars for insets, 60 um. Quantitation of IHC is in Supplementary Fig. S1A. B and C, Tumors were grown in mice either in the absence of exogenously added hormones (placebo), or in the presence of continuous E2, E2+P4, or E2+MPA. Tumor volumes were measured weekly and plotted versus the number of days of incubation ± SEM. Tumor volumes at the final time point were compared using ANOVA followed by a Tukey post-hoc multiple comparison test. Significance (P values) is indicated. n=6–8 tumors per condition. Experiments were performed a minimum of two times with the same statistically significant results. D, UCD4 tumors were grown in mice supplemented with E2 until they reached an average volume of 300 mm3, then either implanted with an MPA pellet, or treated 3 times weekly with tamoxifen (E2+TAM) or vehicle (E2+Veh). Tumor volumes at the final time point were compared using one-way ANOVA followed by a Tukey post-hoc multiple comparison test. n=6 tumors per condition. Significance is indicated; bars represent mean ± SEM.
We next assessed the consequence of co-supplementation with progestins on estrogen dependent growth of UCD4 and UCD65. Established tumors were partitioned into female NSG mice supplemented with placebo, E2 alone, E2+P4, or E2+MPA. UCD4 tumors grew modestly in placebo animals with a 3.5-fold increase in tumor size with E2 (Fig. 1B), while UCD65 tumors grew robustly only the presence of estrogens with no observable residual tumors in placebo treated animals (Fig. 1C). Both P4 or MPA given chronically at tumor implantation together with E2 significantly suppressed E2-dependent growth of both tumors (1.9- and 2.0-fold decrease for P4 and MPA in UCD4; 2.1- and 2.5-fold decrease for P4 and MPA in UCD65). MPA given after tumor establishment in UCD4 significantly reduced E2-dependent tumor growth to a similar degree as tamoxifen (3.1-fold decrease for MPA compared to 2.6-fold decrease for tamoxifen) (Fig. 1D). Final tumor mass confirmed tumor volume data in each experiment (Supplementary Fig. S2A– C). Progestins did not decrease E2-dependent tumor growth in two additional ER+ PDX tumors, both of which have low PR levels (<5% PR+ cells) (Supplementary Fig. S3A and B). We therefore chose to further investigate the mechanisms by which progestins suppress E2-driven tumor growth in UCD4 and UCD65.
Progestins alter mRNA levels of a subset of estrogen regulated genes
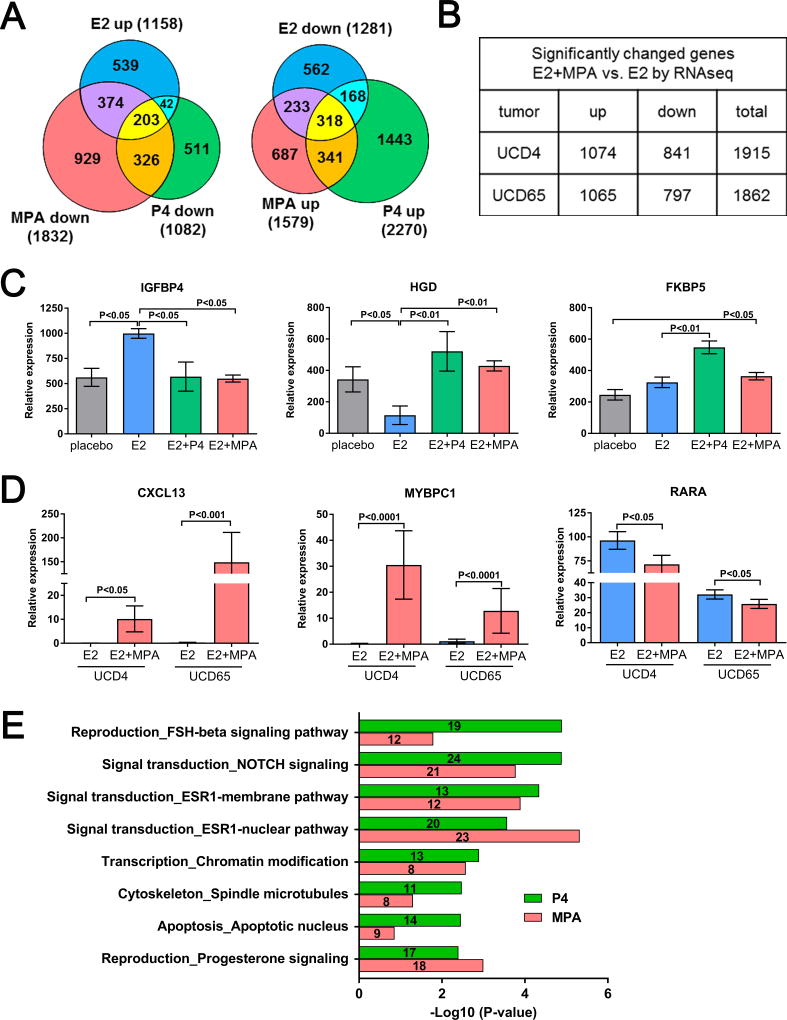
ER and PR crosstalk at the transcriptional level can be synergistic or antagonistic. We therefore determined the extent to which progestins impact E2-regulated genes in PDX tumors. We first utilized UCD4, which forms small tumors in the absence of exogenous estrogens and large tumors in the presence of estrogens, allowing assessment of E2-regulated genes. Gene expression profiles of placebo, E2, E2+P4, or E2+MPA treated UCD4 tumors were determined using Affymetrix Human Gene 1.1 ST Arrays. A total of 1158 mRNAs were significantly upregulated and 1281 mRNAs downregulated in E2 compared to placebo treated tumors (P<0.05) (Fig. 2A). Addition of P4 reversed 21% of E2 up- and 40% of E2 down-regulated transcripts, while MPA reversed 50% and 43% of E2 up- and downregulated transcripts, respectively. The majority of P4 reversed genes were shared by MPA; the additional MPA affected genes are likely due to its higher PR binding affinity, slower rate of metabolism, or androgenic activity. Progestins also altered a significant number of genes independent of E2 regulation, and it is likely these additionally impact tumor phenotype.
Figure 2.
Progestins reverse a subset of estrogen-regulated genes at the transcript level. A, The gene expression profiles of PDX UCD4 grown with placebo, E2, E2+P4, or E2+MPA were determined using Affymetrix human gene 1.1 ST arrays. An n of 3 tumors were profiled for all groups. Venn diagrams depict E2 up- or downregulated genes (E2 vs. placebo, P<0.05) compared to those comparatively up- and downregulated by P4 or MPA (E2+P4 or E2+MPA vs. E2, P<0.05). Total genes in each category are indicated in parentheses. B, Triplicate UCD4 and UCD65 tumors treated with E2 or E2+MPA were analyzed by RNA-seq. The number of genes significantly increased (up) or decreased (down) by the addition of MPA and the sum of genes changed in both directions (total) are indicated (P<0.05). C, Representative microarray genes in UCD4 that were E2-regulated and increased or decreased by P4 and MPA, or increased by P4 and MPA independent of E2. D, Representative RNA-seq genes increased or decreased in UCD4 and UCD65 tumors with E2+MPA compared to E2 only (P<0.05). E, Metacore enriched gene process networks in UCD4 tumors co-treated with E2 plus either P4 and MPA vs. E2 alone.
UCD4 and UCD65 tumors treated with E2 alone or E2+MPA were subsequently assessed in triplicate by directional mRNAseq (Fig. 2B). Compared to E2 alone, the addition of MPA significantly altered a similar number of transcripts in both tumors. Collectively, these data indicate that progestins significantly affect estrogen gene regulation at the transcript level in breast cancer PDX. Relative mRNA expression levels (by microarray) of representative genes that were E2-dependent and increased or decreased with progestins, or were altered only by progestins in UCD4 are depicted in Fig 2C. Genes with increased or decreased mRNAs with E2+MPA compared to E2 alone in UCD4 and UCD65 (by RNA-seq) are depicted in Fig 2D. Among E2-regulated mRNAs altered by progestins include those involved in ER and PR signaling, Notch signaling, and apoptosis (Fig. 2E).
Progesterone modestly alters ER binding sites in chronically treated solid tumors
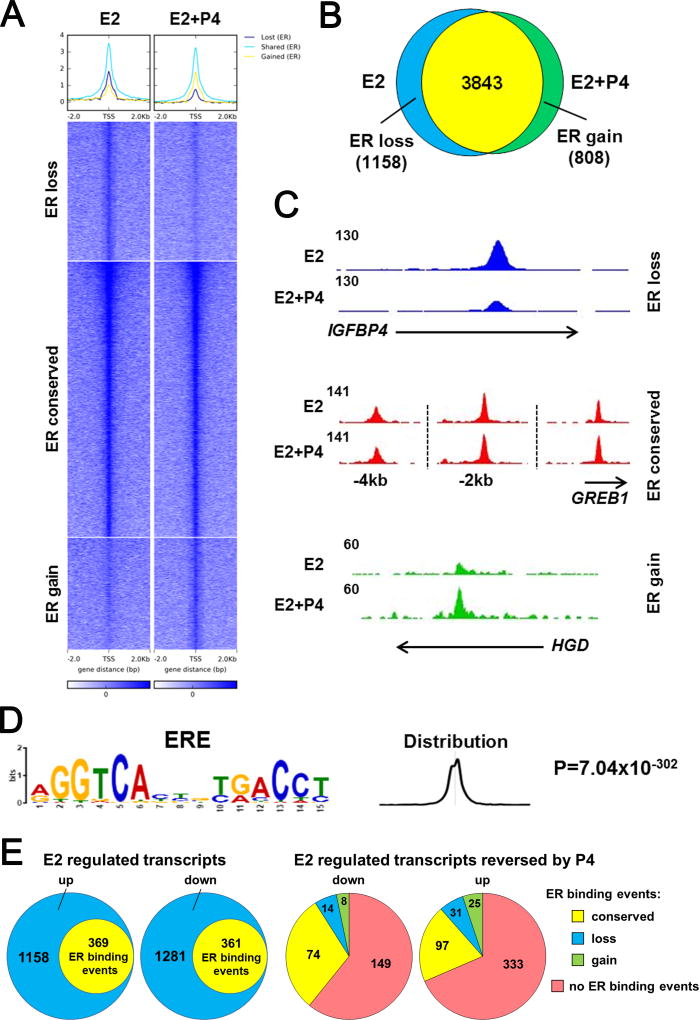
To explore the hypothesis that progestins redirect global ER binding events in PDX tumors as a means to disrupt tumor growth akin to that observed in breast cancer cell lines (21, 22), we performed ChIP-seq for ER in triplicate samples of UCD4 treated with E2 or E2+P4. ER bound to a similar number of genomic locations in E2 and E2 plus P4 tumors (5001 and 4651 binding sites, respectively) (Fig. 3A and B). The majority (76.8%) of the ER peaks were conserved between both hormone treatments. The addition of P4 caused a loss of ER binding at 1158 sites (23.2% of total E2/ER binding sites) and a gain of ER binding at 808 sites (17.4% of E2+P4/ER sites) (Fig. 3A and 3B)). Representative genes with conservation, loss, or gain of ER peaks with P4 treatment are indicated in Fig. 3C. The main sequence associated with ER binding events in all three groups (ER conserved, lost, or gained with P4) was a consensus estrogen response element (Fig. 3D). One third of genes regulated by E2 at the transcript level contained ER binding peaks (nearest gene in a window of 2.0 kb, Fig. 3E). The majority of E2 regulated genes affected by P4 at the transcript did not contain ER binding events. Among those that did have ER binding events, most were conserved and <10% showed a gain or loss with P4 (Fig. 3E). Taken together, these data suggest that redirection of ER binding sites by P4 is likely not the only mechanism by which tumor growth rate is decreased.
Figure 3.
Progesterone modestly alters the ER cistrome. ChIP-seq was performed for ER in triplicate UCD4 tumors grown in mice supplemented with E2 or E2+P4. A, Heat map depicts intensity of ER binding events at loci that did not change between E2 and E2+P4 (ER conserved), that were lost with P4 (ER loss), and that were gained with P4 (ER gain). The heat map is shown in a horizontal window of ± 2 kb. B, Venn diagram depicts overlap between ER binding events in E2 and E2+P4 treated tumors. C, Representative ER peaks that were conserved, lost, or gained in E2+P4 compared to E2 tumors. D, The major enriched motif in all three groups (ER conserved, ER loss, and ER gain) resembles a classic estrogen responsive element (ERE). E, Comparison of E2 regulated transcripts (by microarray) with ER binding events (ChIP-seq). Left two circles, number of E2 regulated genes (placebo vs. E2, up or down) vs. number of genes with ER binding. Right two circles, number of E2 regulated genes reversed by P4 (down or up) that showed no ER binding events, or conservation (E2 and E2+P4), loss (E2 only) or gain (E2+P4 only) of ER binding.
PR genomic binding is distributed between ER co-occupied and ER unoccupied sites
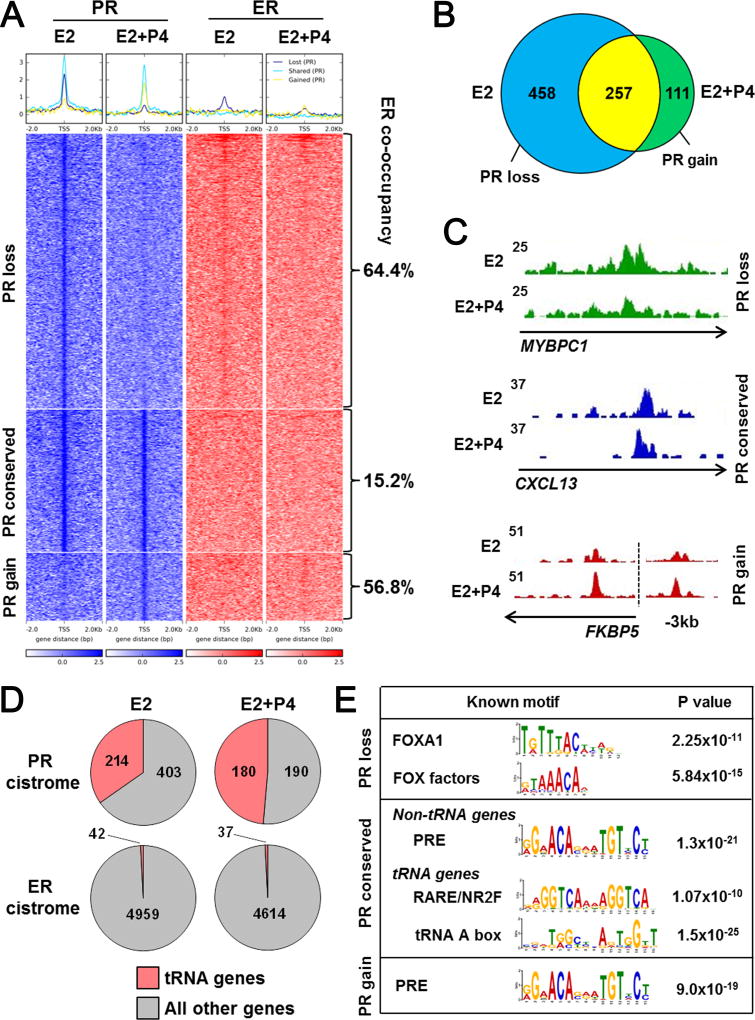
We speculated that progestins influence E2-dependent growth through additional mechanisms that don’t involve changes in ER binding events. To assess PR occupation of chromatin, we performed ChIP-seq for PR in triplicate in the same E2 and E2+P4 treated UCD4 tumors utilized for ER ChIP-seq. PR binding events were less robust than ER and showed a net loss of binding with E2 vs. E2+P4 (715 and 368 binding sites respectively) (Fig. 4A and B). Loss of PR binding with P4 could be partially due to PR downregulation (Fig. 1A). PR binding events were categorized as those lost, conserved, or gained with P4 treatment. Notably, ER co-occupied a majority of PR binding sites that were lost (64.4%) and gained (56.8%), and ER was similarly lost and gained at these sites (Fig. 4A). The presence of PR at binding sites in the E2 only group suggests that PR is localized on chromatin through direct or indirect interaction in the absence of exogenously added ligand, but that P4 is required for potent transcriptional regulation (Fig. 2). Representative genes with conservation, loss, or gain of PR peaks with P4 treatment are indicated in Fig. 4C.
Figure 4.
The PR cistrome is distributed between ER co-occupied binding sites and RNA Pol III-regulated genes. ChIP-seq was performed for PR in triplicate UCD4 tumors grown in mice supplemented with E2 or E2+P4. A, Heat map depicts intensity of PR binding events at genomic loci with loss (PR loss), conservation (PR conserved), or gain (PR gain) in tumors treated with E2+P4 compared to E2 alone (blue). ER binding events at the same loci are depicted on the right (red). Percent of ER co-occupancy with PR is indicated for each group. The heat map is shown in a horizontal window of ± 2 kb. B, Venn diagram depicts overlap between PR binding events in E2 and E2+P4 treated tumors. C, Representative PR peaks that were conserved, lost, or gained with E2+P4 compared to E2 alone. D, Pie charts depict number of total PR or ER binding sites localized near tRNA vs. non-tRNA genes in E2 and E2+P4 treated tumors. E, Enriched sequence motifs in the PR loss, conserved, and gain groups.
Interestingly, PR binding events occurred at a large fraction (214 and 180 in E2 and E2+P4 tumors, respectively) of tRNA genes that are transcribed by Pol III (Fig 4D). PR binding at tRNAs was found almost exclusively in the conserved group, occurring under both E2 and E2+P4 conditions. By contrast, less than 1% of ER binding events occurred near tRNA genes. Motif enrichment analysis also revealed significant differences in PR binding patterns across the lost, conserved, and gained binding site cohorts (Fig. 4E). PR binding sites in the presence of E2 that were lost with P4 tended to be near FOX factor sequence motifs, suggesting that PR is indirectly associated with chromatin and is redirected upon P4 addition. This is supported by the P4-mediated gain in PR binding events at sequences resembling a progesterone response element (PRE). Interestingly, motif sequences near PR binding differed in the conserved group between non-tRNA and tRNA genes. While PR was present at non-tRNA genes near a PRE, PR binding at tRNA genes was not associated with a PRE, but instead with a motif (AGGTCANNAGGTCA) typical of retinoic acid receptor (RAR) and other nuclear receptor 2 family (NR2F) members (Fig. 4E). These data suggest that PR genomic binding patterns in solid tumors is complex. Liganded PR are redirected from non-PRE associated to PRE-associated chromatin binding sites, often in conjunction with ER, at a majority of loci. However, constitutive PR binding at tRNA sites occurs in the absence of ER or PRE-like sequences and may involve crosstalk with other transcription factors at these sites.
PR associates with RNA Polymerase III subunits in breast tumors
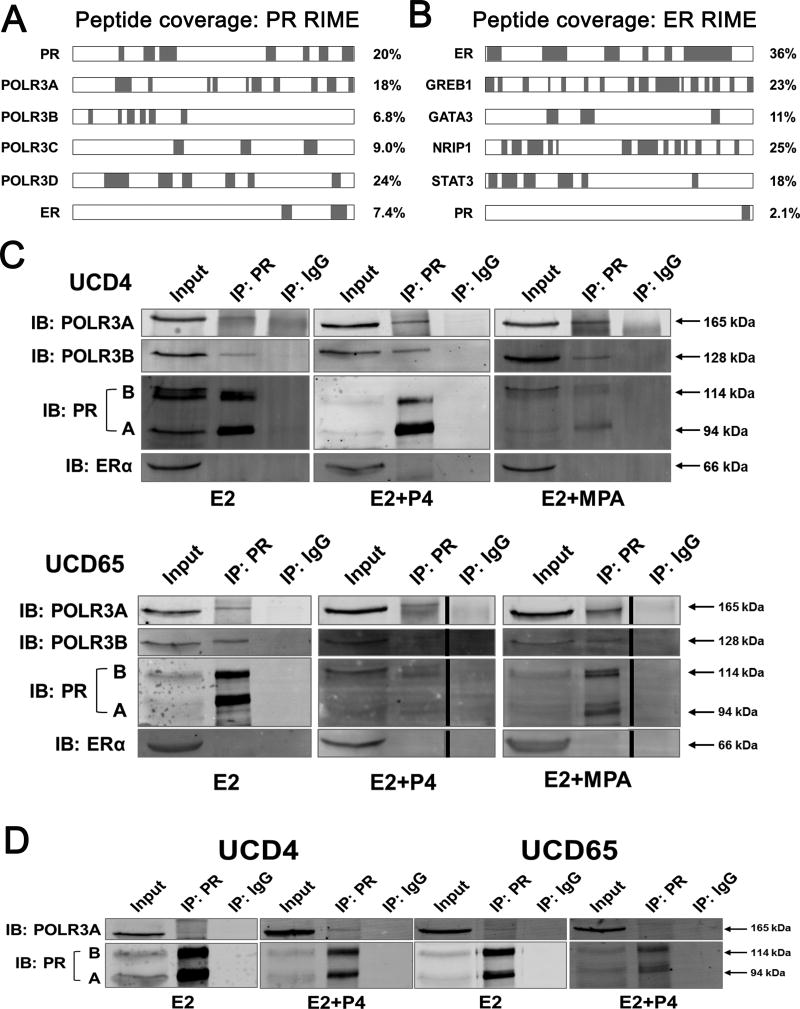
We speculated that the diverse actions of PR may be due to unique protein-protein interactions in breast cancer PDX. To assess this, we determined the global PR and ER interactome in duplicate samples of UCD4 and UCD65 tumors treated with E2+MPA using rapid immunoprecipitation of endogenous proteins followed by mass spectrometry (RIME). Interestingly, in both UCD4 and UCD65 tumors, multiple subunits of the Pol III holoenzyme immunoprecipitated with PR (Fig. 5A and Supplementary Table 3). PR also demonstrated a detectable interaction with ER. In contrast, ER was not associated with any Pol III subunits, but instead was associated with multiple coactivators and transcription factors previously described by Mohammed et al in MCF7 cells (i.e. GREB1, GATA3, NRIP1, STAT3) (Fig. 5B) (32); this also included an association with PR, albeit one that was less robust than the other coactivators.
Figure 5.
PR is associated with the RNA Pol III complex in ER+PR+ breast cancer PDX. A and B, PR and ER associated proteins were determined by RIME for UCD65 tumors grown in mice supplemented with E2 or E2+MPA. Peptide coverage (highlighted in green) of PR (A) or ER (B) associated proteins with percent coverage indicated. C, Co- IP of PR with RNA Pol III subunits POLR3A and POLR3B in UCD4 and UCD65 tumors treated with E2, E2+P4, or E2+MPA. IP was performed with PR Ab sc-7208, and immunoblots (IB) performed with antibodies to PR (A and B isoforms indicated), POLR3A, POLR3B, or ER(α). Molecular weights are indicated. D, Co-IP of PR with POLR3A in UCD4 and UCD65 E2 and E2+P4 treated tumors using IP with a different PR antibody (mAb F-4).
To confirm the interaction between PR and Pol III, we performed co-immunoprecipitation (IP) on lysates from E2, E2+P4, and E2+MPA treated UCD4 and UCD65 tumors using three different PR antibodies. In E2 treated tumors, IP for PR confirmed an interaction between PR and the POLR3A and POLR3B subunits of the Pol III holoenzyme in both UCD4 and UCD65 tumors (Fig. 5C). This was reproduced using two additional PR antibodies to confirm the interaction (Fig. 5D, Supplementary Fig. S4). We could not detect a PR-ER association in UCD4 or UCD65 tumors even though we could observe this interaction in T47D cells under the same conditions (Supplementary Fig. S5). Overall, these data demonstrate that PR associates with the Pol III complex in PDX breast tumors through direct or indirect interactions, and implies PR could influence Pol III activity and tumor growth.
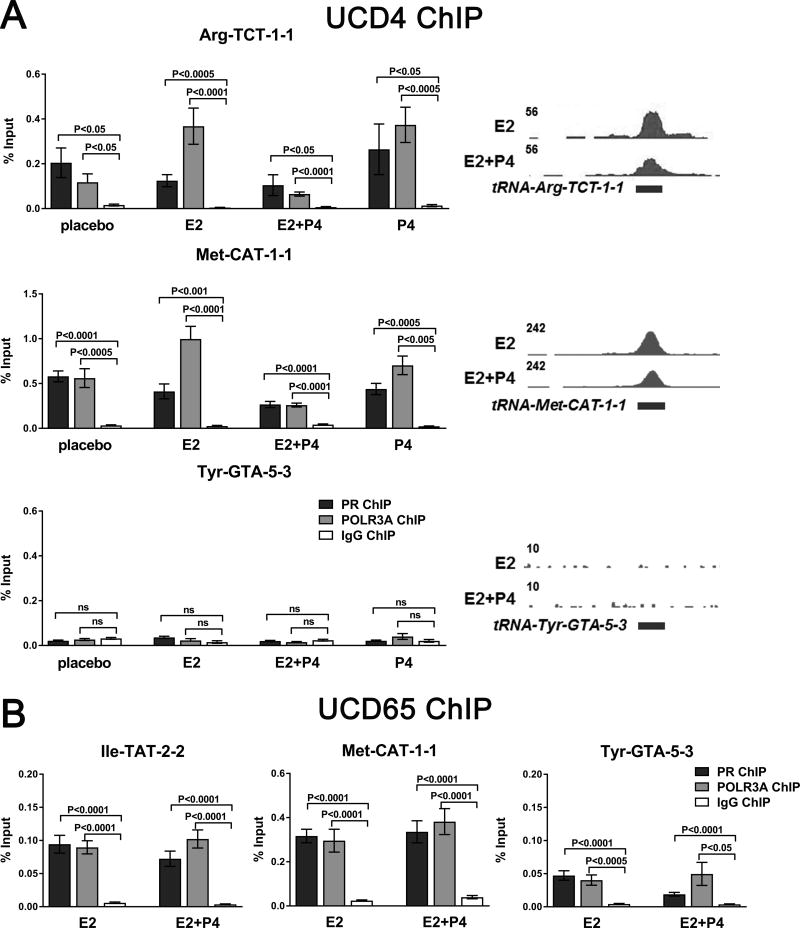
PR co-occupies tRNAs with POLR3A and progestins decrease tRNA levels
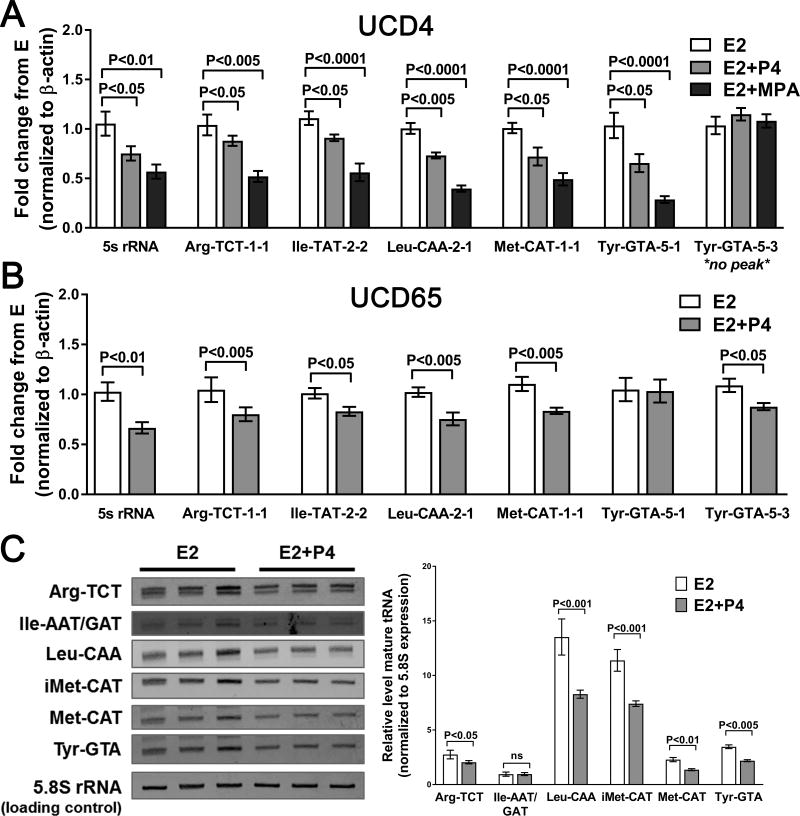
RNA Pol III transcription provides critical translational machinery necessary for robust tumor growth (33). To explore the hypothesis that progestins decrease Pol III-mediated transcription, we evaluated POLR3A and PR co-occupancy at several tRNA genes, and assessed specific tRNA levels under different hormone conditions. ChIP for POLR3A and PR was performed in UCD4 tumors treated with placebo, P4, E2, or E2 plus P4 and UCD65 tumor samples treated with E2 or E2 plus P4. In UCD4, PR and POLR3A were present at two tRNA genes identified as having PR peaks via ChIP-seq (Fig. 6A), and this association occurred under all hormone conditions. Neither PR nor POLR3A were present at a tRNA gene that was absent for PR by ChIP-seq. In UCD65, PR and POLR3A co-occupied at all three tRNA genes tested (Fig. 6B). Pre-tRNA levels were measured by qPCR in UCD4 and UCD65 tumors treated with E2, E2+P4, or E2+MPA (UCD4 only). In UCD4, levels of five individual pre-tRNAs that showed PR occupancy were decreased by both progestins, with a trend towards more robust reduction with MPA compared to P4 (Fig. 7A). By contrast, levels of a pre-tRNA unoccupied by PR by ChIP-seq (Tyr-GTA-5-3) were unchanged in the presence of progestins. Five of the six pre-tRNAs tested were also downregulated in E2+P4 compared to E2 treated UCD65 tumors (Fig. 7B). Progestins also decreased 5S rRNA in both tumors, suggesting a potential broader influence on Pol III directed transcription. Levels of six mature tRNA families were measured by Northern blot. Five mature tRNAs decreased in E2+P4 compared to E2 treated tumors (Fig. 7C). Collectively, these data indicate that progestins suppress expression of tRNA genes with nearby PR binding events in PDX tumors, and could provide a mechanism, independent of direct ER transcriptional antagonism, by which PR modulates tumor cell growth.
Figure 6.
Co-localization of PR and POLR3A at tRNA genes in tumors independent of hormone treatment. A and B, ChIP-qPCR analysis of PR and POLR3A at three tRNA genes in UCD4 tumors grown in mice supplemented with placebo, E2, E2+P4, or P4 (A) and UCD65 tumors grown in mice supplemented with E2 or E2+P4 (B). ChIP was performed from three independent biological replicates. Peaks at the corresponding loci are indicated. Bars represent mean ± SEM. Significance was determined comparing each ChIP to IgG control using a Student’s t-test.
Figure 7.
P4 decreases pre- and mature tRNA levels in ER+PR+ PDX breast tumors. A and B, qPCR analysis of primary tRNA levels in UCD4 (A) and UCD65 (B) tumors grown in mice supplemented with E2, E2+P4, or E2+MPA (UCD4 only). Three independent biological replicates were used for analysis. Bars represent mean ± SEM. Significance was determined using one-way ANOVA followed by Tukey post-hoc multiple comparison tests (A) or Students t-test (B). C, Northern blot analysis of mature tRNA families (i.e. all Arg-TAT species) in UCD 4 tumors grown in mice supplemented with E2 or E2+P4. Three independent biological replicates were used. Quantified was normalized to RNA Pol I-regulated 5.8S rRNA. Quantification is graphed on the right as mean ± SEM. Significance was determined using a Student’s t-test for each tRNA group.
Discussion
Here we used solid tumor PDX models in which progestins suppress estrogen stimulated breast cancer growth to investigate the underlying mechanisms. This occurs in a bimodal fashion: First, liganded PR potently impacts ER transcriptional activity; however, redirection of ER was minimal compared to that seen in breast cancer cell lines (21, 22). Second, we show that PR exerts its own pleiotropic effects independent of ER. These occur by constitutive localization of unliganded PR at Pol III transcribed tRNA genes, followed by their negative regulation upon progestin treatment (P4 or MPA). Thus, we provide evidence that PR employs both direct and indirect mechanisms to hinder the growth-promoting functions of ER. The association of PR with the Pol III complex implies that PR, and perhaps other nuclear receptors, have a broader role in regulating transcription beyond that of Pol II directed genes. Importantly, alterations in translational control can trump elevated transcription, providing a powerful prospective new role for PR in global regulation of cancer cell growth.
There is currently revived interest in utilizing the natural hormone P4 or new synthetic PR ligands in conjunction with endocrine therapies in ER+ breast cancer (4). This is based on two premises: First, the historical efficacy of synthetic progestins, mostly MPA or its derivatives, in reducing breast tumor growth (reviewed in 14). Second, recent genome-wide studies describing that liganded PR impinges on ER mitogenic chromatin interactions, leading to a growth inhibitory phenotype (21, 22). However, the antithesis to these notions are longstanding observations that P4 and progestins exert autonomous proliferative effects in breast cancers under some contexts (reviewed in 3), and more recent discoveries that P4 expands cancer stem cells (reviewed in (5)). Progestin-mediated proliferation tends to occur when progestins are acting in the absence of estrogens, and is especially notable in 3D models (8–10). Unliganded PR may cooperate with ER to induce proliferative signals (20). P4 expansion of cancer stem cells occurs in the absence or presence of estrogens (9, 10, 24). Other reservations are based on the surprising observation that synthetic progestins (mostly MPA) in combination with estrogens in postmenopausal hormonal therapies are tumorigenic in the breast (15, 16). However, use of the natural hormone P4 in postmenopausal therapies has not been statistically linked to breast tumor incidence (discussed in 4). At present, a precise delineation of the mechanisms by which P4 is growth-suppressive vs. growth-stimulating, and the resulting phenotype of P4-treated tumors, is still needed to advise appropriate clinical use.
Our data in solid tumors underscores the complexity of PR action under different contexts. Here we tested conditions under which tumors were chronically exposed to estrogens plus progestins (several months) and demonstrate both phenotypic similarities to short term co-treatment with both ligands in cell lines and explant cultures (21, 22), as well as some mechanistic differences. First, PR association with chromatin was more robust in the absence of added ligand (E2 only) (Fig. 4A and B). The majority of these sites were near sequences for FOX pioneering factors, suggesting a possible indirect association (Fig. 4E). In the presence of exogenous P4, PR was redirected to more conventional PREs in a cohort of genes. Second, the majority of the PR cistrome was co-occupied by ER, confirming significant cross-talk between the two receptors in gene regulation. Interestingly, only a small fraction of E2 regulated genes that significantly changed with P4 had ER or PR binding sites (Fig. 3E), suggesting indirect means of transcript regulation. Third, an ER-PR association was detected (although weakly) in the two PDX tumors tested (by RIME only) similar to that observed in breast cancer cell lines (Fig. 5A and B). Conversely, we could not detect a PR-Pol-III association in T47D cells similar to that observed in our PDX tumors (by co-IP). We speculate that an intermediary factor required for this interaction may be missing in vitro. In support of this theory, ChIP-seq PR binding events were found at only a small subset of tRNA genes (19, or 3% of annotated tRNA genes) in T47D cells (34). Of note, progestins effectively suppressed ER-mediated transcription and growth in tumors with genomic ER alterations (UCD4 contains the ESR1 D538G mutation and UCD65 has amplification/ overexpression of ER), and thus could be beneficial in endocrine refractory and/or ER abnormal tumors that retain PR expression.
Dysregulation of Pol III transcribed tRNAs is linked to increased transformation and cell proliferation in a variety of cancers including breast (35). Pol III and tRNA synthesis can be regulated by a variety of oncogenic signaling pathways (reviewed in 36); however their regulation by nuclear receptors has not been described. There is, however, precedence for steroids affecting tRNA pools. A 1968 paper by O’Malley et al described an estrogen-induced increase in tRNA levels in the chick oviduct (37). In addition, P4 and dihydrotestosterone directly bound to aminoacyl-tRNAs to inhibit protein synthesis in an in vitro system (38). Analysis by tRNA-scan-SE has identified 610 tRNA genes in the hg19 reference human genome (39); these encode 49 redundant tRNA isoacceptors for translating 61 sense codons. We describe the presence of PR at a large proportion (40%) of tRNA genes in a breast tumor (Fig. 4D); its presence coincided with a P4-mediated reduction in expression of select pre- and mature tRNAs (Fig. 7). Interestingly, PR occupied the highest proportion (70%) of initiator methionine (tRNAiMet) loci in UCD4. Overexpression of tRNAiMet increases cell proliferation and metabolic activity of epithelial cells (40). Thus, the reduction of mature tRNAiMet (Fig. 7C) would prospectively impede proliferation through stalling mRNA translation at the ribosome. In fact, codon usage can be related to specific transcriptional programs including cellular proliferation in cancer cells (41, 42), such that alterations in tRNA pools can support selective translation and potentially also affect mRNA stability.
We therefore speculate that a progestin mediated alteration in tRNA pools could affect specific gene sets at the translational level. That a typical PRE sequence was not well represented at tRNA loci implicates that PR is associated through other mechanisms. The presence of a direct repeat (DR)2 RARE near the PR bound tRNA genes is intriguing, and we speculate that PR may be associated through RAREs or through interaction with RARs. There is some data to support this theory. RARα is a common and necessary co-factor at many ER transcribed genes in breast cancer cells (43). Furthermore, we have observed co-recruitment of PR and RARα at sequences resembling PREs and RAREs in breast cancer cells (44). Our results also imply that PR has a general impact on Pol III transcription and could possibly affect ribosome function since 5S rRNA abundance was depressed by P4 and MPA (Fig. 7A and B).
In summary, we provide provocative new insight into PR action in solid breast tumors. Our data describe PR autonomous (of ER) influence on solid tumors through interference with RNA Pol III directed transcription; in addition to corroborating recent works that describe PR repositioning of ER in breast cancer genomes. This opens up exciting possibilities for PR targeted therapies. However, targeting PR can be tricky. Analysis of TCGA data has revealed loss of heterozygosity at the PR locus in 20–40% of ER+ breast tumors (21, 45). Concordantly, up to half of ER+ breast cancers show loss or underexpression of PR (46). Still, we speculate that ~25% of ER+ tumors would have adequate PR levels to confer tumor suppressive effects. There are currently no biomarkers that delineate which ER+PR+ breast cancers may be P4 responsive; and indeed P4 could be harmful in some situations underscoring this gap in our knowledge. While this study focused on PR-ER interactions, it is becoming more evident that individual steroids and steroid receptors rarely act in isolation. AR and GR also modulate ER action within ER+ breast cancers (47–49) and there is a growing interest in co-targeting multiple nuclear receptors in breast cancer (50). Therefore, a global understanding of the collective cacophony of steroid receptor activity at both the transcription and translation level is necessary to pinpoint the subset of tumors appropriate for steroidal intervention, and which combinations of steroids/antisteroids are required to achieve therapeutic efficacy.
Supplementary Material
Acknowledgments
We thank the University of Colorado Cancer Center Genomics and Microarray, Research Histology, and Biorepository Cores supported by P30CA046934 for their technical assistance and services. We also thank the Colorado Genetics Laboratory for their services. We thank Dean Edwards for antibody advice and Kate Horwitz for critical review of the manuscript.
Financial support: This work was supported by NIH 1F32CA177081 (J. Finlay-Schultz) and NIH R01 CA140985 (C.A. Sartorius), and the Breast Cancer Research Foundation (BCRF-16-072; C.A. Sartorius co-PI).
Footnotes
The authors declare no conflict of interest.
References
- 1.Horwitz KB, McGuire WL. Predicting response to endocrine therapy in human breast cancer: a hypothesis. Science. 1975;189:726–7. doi: 10.1126/science.168640. [DOI] [PubMed] [Google Scholar]
- 2.Jacobsen BM, Horwitz KB. Progesterone receptors, their isoforms and progesterone regulated transcription. Mol Cell Endocrinol. 2012;357:18–29. doi: 10.1016/j.mce.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagan CR, Lange CA. Molecular determinants of context-dependent progesterone receptor action in breast cancer. BMC Med. 2014;12:32. doi: 10.1186/1741-7015-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll JS, Hickey TE, Tarulli GA, Williams M, Tilley WD. Deciphering the divergent roles of progestogens in breast cancer. Nat Rev Cancer. 2017;17:54–64. doi: 10.1038/nrc.2016.116. [DOI] [PubMed] [Google Scholar]
- 5.Finlay-Schultz J, Sartorius CA. Steroid Hormones, Steroid Receptors, and Breast Cancer Stem Cells. J Mammary Gland Biol Neoplasia. 2015;20:39–50. doi: 10.1007/s10911-015-9340-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Musgrove EA, Lee CS, Sutherland RL. Progestins both stimulate and inhibit breast cancer cell cycle progression while increasing expression of transforming growth factor alpha, epidermal growth factor receptor, c-fos, and c-myc genes. Mol Cell Biol. 1991;11:5032–43. doi: 10.1128/mcb.11.10.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groshong SD, Owen GI, Grimison B, Schauer IE, Todd MC, Langan TA, et al. Biphasic regulation of breast cancer cell growth by progesterone: role of the cyclin-dependent kinase inhibitors, p21 and p27(Kip1) Mol Endocrinol. 1997;11:1593–607. doi: 10.1210/mend.11.11.0006. [DOI] [PubMed] [Google Scholar]
- 8.Faivre EJ, Lange CA. Progesterone receptors upregulate Wnt-1 to induce epidermal growth factor receptor transactivation and c-Src-dependent sustained activation of Erk1/2 mitogen-activated protein kinase in breast cancer cells. Mol Cell Biol. 2007;27:466–80. doi: 10.1128/MCB.01539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay-Schultz J, Cittelly DM, Hendricks P, Patel P, Kabos P, Jacobsen BM, et al. Progesterone downregulation of miR-141 contributes to expansion of stem-like breast cancer cells through maintenance of progesterone receptor and Stat5a. Oncogene. 2015;34:3676–87. doi: 10.1038/onc.2014.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodman CR, Sato T, Peck AR, Girondo MA, Yang N, Liu C, et al. Steroid induction of therapy-resistant cytokeratin-5-positive cells in estrogen receptor-positive breast cancer through a BCL6-dependent mechanism. Oncogene. 2016;35:1373–85. doi: 10.1038/onc.2015.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang Y, Benakanakere I, Besch-Williford C, Hyder RS, Ellersieck MR, Hyder SM. Synthetic progestins induce growth and metastasis of BT-474 human breast cancer xenografts in nude mice. Menopause. 2010;17:1040–7. doi: 10.1097/gme.0b013e3181d3dd0c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen DX, Xu YF, Mais DE, Goldman ME, McDonnell DP. The A and B isoforms of the human progesterone receptor operate through distinct signaling pathways within target cells. Mol Cell Biol. 1994;14:8356–64. doi: 10.1128/mcb.14.12.8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdel-Hafiz H, Takimoto GS, Tung L, Horwitz KB. The inhibitory function in human progesterone receptor N termini binds SUMO-1 protein to regulate autoinhibition and transrepression. J Biol Chem. 2002;277:33950–6. doi: 10.1074/jbc.M204573200. [DOI] [PubMed] [Google Scholar]
- 14.Santen RJ, Manni A, Harvey H, Redmond C. Endocrine treatment of breast cancer in women. Endocr Rev. 1990;11:221–65. doi: 10.1210/edrv-11-2-221. [DOI] [PubMed] [Google Scholar]
- 15.Chlebowski RT, Anderson GL, Gass M, Lane DS, Aragaki AK, Kuller LH, et al. Estrogen plus progestin and breast cancer incidence and mortality in postmenopausal women. JAMA. 2010;304:1684–92. doi: 10.1001/jama.2010.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beral V, Reeves G, Bull D, Green J. Breast cancer risk in relation to the interval between menopause and starting hormone therapy. J Natl Cancer Inst. 2011;103:296–305. doi: 10.1093/jnci/djq527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng ZY, Bay BH, Aw SE, Lin VC. A novel antiestrogenic mechanism in progesterone receptor-transfected breast cancer cells. J Biol Chem. 2005;280:17480–7. doi: 10.1074/jbc.M501261200. [DOI] [PubMed] [Google Scholar]
- 18.Ballare C, Uhrig M, Bechtold T, Sancho E, Di Domenico M, Migliaccio A, et al. Two domains of the progesterone receptor interact with the estrogen receptor and are required for progesterone activation of the c-Src/Erk pathway in mammalian cells. Mol Cell Biol. 2003;23:1994–2008. doi: 10.1128/MCB.23.6.1994-2008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giulianelli S, Vaque JP, Soldati R, Wargon V, Vanzulli SI, Martins R, et al. Estrogen receptor alpha mediates progestin-induced mammary tumor growth by interacting with progesterone receptors at the cyclin D1/MYC promoters. Cancer Res. 2012;72:2416–27. doi: 10.1158/0008-5472.CAN-11-3290. [DOI] [PubMed] [Google Scholar]
- 20.Daniel AR, Gaviglio AL, Knutson TP, Ostrander JH, D'Assoro AB, Ravindranathan P, et al. Progesterone receptor-B enhances estrogen responsiveness of breast cancer cells via scaffolding PELP1- and estrogen receptor-containing transcription complexes. Oncogene. 2015;34:506–15. doi: 10.1038/onc.2013.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohammed H, Russell IA, Stark R, Rueda OM, Hickey TE, Tarulli GA, et al. Progesterone receptor modulates ERalpha action in breast cancer. Nature. 2015;523:313–7. doi: 10.1038/nature14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singhal H, Greene ME, Tarulli G, Zarnke AL, Bourgo RJ, Laine M, et al. Genomic agonism and phenotypic antagonism between estrogen and progesterone receptors in breast cancer. Sci Adv. 2016;2:e1501924. doi: 10.1126/sciadv.1501924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabos P, Finlay-Schultz J, Li C, Kline E, Finlayson C, Wisell J, et al. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res Treat. 2012;135:415–32. doi: 10.1007/s10549-012-2164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sartorius CA, Harvell DM, Shen T, Horwitz KB. Progestins initiate a luminal to myoepithelial switch in estrogen-dependent human breast tumors without altering growth. Cancer Res. 2005;65:9779–88. doi: 10.1158/0008-5472.CAN-05-0505. [DOI] [PubMed] [Google Scholar]
- 25.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011:17. [Google Scholar]
- 26.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li QH, Brown JB, Huang HY, Bickel PJ. Measuring Reproducibility of High-Throughput Experiments. Ann Appl Stat. 2011;5:1752–79. [Google Scholar]
- 28.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–5. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthews SB, Sartorius CA. Steroid hormone receptor positive breast cancer patient-derived xenografts. Horm Cancer. 2016 doi: 10.1007/s12672-016-0275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammed H, D'Santos C, Serandour AA, Ali HR, Brown GD, Atkins A, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–9. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–16. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- 34.Clarke CL, Graham JD. Non-overlapping progesterone receptor cistromes contribute to cell-specific transcriptional outcomes. PLoS One. 2012;7:e35859. doi: 10.1371/journal.pone.0035859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grewal SS. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochim Biophys Acta. 2015;1849:898–907. doi: 10.1016/j.bbagrm.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 36.White RJ. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008;24:622–9. doi: 10.1016/j.tig.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 37.O'Malley BW, Aronow A, Peacock AC, Dingman CW. Estrogen-dependent increase in transfer RNA during differentiation of the chick oviduct. Science. 1968;162:567–8. doi: 10.1126/science.162.3853.567. [DOI] [PubMed] [Google Scholar]
- 38.Chin RC, Kidson C. Selective associations of hormonal steroids with aminoacyl transfer RNAs and control of protein synthesis. Proc Natl Acad Sci U S A. 1971;68:2448–52. doi: 10.1073/pnas.68.10.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 2009;37:D93–7. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavon-Eternod M, Gomes S, Rosner MR, Pan T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA. 2013;19:461–6. doi: 10.1261/rna.037507.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–92. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Truitt ML, Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2016;16:288–304. doi: 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, et al. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24:171–82. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fettig LM, McGinn O, Finlay-Schultz J, LaBarbera DV, Nordeen SK, Sartorius CA. Crosstalk between progesterone receptors and retinoic acid receptors in regulation of cytokeratin 5 positive breast cancer cells. Oncogene. 2017 doi: 10.1038/onc.2017.204. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knutson TP, Lange CA. Tracking progesterone receptor-mediated actions in breast cancer. Pharmacol Ther. 2014;142:114–25. doi: 10.1016/j.pharmthera.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui X, Schiff R, Arpino G, Osborne CK, Lee AV. Biology of progesterone receptor loss in breast cancer and its implications for endocrine therapy. J Clin Oncol. 2005;23:7721–35. doi: 10.1200/JCO.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 47.Peters AA, Buchanan G, Ricciardelli C, Bianco-Miotto T, Centenera MM, Harris JM, et al. Androgen receptor inhibits estrogen receptor-alpha activity and is prognostic in breast cancer. Cancer Res. 2009;69:6131–40. doi: 10.1158/0008-5472.CAN-09-0452. [DOI] [PubMed] [Google Scholar]
- 48.West DC, Pan D, Tonsing-Carter EY, Hernandez KM, Pierce CF, Styke SC, et al. GR and ER coactivation alters the expression of differentiation genes and associates with improved ER+ breast cancer outcome. Mol Cancer Res. 2016;14:707–19. doi: 10.1158/1541-7786.MCR-15-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D'Amato NC, Gordon MA, Babbs B, Spoelstra NS, Carson Butterfield KT, Torkko KC, et al. Cooperative dynamics of AR and ER activity in breast cancer. Mol Cancer Res. 2016;14:1054–67. doi: 10.1158/1541-7786.MCR-16-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sikora MJ. Family matters: collaboration and conflict among the steroid receptors raises a need for grouptherapy. Endocrinology. 2016;157:4553–60. doi: 10.1210/en.2016-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.