Abstract
Recently, the combination of androgen deprivation therapy (ADT) and radiation therapy (RT) has been shown to block the androgen receptor (AR)-driven DNA damage response (DDR) and enhance RT-mediated cell kill of prostate cancer (PCa). Since ADT may induce expression of AR splice variants (ARVs) we hypothesized that ARVs can drive DDR and mediate resistance to combined ADT and RT. Herein, we demonstrate that ARVs can increase the clonogenic survival of PCa cells following RT in an ADT-independent manner. RT induces the interaction between ARVs and a DDR driver, the DNA-dependent protein kinase catalytic subunit (DNA-PKc). Pharmacological inhibition of DNA-PKc blocks its interaction with ARVs and results in persistence of DNA damage, increased tumor cell kill and improved PCa cell survival following RT. These results indicate that combinatorial targeting of DNA-PKc with ADT and RT may be an effective strategy for overcoming radioresistance when treating clinically localized PCa.
Keywords: Prostate cancer, radiation resistance, androgen receptor variants, DNA damage, DNA-PKc
Introduction
The combination of radiation therapy (RT) and androgen deprivation therapy (ADT) is superior to RT alone for treatment of patients with clinically localized, intermediate-risk (PSA 10–20ng/ml, cT2 and Gleason sum = 7) and high-risk (PSA ≥20ng/ml, ≥cT3 and Gleason sum ≥8) prostate cancer (PCa) (1,2). Clinically, patients receive 2–3 months of initial ADT, followed by combined RT and ADT. High-risk patients may then receive an additional 2–3 years of ADT.
Biologically, ADT abrogates the ability of the androgen receptor (AR) to promote the cellular DNA damage repair (DDR) (3,4). When AR signaling is blocked by ADT, PCa cells cannot efficiently activate DDR. Thus, the combination of ADT and RT enhances DNA damage and lethality of RT (3–5).
Despite use of combined ADT and RT, biochemical disease recurrence is common (up to 50% of high-risk PCa patients)(6). Recurrence is associated with a higher likelihood of metastatic progression and death from PCa (7). While recurrence may be a function of advanced disease at the time of RT, the finding of viable radioresistant PCa in salvage prostatectomy specimens suggests that some PCas may be intrinsically resistant to ADT + RT (8). These data suggest that an understanding of molecular mechanisms of radioresistance may enable more effective RT therapy for localized PCa.
Emerging evidence supports a role for AR variants (ARVs) in the development of resistance of PCa to ADT and second-generation AR-targeting therapies (9–11). ARVs lack the AR ligand-binding domain (LBD) and are not responsive to drugs targeting AR-LBD. ARVs may be formed by AR splicing (plastic) that can be induced by ADT (10), and/or AR gene rearrangements (permanent) (11). ARV expression serves as a predictive biomarker for response to enzalutamide or abiraterone acetate (12). ARVs are nuclear, constitutively active, bind to similar androgen response elements (AREs), as that engaged by full length AR (ARfl) (13), and interact with similar coregulators as ARfl (14–16). Thus, ARVs may substitute for ARfl, when the ARfl is effectively blocked by ADT (13).
We reasoned that the regimen of 2–3 months of initial ADT prior to RT was sufficient to induce ARVs in PCa prior to RT (10,17). Evaluation of the primary prostate tissues from the CALGB 90203 study indicates that neoadjuvant ADT induces both ARfl and ARV expression (M. Gleave, personal communications). Using short term (48h) ex vivo cultures of primary prostate tissues, we noted that enzalutamide was able to induce significantly more AR transcripts containing the AR DBD (reflecting both ARfl and ARV expression) than AR LBD (ARfl expression only) (Fig. S1A, B). Taken together, these data suggest that ADT may induce ARVs in human PCa tissues. We reasoned that these induced ARVs may drive DDR and represent a potential mechanism of resistance to combined ADT and RT.
Materials and Methods
Cell lines and culture condition
22Rv1, VCaP, C4-2 and PC3 cell lines were obtained from American Type Culture Collection (ATCC). R1-D567 and R1-AD1 have been described (13). Provenance of all cell lines is verified by microsatellite genotyping using the PowerPlex 1.2 microsatellite detection kit (Promega) and the fingerprint library maintained by ATCC. NU7441 was obtained from Selleckchem (Houston, TX). Cells were irradiated using a 137Cs source (Mark 1–68 irradiator; JL Shepherd and Associates, San Fernando, CA) at a dose rate of 3.47 Gy/min.
Small interfering RNA (siRNA)
AR siRNAs and DNA-PKc smart pool were purchased from Dharmacon.
ChIP-seq
Genome-wide binding of RNA pol II (Biolegend, #Cat, 920101) and AR in R1-D567 cells was studied by Chromatin Immunoprecipitation- (ChIP) followed by high-throughput DNA sequencing (ChIP-seq). Following 1% formaldehyde cross-linking for 8m, 1/10th volume of 1.25 M glycine was added for 5m at RT. Chromatin was sheared to 300bp (Bioruptor Pico, Diagenode). ChIP antibodies were tagged to protein G magnetic beads with IgG and input DNA as controls. Protein bound DNA was washed, reverse crosslinked, and eluted using the IPure kit (Diagenode). Purified DNA was analyzed by fluorometry (Qubit 3.0, Invitrogen) and 40% of ChIP-DNA was used for sequencing library using KAPA HyperPrep kit (KAPA Biosystems) and TruSeq adapters (Integrated DNA Technologies). PCR products were selected using Ampure beads (Beckman Coulter). Quality check of purified libraries was carried out using Bioanalyzer 2100 (Agilent) and sequenced on Illumina NextSeq 500 Sequencer with read length of single-end 75bp.
Immunoprecipitation-mass spectrometric (IP-MS)
Following treatment of R1-D567 cells with 2Gy RT, cell lysates were incubated with Dynabeads Protein G (Invitrogen) pre-coupled with AR antibody (Sigma) or an irrelevant IgG control, eluted at 95°C, separated by SDS-PAGE, proteolytically digested overnight with trypsin (Promega) following reduction and alkylation with DTT and iodoacetamide (Sigma–Aldrich). Samples then underwent solid-phase extraction cleanup with Oasis HLB plates (Waters) and were analyzed by LC/MS/MS, using an Orbitrap Fusion Lumos (Thermo Electron) coupled to an Ultimate 3000 RSLC-Nano liquid chromatography systems (Dionex). MS operated in positive ion mode with a source voltage of 2.2 kV and capillary temperature of 275°C. MS scans were acquired at 120,000 resolution and up to 10 MS/MS spectra were obtained in the ion trap for each full spectrum acquired using higher- energy collisional dissociation (HCD) for ions with charge 2–7. Dynamic exclusion was set for 25s. Raw MS data files were converted to a peak list format and analyzed using the central proteomics facilities pipeline (CPFP), version 2.0.3(18). Label-free quantitation of proteins across samples was performed using SINQ normalized spectral index Software (18).
Western blots and antibodies
Western blotting was performed as described previously, using antibodies to AR (Santa Cruz, N-20, #sc-816, or Santa Cruz, 441, #sc-7305), or AR-V7 (Precision Antibody, #AG10008, 1:1000), phosphorylated histone H2AX (γ-H2AX) (Millipore, 05-636; 1:1,000), actin (Sigma-Aldrich, A2066; 1:5,000), pS2056-DNA-PKc (Abcam, ab18192; 1:400), DNA-PKc (Abcam, ab32566; 1:1,000), and horseradish peroxidase-conjugated donkey anti-rabbit and sheep anti-mouse, AlexaFluor 594 anti-mouse and AlexaFluor 488 anti-rabbit antibodies.
Clonogenic survival assay
Cells were treated with enzalutamide for 24h and then treated with increasing doses of radiation (0, 1, 2, and 3 Gy). Colony formation assay were performed as described previously (3). Colony formation assay was also performed using siRNAs. After 72 h, cells were plated for colony formation assay and Western blot analysis.
Double-strand break repair assay
The number of phospho-γH2AX foci (green) and 53BP1 (red) was determined at each time point (average of 200 nuclei), and the percentage of foci remaining was plotted against time to obtain DSB repair kinetics. Data is represented as mean ± SEM.
Comet assay
Cells were subjected to radiation (10Gy, 30 min recovery), harvested and re-suspended in ice cold PBS (without Mg2+ and Ca2+). Amount of damaged DNA was visualized by single-cell agarose gel electrophoresis under alkaline conditions following the manufacture’s protocol. 100ul/well of diluted Vista Green DNA dye were applied onto slides. Comet images were taken by EVOS fluorescence microscopy (Thermo Fisher) using a FITC filter.
Quantitative immunofluorescence of γ-H2AX and 53BP1
Cells grown on coverslips were incubated with rabbit polyclonal anti-H2AX (p-S139) antibody (Cell Signaling Technology) followed by incubation with biotinylated swine anti-rabbit IgG (Dako, Glostrup, Denmark) and streptavidin-conjugated fluorescein-isothiocyanate (Dako). Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPi), analyzed on a Nikon microscope (40x magnification) equipped with a CCD camera and processed using ImageJ software (NIH, Bethesda, MD, USA). Exposure time, binning, microscope settings and light source intensity were kept constant. Nuclei were segmented on the basis of DAPI staining and then signal-integrated density of γ-H2AX and 53BP1 staining quantified using ImageJ software. More than 200 γ-H2AX-positive and 53BP1-positive cells were analyzed per experiment per condition.
Proximity ligation assay (PLA)
Cells grown on coverslips were fixed with methanol at −20°C for 10m followed by a 5m extraction in 0.3% Triton X-100 in PBS. In-situ proximity ligation was performed using a Duolink Detection Kit in combination with anti-mouse PLUS and anti-rabbit MINUS PLA probes, according to manufacturer’s instructions (Sigma-Aldrich Duolink). Nuclear foci were imaged using a Nikon E600 Eclipse microscope equipped with a 40× lens. The number of nuclear foci/cell was quantified using ImageJ. More than 50 cells were analyzed per experiment per condition.
Quantitative reverse transcription-PCR
Total RNA was isolated using RNeasy mini kit according to manufacturer’s instructions (Qiagen, Valencia, CA) and reverse-transcribed using the cDNA synthesis kit (Bio-Rad, Hercules, CA). Quantitative PCR used iQ SYBR Green Supermix (Bio-Rad) on an iCycler thermal cycler (Bio-Rad) with a denaturation step at 95°C for 3m followed by 40 cycles of amplification at 95°C for 30s, 60°C for 30s, and 72°C for 60s. Primer sequences are in Figure S1C. Fold changes in mRNA expression levels were calculated using the comparative CT method.
Tumor growth
Male athymic nude mice (nu/nu, 5–6 weeks old) were injected (1 × 106 cells in 100μL 50% Matrigel,) subcutaneously into the right posterior flanks. Mice were castrated when tumor size reached 200mm3. Treatment groups (5 animals per group) included untreated control (treated with 0.9% saline), those treated with radiation (2 Gy/d, 2d), or combined treatment with NU7441 and IR. NU7441 was administered i.p. 2 h before focal IR. All experiments were conducted under Institutional Animal Care and Use Committee of UTSW approved guidelines for animal welfare.
Data analysis
Numerical data were analyzed using parametric and nonparametric statistical tests, namely student’s t test and split-plot (mixed-design) ANOVA test. Significance thresholds are indicated in figure legends.
Results
ARV can mediate DDR following RT
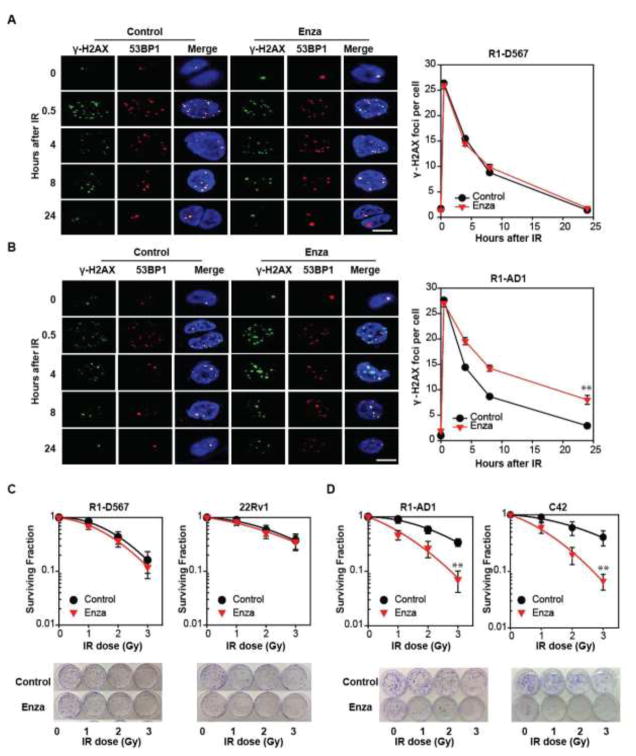
To evaluate the ability of either ARfl or ARVs alone to mediate DDR, we used transcription activator-like effector nuclease genetically-engineered R1-AD1 and R1-D567 cell lines derived from CWR-R1 PCa cells (13,19) (Fig. S2A). Whereas R1-AD1 cells expresses only ARfl, R1-D567 cells only express the constitutively active ARV, ARv567es (due to genomic deletion of AR exons 5–7) (19). Following ionizing radiation (IR), we noted induction of DSBs in both cell lines, as evidenced by foci of γ-H2AX and 53BP1. Similar DNA repair kinetics were noted in both R1-AD1 and R1-D567 cells, with complete resolution of IR-induced γ-H2AX and 53BP1 foci within 24h (Fig. 1A). The combination of enzalutamide and IR delayed and decreased DNA repair in R1-AD1 cells (30%–40% γ-H2AX foci at 24h) (Fig. 1B) but not in R1-D567 cells (<5% γ-H2AX foci at 24h) (Fig. 1A). Depletion of ARfl or ARv567es, using siRNA (Fig. S2B), altered DNA repair kinetics in R1-AD1 and R1-D567 cells, respectively (Fig. S2 C). Our use of enzalutamide rather than ADT in PCa cells is both to achieve a more complete AR blockade and to reflect the evolving clinical paradigm with earlier use of second-generation anti-androgens. Overall, these data suggest that a cell line expressing ARVs alone can mediate DDR and that ARV-mediated DDR is not affected by enzalutamide.
Fig. 1.
ARV mediates DDR and clonogenic survival. (A) R1-D567 and (B) R1-AD1 cells in the presence of vehicle (DMSO) or Enza (5μM), were subjected to IR (2Gy), fixed and stained with γ-H2AX (green) and 53BP1 (red) antibodies. Foci imaged by confocal microscopy were plotted as average foci per cell. (C) R1-D567, R1-AD1, C4-2 and 22Rv1 cells were cultured with vehicle or Enza for 24 hours, then treated with escalating doses of IR. Following fixation and staining 14 days later, colonies greater than 50 cells were counted. **p<0.01.
Enzalutamide does not influence clonogenic survival in PCa cells that express ARVs
Following IR, clonogenic survival of R1-D567 cells, was not affected by enzalutamide (Fig. 1C). Importantly, clonogenic survival of 22Rv1 cells, which express both ARfl and ARv7 (Fig. S2A), was not affected neither by enzalutamide (Fig. 1C) nor by CRISPR-mediated knockdown of ARfl or ARv7 (Fig. S2D, S2E). In contrast, the combination of IR and enzalutamide decreased clonogenic survival of both R1-AD1 and C4-2 cells (which express only ARfl), in comparison to IR alone (Fig. 1D). These data validate prior findings of a synergism between ADT and RT in mediating DNA repair in PCa cells expressing ARfl and suggest that PCa cells that express ARVs either alone or in combination with ARfl do not exhibit this same synergism.
When R1-D567 subcutaneous xenografts were subject to short-course RT, their rate of growth slowed down initially but rebounded quickly (Fig. S3A). A similar profile of response was noted with R1-AD1 subcutaneous xenografts (Fig. S3B). We observe that R1-D567 xenografts grow faster than matched R1-AD1 xenografts; this difference become even more striking upon IR treatment. This suggests that ARV’s may confer enhanced radioresistance as compared to ARfl. Using siRNA, we noted that decreased expression of either ARfl or ARv7 had minimal impact on clonogenic survival following IR in 22Rv1 cells (Fig. 2A). Knockdown of both ARfl and ARv7 was needed to enhance the effect of IR in clonogenic survival studies in 22Rv1 cells (Fig. 2A). These data indicate that inhibition of both forms of AR are needed to effectively increase RT-mediated cell kill in 22Rv1 cells.
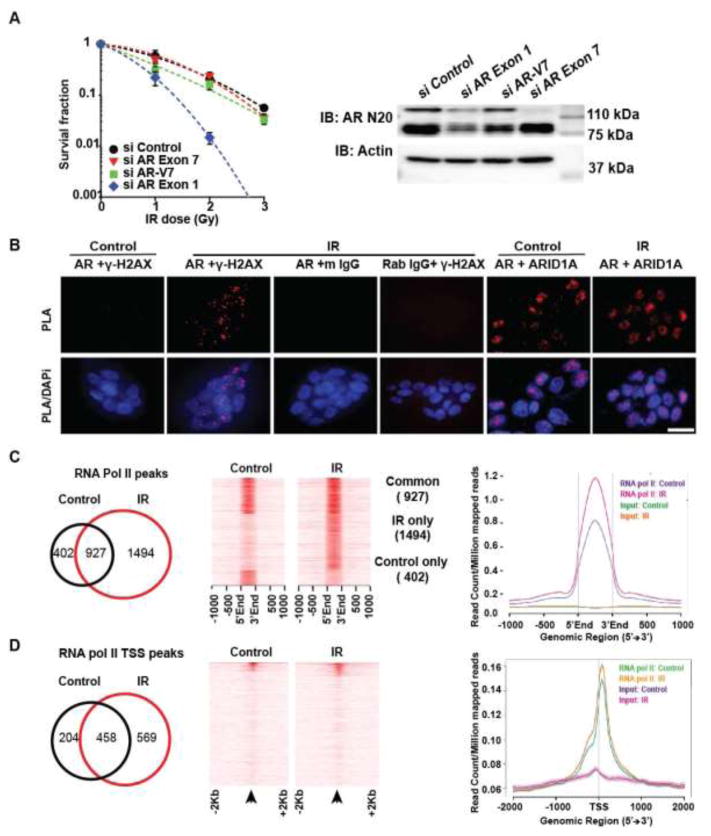
Fig. 2.
ARVs are necessary for DDR and are recruited to DNA damage sites: (A) Clonogenic survival of 22Rv1 following knockdown of ARfl alone, ARv7 alone or both ARfl and ARv7 demonstrates that either ARVs or ARfl alone can mediate DDR. Western blot shows ARfl and ARV protein expression in 22Rv1 cells following siRNA knockdown (right), with β-actin control. (B) PLA shows IR-induced interaction between ARV and γ-H2AX in R1-D567 cells. (controls: AR and mIgG, AR and ARID1A and γ-H2AX and rIgG). All irradiated cells (2 Gy) contained >5 dots each. (C) Venn diagram (left) and heatmap (middle) and gene body plots (right) shows the increase in RNA polymerase II-enriched ChIP-seq peaks in R1-D567 upon IR treatment. (D) Venn diagram (left) and heatmap (middle) and gene body plots (right) shows the increase in RNA polymerase II transcription start site (TSS) in R1-D567 upon IR treatment.
ARV localizes to IR-induced DNA damage
Proximity ligation assays (PLA) indicated that ARv567es was recruited within minutes of IR to sites of DNA damage, as evidenced by its interaction with γ-H2Ax foci in R1-D567 cells (Fig. 2B). Every irradiated (2Gy) R1-D567 cells displayed >5 foci/cell. Double immunofluorescence staining indicated overlap between most γ-H2AX foci and ARv567es staining within the nucleus of R1-D567 cells (Fig. S4A). No change was noted in AR interaction with a known coregulator ARID1A following IR (Fig. 2B). Similarly, PLA studies in 22Rv1 cells, indicate that both ARv7 and ARfl were recruited to DNA-damage sites (γ-H2AX) upon IR (Fig. S4B). These data suggest that ARfl and ARV can be recruited to sites of DNA damage.
IR induces global gene expression in R1-D567 cells
Evaluation of global RNA-polymerase II (Pol II) binding via ChIP-Seq analyses indicated that IR dramatically increased Pol II promoter occupancy in R1-D567 cells (Fig. 2C, D). Quantitative RT-PCR validated RT-induced expression of genes involved in homologous recombination (HR) and non-homologous end joining (NHEJ) pathways (Fig. S4C). Pathway analysis of genes differentially occupied by Pol II upon IR treatment indicated up-regulation of steroid hormone receptor pathways involving AR, glucocorticoid (GR), estrogen and aldosterone (Fig. S4C). Interestingly, IR treatment did not increase the levels of AR at 16 hours. Our integrative analyses suggested that IR treatment up-regulates androgen signaling at a pathway level rather than a gene level, as no single component of the pathway was drastically up-regulated (Fig. S5B). Rather, moderate up-regulation of a subset of individual components contributed to increase in the pathway flux (Fig. S5C). These data also explain why multiple steroid hormone receptor pathways (e.g. AR and GR) could operate temporally during longitudinal progression of PCa in response to enzalutamide.
We were intrigued by our observation that AR in R1-D567 cells bound to <100 sites in the genome. A majority of AR bound sites in R1-D567 cells were associated with ARE motifs, indicating true AR binding sites. However, evaluation of AR ChIP-seq indicated a decrease in ARv567es binding upon IR treatment in R1-D567 cells (Fig. S5A). These data indicate, while IR globally induces transcription in R1-D567 cells, that the ARVs are unlikely to be the primary drivers of this transcriptional programme.
IR enhances the interaction between ARV and DNA-PKc
Since ARV localized to the sites of DNA damage, we hypothesized that evaluation of the ARV interactome could elucidate its mechanisms of action. Using unbiased IP-MS analysis, we identified DNA-PKc as the top-binder of ARv567es in R1-D567 cells regardless of IR treatment (Fig. 3A). These data were supported by co-IP studies demonstrating that IR enhanced interaction between endogenous ARfl or ARv567es and DNA-PKc in R1-AD1 and R1-D567 cells respectively (Fig. 3B). PLA also indicated increased interaction between DNA-PKc and ARfl or ARVs upon IR in PCa cells (Fig. 3C). Mapping studies indicated differential IR-induced interactions between ARfl and ARVs with DNA-PKc: the WHTLF domain appears to be critical for ARV-DNA-PKc interaction, while the AR-LBD may also play a role in ARfl interaction with DNA-PKC (Fig. S6A, B, and C). These data are supported by the ARΔ1 mutant to interact with DNA-PKc and prior mapping studies indicating AR-LBD and DNA-PKc interaction (20). Importantly, enzalutamide abolished IR induced interaction between ARfl and DNA-PKc but not ARVs and DNA-PKc interaction (Fig. 3C). Further, IR-mediated induction of ARv567es interaction with DNA-PKc was blocked by a selective DNA-PKc inhibitor, NU7441 (Fig. 3D). In contrast, the interaction between ARv567es and ARID1A was not influenced by IR or NU7441 (Fig. 3D). Taken together, these data indicate that IR enhances the interaction between DNA-PKc and ARfl or ARV.
Fig. 3.
IR increases the interaction between ARV and DNA-PKc. (A) IP-MS analysis of proteins interacting with ARv567es in R1-D567 cells shows that PRKDC is the top binder to ARVs: a list of the top AR binders is tabulated. Ability of IR (10 Gy) to enhance interaction between ARV or ARfl and DNA-PKc is demonstrated by Co-IP (B) and PLA (C) at 16 hours after IR. The interaction between ARV and DNA-PKc in R1-D567 cells can be blocked by pretreatment with NU7441 (D).
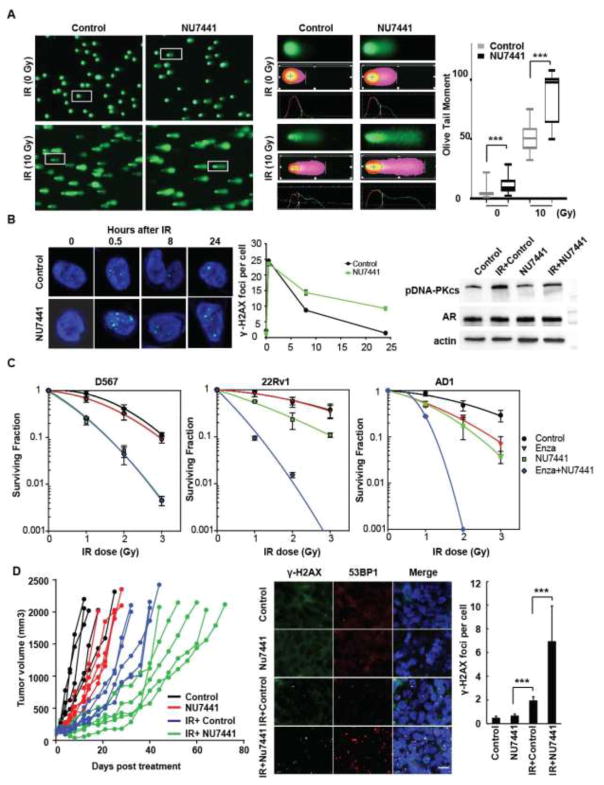
DNA-PKc inhibition enhances IR-induced DNA damage in PCa cells
Comet assays indicated that pretreatment with NU7441 significantly increased DNA fragmentation following IR in R1-D567 cells (Fig. 4A). These data were supported by persistence of residual DSBs after IR in NU7441-treated R1-D567 cells, as evaluated by γ-H2AX and 53BP1 staining (Fig. 4B, shown for γ-H2AX). Mechanistic evaluation indicated that DNA-PKc inhibition with either chemical inhibition (Fig. S7A, B, and C) or siRNA mediated knockdown blocked NHEJ repair (Fig. S7D, E). Importantly, NU7441 enhanced the effect of IR and enzalutamide on PCa cell survival, as shown by clonogenic survival assay in R1-D567, 22Rv1 and R1-AD1 cells (Fig. 4C). The combination of IR, NU7441 and enzalutamide was more potent than IR and enzalutamide or IR and NU7441 in R1-AD1 and 22Rv1 cells. In R1-D567 cells, NU7441 was not additive with enzalutamide but potently decreased survival after IR (Fig. S8A). Thus, DNA-PKc inhibition was associated with increased IR-induced DNA damage and decreased cell survival in PCa cells regardless whether they express ARfl alone (Fig. S8B), ARV alone (Fig. S8C), or both ARfl and ARVs (Fig. S8D). These in vitro results suggest that DNA-PKc inhibition can enhance the effect of IR on PCa cells, irrespective of ARfl and ARV expression status.
Fig. 4.
DNA-PKc inhibitors block DDR following IR and enhance IR-mediated PCa cell death: Pretreatment with NU7441 enhances the IR-induced olive tail moment in comet assays (A) and alters kinetics of DNA repair, as evidenced by persistence of γ-H2Ax foci at 24h in R1-D567 cells (p<0.0001) and decreased IR induced phospho-DNA-PKc at 1 h after IR (B). (C) Clonogenic survival assays indicate that NU7441 pretreatment dramatically decreases the survival of R1-AD1, R1-D567 and 22Rv1 cells (p≤ 0.02). (D) Pretreatment of R1-D567 xenografts (200 mm3) with NU7441 prior to a single IR dose (2Gy) decreased tumor growth more than IR alone (each xenograft is represented by a separate line, with each measurement of tumor volume represented by a dot. Groups of mice receiving the same therapy are grouped by color. Tumor growth rates after NU7441, IR or both all significantly differed from controls when assessed by split-plot ANOVA (p = 0.009, 0.003 and <0.001, respectively). Pretreated R1-D567 xenografts shows persistence of γ-H2AX and 53BP1 foci at 24h after IR. (Scale bar- 40 μm; blue- DAPI; green- γ-H2AX; red- 53BP1)(p < 0.001; n = 3).
To validate these findings in vivo, we established subcutaneous R1-D567 xenografts in Nu/Nu mice. When tumors reached 200mm3, mice were castrated and then were treated with 2 daily doses of NU7441 or vehicle, followed by 2Gy IR. The combination of IR and NU7441 was more effective in slowing the R1-D567 tumor growth than IR alone (Fig. 4D). While DNA DSBs were almost completely repaired within 24h in the vehicle-treated irradiated tumors, NU7441–treated irradiated tumors still had significant foci of DDR, suggesting that DNA damage was still present (Fig. 4D). These data support our hypothesis that inhibition of DNA-PKc can effectively synergize with IR in ARV-expressing PCa cells. Overall, our data support a role for combined IR and AR and DNA-PKc inhibition to optimize outcomes of patients undergoing RT (Fig. S8 B, C, and D).
Discussion
In PCa cells that express AR, RT induced DDR can be driven by ARfl-mediated signaling. Combined ADT and RT decrease DDR and increase RT-mediated cell kill. In this study, we demonstrate using genetically engineered PCa cells, that ARVs mediate DDR in response to IR. We have shown that in PCa cells that express ARVs, the combination of RT and ADT is not more effective than RT in blocking DDR. These data imply that the expression of ARVs in PCa cells may lead to resistance to combined RT and ADT. Since ADT can rapidly induce ARVs in PCa cells, our study identifies a potential molecular mechanism of resistance to the current paradigm of upfront ADT followed by RT and a therapeutic strategy to abrogate this.
Importantly, we show that while IR globally induces transcription in R1-D567 cells, that the ARVs are unlikely to be the primary drivers of this transcriptional program. Our data suggests that the observed effects of ARVs in terms of clonogenic cell survival and radiation resistance are likely due to a direct physical interaction with DNA-PKc. In contrast, we have evidence indicating that ARfl influences DNA repair both directly through its interaction with DNA-PKc and indirectly through its control of the expression of DNA repair genes. Thus, in PCa cells expressing both ARfl and ARV (e.g. 22Rv1), Enzalutamide will inhibit the transcriptional regulation and perhaps DNA repair by ARfl, but not the DNA repair functions mediated by ARVs. Our data suggest that transcriptional regulation by AR may be less important to enzalutamide resistance in metastatic CRPC, with this increased DNA repair function being more critical. We speculate that the role of AR amplification in CRPC is actually to promote DNA repair as well as regulate transcription.
Overall, our data indicate that ARVs and ARfl interact with DNA-PKc and that IR enhances this interaction. We show that DNA-PKc is critical for these DNA repair mechanisms following IR and that the combined blockade of AR and DNA-PKc enhances IR-mediated cell kill, regardless of whether the PCa cell expresses ARfl alone, ARVs alone or ARfl and ARVs together. DNA-PKc inhibitors block AR-DNA-PK interaction, disrupt DNA repair and enhance RT-mediated cell kill. These data establish the importance of DNA-PKc in IR-mediated DDR and establish the rationale for a potential clinical trial combining drugs targeting AR and DNA-PKc in combination with RT for patients with clinically localized PCa. While the optimal multimodal strategy incorporating antiandrogens, DNA-PKc blockade and RT needs to be clinically validated, our findings would suggest the incorporation of the DNA-PKc antagonist immediately prior to radiotherapy, to enhance cell kill.
Acknowledgments
Financial Support: This work was supported by the following grants: W81XWH-15-1-0128 (Y. Yin), W81XWH-14-1-0556 (Y. Yin), W81XWH-13-2-0093 (G. V. Raj), and W81XWH-12-1-0288 (G. V. Raj) from the Department of Defense, 1R01CA200787-01(G. V. Raj), R01CA174777 (S.M. Dehm), RO1CA149461, RO1CA197796 and R21CA202403 (S. Burma), R01CA166677 (B. P. Chen), UL1TR001105 (C. Xing), and R00CA160640 (R. S. Mani) from the NIH, NNX16AD78G from the National Aeronautics and Space Administration (S. Burma) and the Mimi and John Cole Prostate Cancer Fund (G. V. Raj).
We thank K. Luby-Phelps for confocal imaging.
Footnotes
Conflict of interest: The authors declare that they have no competing interests.
References
- 1.Bolla M, Gonzalez D, Warde P, Dubois JB, Mirimanoff RO, Storme G, et al. Improved survival in patients with locally advanced prostate cancer treated with radiotherapy and goserelin. N Engl J Med. 1997;337:295–300. doi: 10.1056/NEJM199707313370502. [DOI] [PubMed] [Google Scholar]
- 2.Jones CU, Hunt D, McGowan DG, Amin MB, Chetner MP, Bruner DW, et al. Radiotherapy and short-term androgen deprivation for localized prostate cancer. N Engl J Med. 2011;365:107–18. doi: 10.1056/NEJMoa1012348. [DOI] [PubMed] [Google Scholar]
- 3.Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, et al. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer discovery. 2013;3:1254–71. doi: 10.1158/2159-8290.CD-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polkinghorn WR, Parker JS, Lee MX, Kass EM, Spratt DE, Iaquinta PJ, et al. Androgen receptor signaling regulates DNA repair in prostate cancers. Cancer discovery. 2013;3:1245–53. doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spratt DE, Evans MJ, Davis BJ, Doran MG, Lee MX, Shah N, et al. Androgen Receptor Upregulation Mediates Radioresistance after Ionizing Radiation. Cancer Res. 2015;75:4688–96. doi: 10.1158/0008-5472.CAN-15-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bolla M, Van Tienhoven G, Warde P, Dubois JB, Mirimanoff RO, Storme G, et al. External irradiation with or without long-term androgen suppression for prostate cancer with high metastatic risk: 10-year results of an EORTC randomised study. Lancet Oncol. 2010;11:1066–73. doi: 10.1016/S1470-2045(10)70223-0. [DOI] [PubMed] [Google Scholar]
- 7.Jones JS. Radiorecurrent prostate cancer: an emerging and largely mismanaged epidemic. Eur Urol. 2011;60:411–2. doi: 10.1016/j.eururo.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Stephenson AJ, Eastham JA. Role of salvage radical prostatectomy for recurrent prostate cancer after radiation therapy. J Clin Oncol. 2005;23:8198–203. doi: 10.1200/JCO.2005.03.1468. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Van der Steen T, Tindall DJ. Are androgen receptor variants a substitute for the full-length receptor? Nat Rev Urol. 2015;12:137–44. doi: 10.1038/nrurol.2015.13. [DOI] [PubMed] [Google Scholar]
- 10.Yu Z, Chen S, Sowalsky AG, Voznesensky OS, Mostaghel EA, Nelson PS, et al. Rapid induction of androgen receptor splice variants by androgen deprivation in prostate cancer. Clin Cancer Res. 2014;20:1590–600. doi: 10.1158/1078-0432.CCR-13-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Henzler C, Li Y, Yang R, McBride T, Ho Y, Sprenger C, et al. Truncation and constitutive activation of the androgen receptor by diverse genomic rearrangements in prostate cancer. Nature communications. 2016;7:13668. doi: 10.1038/ncomms13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan SC, Selth LA, Li Y, Nyquist MD, Miao L, Bradner JE, et al. Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peacock SO, Fahrenholtz CD, Burnstein KL. Vav3 enhances androgen receptor splice variant activity and is critical for castration-resistant prostate cancer growth and survival. Mol Endocrinol. 2012;26:1967–79. doi: 10.1210/me.2012-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He B, Lanz RB, Fiskus W, Geng C, Yi P, Hartig SM, et al. GATA2 facilitates steroid receptor coactivator recruitment to the androgen receptor complex. Proc Natl Acad Sci U S A. 2014;111:18261–6. doi: 10.1073/pnas.1421415111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G, Sprenger C, Wu PJ, Sun S, Uo T, Haugk K, et al. MED1 mediates androgen receptor splice variant induced gene expression in the absence of ligand. Oncotarget. 2015;6:288–304. doi: 10.18632/oncotarget.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16–22. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trudgian DC, Mirzaei H. Cloud CPFP: a shotgun proteomics data analysis pipeline using cloud and high performance computing. J Proteome Res. 2012;11:6282–90. doi: 10.1021/pr300694b. [DOI] [PubMed] [Google Scholar]
- 19.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KA, et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci U S A. 2013;110:17492–7. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodwin JF, Kothari V, Drake JM, Zhao S, Dylgjeri E, Dean JL, et al. DNA-PKcs-Mediated Transcriptional Regulation Drives Prostate Cancer Progression and Metastasis. Cancer cell. 2015;28:97–113. doi: 10.1016/j.ccell.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]