Abstract
The teosinte branched1/cycloidea/proliferating cell factor (TCP) gene family is a plant-specific transcription factor that participates in the control of plant development by regulating cell proliferation. However, no report is currently available about this gene family in turnips (Brassica rapa ssp. rapa). In this study, a genome-wide analysis of TCP genes was performed in turnips. Thirty-nine TCP genes in turnip genome were identified and distributed on 10 chromosomes. Phylogenetic analysis clearly showed that the family was classified as two clades: class I and class II. Gene structure and conserved motif analysis showed that the same clade genes have similar gene structures and conserved motifs. The expression profiles of 39 TCP genes were determined through quantitative real-time PCR. Most CIN-type BrrTCP genes were highly expressed in leaf. The members of CYC/TB1 subclade are highly expressed in flower bud and weakly expressed in root. By contrast, class I clade showed more widespread but less tissue-specific expression patterns. Yeast two-hybrid data show that BrrTCP proteins preferentially formed heterodimers. The function of BrrTCP2 was confirmed through ectopic expression of BrrTCP2 in wild-type and loss-of-function ortholog mutant of Arabidopsis. Overexpression of BrrTCP2 in wild-type Arabidopsis resulted in the diminished leaf size. Overexpression of BrrTCP2 in triple mutants of tcp2/4/10 restored the leaf phenotype of tcp2/4/10 to the phenotype of wild type. The comprehensive analysis of turnip TCP gene family provided the foundation to further study the roles of TCP genes in turnips.
Keywords: TCP, transcription factors, expression analysis, turnip, cell proliferation
Introduction
Teosinte branched1/cycloidea/proliferating cell factor (TCP) gene family is a plant-specific transcription factor that regulates plant growth by controlling cell proliferation. TCP gene family has been identified in many plant species. For instance, Arabidopsis has 24 TCP genes, Oryza sativa has 28 TCP genes, tomato has 30 TCP genes, Populus euphratica has 33 TCP genes, Populus trichocarpa has 36 TCP genes, Citrullus lanatus has 27 TCP genes, and Prunus mume has 19 TCP genes (Martin-Trillo and Cubas, 2010; Parapunova et al., 2014; Ma X. et al., 2016; Shi et al., 2016; Zhou et al., 2016). The TCP domain contains a 59-amino-acid basic helix–loop–helix (bHLH) motif involved in DNA binding and protein–protein interaction (Martin-Trillo and Cubas, 2010). On the basis of the TCP domains, the members of the TCP family can be grouped into two subfamilies: class I (PCF or TCP-P class) and class II (TCP-C class) (Kosugi and Ohashi, 2002; Navaud et al., 2007; Martin-Trillo and Cubas, 2010). The difference between the two is a four-amino-acid deletion in the TCP domain in class I compared with class II.
Class I TCP genes are assumed to stimulate cell proliferation and leaf development, based mainly on the expression of rice PCF1/PCF2 and AtTCP20 in meristematic tissuses (Kosugi and Ohashi, 1997; Li et al., 2005). In Arabidopsis, loss-of-function tcp15 mutant did not show any significant differences in comparison with wild-type plants. TCP15 fusion with SRDX repression domain elucidated that TCP15 regulated plant development via auxin response (Uberti-Manassero et al., 2012). tcp14 tcp15 double mutants displayed shortened internode length, altered leaf shape, and severe reduction in seed germination capability compared with wild type (Kieffer et al., 2011; Resentini et al., 2015). Moreover, AtTCP9 acts repeatedly with AtTCP20 in regulating leaf senescence via the jasmonate signaling pathway (Danisman et al., 2012). However, pentuple mutant tcp8 tcp15 tcp21 tcp22 tcp23 exhibited upregulated expression levels of SHOOT-MERISTEMLESS, BP, and CYCA1:1 and resulted in large leaf blades (Aguilar Martinez and Sinha, 2013).
Class II can be further divided into subclades: CIN and CYC/TB1 (Martin-Trillo and Cubas, 2010). Class II usually prevented cell proliferation and differentiation during the development of leaf blades. In Arabidopsis, CIN-type genes TCP2, TCP3, TCP4, TCP10, and TCP24 are targets of miR319a. jaw-D (overexpression of miR319a) plants resulted in large and crinkled leaves (Palatnik et al., 2003). Single loss-of-function miR319a-targeted TCPs had slight developmental phenotypes. Double mutants (tcp2 tcp4) and triple mutants (tcp2 tcp4 tcp10) showed less increase in leaf size with some crinkled signs than jaw-D plants. miR319a-targeted TCP transcription factors negatively regulated leaf growth and positively regulated leaf senescence via mediating LOX2 gene expression (Schommer et al., 2008). miR319a-targeted TCP4 is required for proper petal growth and development (Nag et al., 2009). miR319a-targeted TCPs interact with ASYMMETRIC LEAVES2 and ensure normal leaf development by repressing the expression of BP and KNAT2 by binding their promoter (Li Z. et al., 2012).
Turnip (Brassica rapa ssp. rapa) is one of the most economically important vegetable crops in the Tibet Plateau. However, no report on the turnip (Brassica rapa ssp. rapa) TCP family exists. The turnip genome has been sequenced and assembled (Lin et al., 2014), providing the basis for determining the turnip family. In this study, genome-wide identification of TCP transcription factors in turnips is performed. Thirty-nine BrrTCP genes were identified in the turnip genome, and their phylogenetic relationship, gene structure, protein motifs, chromosome location, transcript levels in different tissue, and forms of homo- and heterodimer interaction were analyzed. Furthermore, a CIN-type gene, BrrTCP2, was functionally characterized in transgenic wild-type and loss-of-function mutant Arabidopsis. Our findings indicate that the BrrTCP2 plays a vital function in leaf development by modulating cell division.
Materials and methods
Identification of the TCP genes in turnips
The genome sequence of turnips was downloaded from www.bioinformatics.nl/brassica/turnip. To find all TCP genes in turnips, NCBI BLASTn searches against a local database built using nucleic acid sequences were performed using sequences from all 24 known TCPs from Arabidopsis. Subsequently, the Pfam database was used to determine if each candidate TCP sequence was a member of the TCP gene family. To exclude overlapping genes, all candidate TCP genes were aligned using DNAMAN 4.0 (Lynnon Biosoft) and checked manually. All nonoverlapping TCP genes were used for further analysis.
Analysis of conserved motifs
Conserved motifs of BrrTCP proteins were analyzed using MEME (http://meme-suite.org/tools/meme) with the following parameters: (1) the optimum motif width was set from 6 to 200, and (2) the maximum number of motifs was set to identify 20 motifs.
Gene structure, genomic distribution, and divergence time estimation of BrrTCP genes
BrrTCP genes were mapped on chromosomes by confirming their detailed chromosomal positions supplied by the Turnip Genome Database. To illustrate the structure of introns and exons of BrrTCP genes, full-length genome and coding sequences of BrrTCP genes were subjected to online GSDS analysis (http://gsds.cbi.pku.edu.cn/). To determine their physical location, the starting positions of all BrrTCP genes on each chromosome were confirmed based on a local database of the complete sequence of the turnip genome through BLASTn searching. The segmental and tandem duplication regions were obtained from MCscanX. For synteny analysis, synteny block of the turnip gene was visualized using Circos (http://circos.ca/). Synonymous (Ks) and nonsynonymous (Ka) substitution rates were estimated by the codeml program of PAML4 (Yang, 2007). The divergence time (T) of turnip gene pairs was calculated using the formula T = Ks/2λ, where λ represents the divergence rate of 1.5 × 10−8 for Arabidopsis (Gaut et al., 1996).
Plant material and growth conditions
Brassica rapa ssp. rapa “KTRG-B48a” from Xianggelila City, Yunnan Province, China, was used. For root collection, seedlings were grown on a Whatman filter paper and watered with 1/2 MS medium for 2 weeks. For other tissues, plants were grown in the greenhouse (22°C) under long-day conditions (16 h light/day).
Quantitative RT-PCR
Total RNA was extracted using Eastep™ Universal RNA Extraction Kit (Promega, Shanghai, China) from roots of 1-week-old plants, leaves and stems of 8-week-old plants, and floral buds of 10-week-old plants. RNA quality and concentration were assessed using electrophoresis and ND-1000 Spectrophotometer (NanoDrop Technologies, Delaware, USA). Two micrograms of total RNA were reverse transcribed using GoScript™ Reverse Transcription System (Promega). Quantitative RT-PCR (qRT-PCR) was performed with ABI7500 Real-Time PCR System using TransStart® Top Green qPCR SuperMix (TransGen, Beijing, China). BrrACT2 was used as reference gene. The primers are listed in Table S1. Triplet biological replicates were analyzed.
Yeast two-hybrid assays
All BrrTCP ORFs were amplified from seedling cDNA of KTRG-B48a into pENTR vector used primers (Table S2). The entry vectors were subcloned into the pGADT7 prey vector (pDEST-GSDT7) and pGBKT7 bait vector (pDEST-GBKT7) using gateway method according to Bai et al. (2016). The prey vector was transformed into yeast strain Y187, and all bait vectors were transformed into yeast strain Y2H gold and selected on SD plates lacking Leu and Trp. After selection, the yeasts harboring prey and bait plasmids were spotted onto control medium (SD plates lacking Leu and Trp) and test medium (SD plates lacking Leu, Trp, and His). Yeast growth was observed daily in a growth chamber at 28°C for 2–5 days.
Laser confocal microscopy
The entry vectors of BrrTCP were subcloned into pRI101-GFP using gateway method. 35S:GFP-TCPs were introduced into Agrobacterium GV3101. Positive transformants were selected on LB agar medium (50 mg/L kanamycin, 50 mg/L gentamicin, and 50 mg/L rifampicin). After 3 days, a single colony was inoculated into 2 mL LB liquid medium. Target inserts were confirmed through PCR. Nicotiana benthamiana plants were grown at 22°C under 16 h light/8 h dark conditions. One-month-old plants were used for infiltration. Two days before infiltration, 2 mL cultures of the Agrobacterium strains were inoculated from single colonies on plates and grown for 24 h at 28°C. The working cultures were inoculated from the starter culture at a 1:100 ratio. Cells were harvested through centrifugation at 3,000 g, 22°C, for 5 min. Cell pellets were resuspended in infiltration medium (10 mM MgCl2; 10 mM MES, pH 5.7; and 150 μM acetosyringone) with OD600 adjusted to 1. Resuspended cell cultures were kept at room temperature for 3 h before infiltration. Leaf infiltration was conducted by depressing a 1 mL disposable syringe to the surface of fully expanded leaves and slowly depressing the plunger. Leaf discs were collected 72 h after infiltration for measurements using green fluorescent protein (GFP) fluorescence assay. Fluorescence images were obtained under an Olympus FV1000 laser confocal microscope (Olympus, Tokyo, Japan), with excitation at 488 nm and emission at 505–530 nm.
Transformation of Arabidopsis
The 35S:GFP-BrrTCP2 construct was introduced into Agrobacterium GV3101, which was used to transform wild-type Arabidopsis via floral dip (Clough and Bent, 1998). T0 seeds were screened on MS medium containing 30 mg L−1 kanamycin. All transgenic lines overexpressing 35S:GFP-BrrTCP2 were verified by Western blot using GFP antibody. OXBrrTCP2 x tcp2/4/10 plants were obtained by crossing OXBrrTCP2 with tcp2/4/10.
Scanning electron microscopy
The sixth rosette leaves were selected for scanning electron microscopic analysis as previously described by Sun et al. (2013). The samples were observed and photographed under a scanning electron microscope (Gemini Sigma 300/VP SEM, Carl Zeiss, Germany) at an accelerating voltage of 10 kV.
Results
Identification of TCP genes in turnips
The release of the complete turnip genome sequences allowed the genome-wide identification of turnip genes (Lin et al., 2014). In the present study, BLAST was carried out to search BrrTCP in the turnip genome. The obtained sequences were further verified through hidden Markov model. Finally, a total of 39 nonredundant BrrTCPs were identified from turnip genome. The BrrTCP genes were named following the nomenclature of Arabidopsis thaliana depending on protein sequence similarities (Figure 1). Sequence analysis revealed that AtTCP11 and 16 had no orthologs in turnip. AtTCP1, 4, 7, 9, 13, 15, 17, 18, 20, 21, and 24 had more than one ortholog in the turnip genome. Given the lack of standard annotations assigned to the 39 TCPs in turnips, the orthologs were designated as shown in Table 1 based on protein sequence similarities to their orthologs in Arabidopsis.
Figure 1.
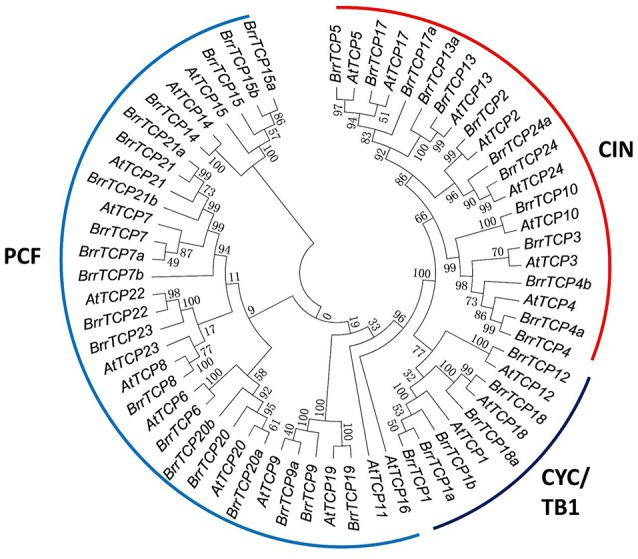
Phylogenetic tree of TCP proteins from turnips and Arabidopsis. The phylogenetic tree was generated using the neighbor-joining (NJ) method implemented in MEGA 7.0 software with JTT model and pairwise gap deletion option. Bootstrap analysis was conducted with 1,000 iterations.
Table 1.
TCP gene family in turnip.
| Gene Name | Accession number | Length(aa) | MW(Da) | pI | Chr. Location |
|---|---|---|---|---|---|
| BrrTCPl | KY607998 | 351 | 39,826.39 | 6.47 | Chr07: 17379980–17381035 |
| BrrTCPla | KY607999 | 347 | 39,385.23 | 6.55 | Chr07: 16762673–16763813 |
| BrrTCPlb | KY608000 | 346 | 39,314.74 | 5.54 | Chr02: 10171599–10172639 |
| BrrTCP2 | KY608001 | 384 | 42,403.17 | 7.47 | Chr03: 23210500–23211639 |
| BrrTCPS | KY608002 | 341 | 37,222.15 | 7.53 | Chr08: 1024759–1025793 |
| BrrTCP4 | KY608003 | 406 | 44,173.60 | 7.34 | Chr05: 20022294–20023514 |
| BrrTCP4a | KY608004 | 407 | 44,425.04 | 8.13 | Chr03: 17113530–17114753 |
| BrrTCP4b | KY608005 | 348 | 38,161.10 | 7.06 | Chr01: 24280209–24281261 |
| BrrTCP5 | KY608006 | 366 | 40,706.24 | 6.52 | Chr03: 21107231–21108331 |
| BrrTCP6 | KY608007 | 224 | 24,641.60 | 8.02 | Chr04: 8105848–8106522 |
| BrrTCP7 | KY608008 | 252 | 27,180.36 | 9.25 | Chr06: 16403133–16407846 |
| BrrTCP7a | KY608009 | 252 | 18,681.04 | 8.06 | Chr02: 25460706–25461236 |
| BrrTCP7b | KY608010 | 219 | 23,770.77 | 8.21 | Chr01: 8925053–8929517 |
| BrrTCP8 | KY608011 | 394 | 41,385.39 | 6.38 | Chr09: 9571412–9572956 |
| BrrTCP9 | KY608012 | 325 | 34,463.24 | 9.68 | Chr05: 2500244–2501218 |
| BrrTCP9a | KY608013 | 318 | 33,838.78 | 9.88 | Chr03: 10874971–10875906 |
| BrrTCPl0 | KY608014 | 348 | 38,671.40 | 6.71 | Chr05: 7091828–7092874 |
| BrrTCPl2 | KY608015 | 336 | 38,324.79 | 8.66 | Chr02: 11017245–11018356 |
| BrrTCP13 | KY608016 | 320 | 35,694.44 | 7.07 | Chr03: 14413642–14414472 |
| BrrTCP13a | KY608017 | 309 | 34,451.40 | 7.56 | Chr05: 23621188–23622581 |
| BrrTCP14 | KY608018 | 466 | 50,250.46 | 6.83 | Chr06: 10402184–10403578 |
| BrrTCPl5 | KY608019 | 321 | 34,030.17 | 7.26 | Chr07: 18427674–18428612 |
| BrrTCP15a | KY608020 | 245 | 25,663.89 | 6.67 | Chr07: 16070535–16071269 |
| BrrTCPl5b | KY608021 | 246 | 25,691.98 | 6.74 | Chr02: 11422832–11423572 |
| BrrTCP17 | KY608022 | 301 | 33,762.21 | 6.83 | Chr02: 25620883–25621956 |
| BrrTCP17a | KY608023 | 246 | 27,372.44 | 6.82 | Chr03: 1428184–1428909 |
| BrrTCP18 | KY608024 | 424 | 48,340.37 | 8.69 | Chr03: 17927181–17929425 |
| BrrTCP18a | KY608025 | 285 | 32,645.80 | 9.46 | Chr01: 22021125–22022887 |
| BrrTCP19 | KY608026 | 281 | 30,182.56 | 5.52 | Chr02: 8815586–8816431 |
| BrrTCP20 | KY608027 | 200 | 22,050.51 | 5.48 | Chr02: 22055944–22058191 |
| BrrTCP20a | KY608028 | 311 | 32,789.26 | 7.68 | Chr06: 22527874–22528797 |
| BrrTCP20b | KY608029 | 278 | 29,658.68 | 6.38 | Chr09: 1164212–1165048 |
| BrrTCP21 | KY608030 | 234 | 24,508.21 | 8.81 | Chr10: 16190790–1619500 |
| BrrTCP21a | KY608031 | 234 | 24,261.29 | 10.20 | Chr03: 1493264–1493968 |
| BrrTCP21b | KY608032 | 229 | 23,872.73 | 9.55 | Chr02: 2637716–2638405 |
| BrrTCP22 | KY608033 | 374 | 39,116.38 | 8.95 | Chr02: 12405545–12406669 |
| BrTCP23 | KY608034 | 356 | 37,152.33 | 7.64 | Chr07: 19406027–19407097 |
| BrrTCP24 | KY608035 | 319 | 35,402.64 | 7.93 | Chr09: 22077911–22078867 |
| BrrTCP24a | KY608036 | 313 | 34,970.16 | 7.44 | Chr08: 16381466–16383191 |
Information of the BrrTCP gene family members is shown in Table 1. The ORF length of BrrTCP genes varied from 603 to 1,401 bp, encoding polypeptides of 200–466 amino acids, with a predicted molecular mass of 22.05–50.25 kDa. The theoretical pI ranged from 5.48 to 10.20. Genomic localization of each BrrTCP in turnips is shown in Figure 2. These genes were distributed in all 10 turnip chromosomes. The maximum number of BrrTCP genes per chromosome was found for chromosome A02 with 9 TCP genes. Eight genes were located at chromosome 3, and five and four genes were located at chromosomes 7 and 5, respectively. Chromosomes 4 and 10 had the minimum BrrTCP genes, only one each. Three genes each were located on chromosomes 1, 6, and 9. Chromosome 8 contained two BrrTCP genes. A total of seven pairs of putative BrrTCP paralog proteins were produced by segmental duplication, which were distributed in different chromosomes. These results indicated the large-scale segmental duplication events involved in the expansion of BrrTCP gene family in turnips.
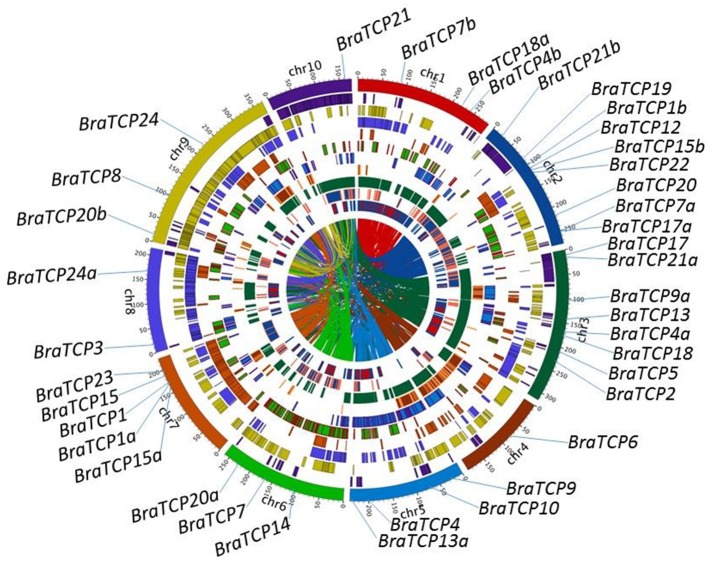
Figure 2.
BrrTCPs' chromosome distributions, synteny block, and the turnip genome duplication event caused paralogous relationships. Chromosomes are shown in different colors and in the outer circle, where the numbers represent the chromosome length in 100 Kb. The BrrTCP genes are marked on the approximate positions with specific color lines on the circle. Filled blocks in different colors denote the syntenic relationships of turnip TCP genes.
The divergence time (T) of seven pairs of BrrTCP paralog proteins was estimated by measuring the synonymous (Ks) and nonsynonymous (Ka) mutation rates (Table 2). The estimated divergence time (T) for the BrrTCP paralogs was from 10.3067 to 27.8600 million years ago (MYA), with an average duplication time of approximately 18.7862 MYA. Schranz et al. (2006) estimated the time of very early radiation of Brassicaceae at 34 MYA. Comparative physical mapping studies have confirmed genome triplication in a common ancestor of B. oleracea (O'Neill and Bancroft, 2000) and B. rapa (Park et al., 2005) since its divergence from the A. thaliana lineage at least 13–17 MYA (Yang et al., 1999; Town et al., 2006; Beilstein et al., 2010). The divergence time of three pairs of BrrTCP paralogs (BrrTCP9/9a, BrrTCP22/23, and BrrTCP24/24a) occurred precedent to the period of the origin of the B. rapa. The ω values for the BrrTCP paralogs were less than one. However, two pairs of BrrTCPs (BrrTCP1a/1b, ω = 0.3794; BrrTCP24/24a, ω = 0.3646) achieved relatively large ω values, which suggested that they may have evolved rapidly from those of the last common ancestor.
Table 2.
Dates of duplication of duplicated gene pairs.
| Seql | Seq2 | Identity (%) | Ks | Ka | to | T (MYA) |
|---|---|---|---|---|---|---|
| BrrTCPla | BrrTCPlb | 73.24 | 0.3092 | 0.1173 | 0.379366106 | 10.3067 |
| BrrTCP4 | BrrTCP4a | 79.24 | 0.416 | 0.0443 | 0.106490385 | 13.8667 |
| BrrTCP9 | BrrTCP9a | 70.76 | 0.6658 | 0.1743 | 0.261790327 | 22.1933 |
| BrrTCPl5a | BrrTCPl5b | 88.10 | 0.3792 | 0.0404 | 0.106540084 | 23.8867 |
| BrrTCP2l | BrrTCP2la | 86.50 | 0.544 | 0.0441 | 0.081066176 | 18.1333 |
| BrrTCP22 | BrrTCP23 | 71.76 | 0.4577 | 0.1117 | 0.244046319 | 15.2567 |
| BrrTCP24 | BrrTCP24a | 68.13 | 0.8358 | 0.3047 | 0.3645609 | 27.8600 |
Gene structure and conserved motifs
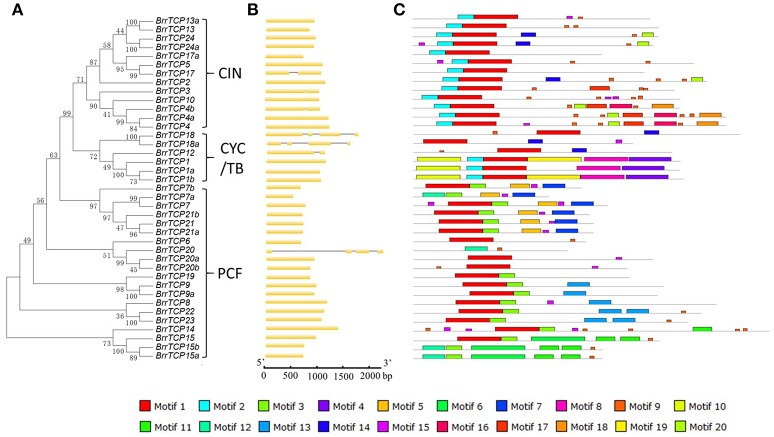
To better understand the diversification of the TCP genes in turnips, the exon/intron organization and conserved motifs of BrrTCPs were analyzed. A new neighbor-joining phylogenetic tree was constructed using the protein sequences of BrrTCPs (Figure 3A). The genome sequences and corresponding coding sequences of TCP genes in turnip analysis revealed that most BrrTCP genes have similar gene structures in the same group (Figure 3B). A total of 32 members of BrrTCP gene family have one exon (82%), 4 genes have two exons (10%), and 3 have four exons (8%). In Arabidopsis, the number of exons ranged from one to four, and 82% of the genes contained only one exon. The TCP genes in turnips exhibited similar gene structure. The MEME online tool was used to predict the conversed motif composition of BrrTCPs (Figure 3C). The number of motifs varied from 2 to 11. The motifs were evaluated using InterProScan for annotation. The results revealed that motifs 1, 2, and 3 were identified as TCP domain. In addition, some motifs only presented at specific subclades, such as motifs 6, 7, 10, and 19 in BrrTCP1, 1a, and 1b; motif 9 in BrrTCP15, 15a, and 15b; and motif 16 in BrrTCP4, 4a, and 4b, suggesting that they may have subclade-specific functions.
Figure 3.
Genomic structure and motif composition of turnip TCPs. (A) Phylogenetic tree of turnip TCP proteins. (B) Genomic structure of turnip gene. Exons and introns are represented with yellow boxes and black lines. (C) The conserved motifs in turnip TCP proteins were identified using MEME. Each motif is represented with a specific color.
In Arabidopsis, miR319 controls jasmonate biosynthesis and senescence by cleaving TCP transcription factors (Schommer et al., 2008) and petal development (Nag et al., 2009). The AtmiR319-targeted TCP genes, namely, AtTCP2, AtTCP3, AtTCP4, AtTCP10, and AtTCP24, all belong to the CIN clade. Similarly, four TCP genes in turnips contain miR319 binding sites (Figure 4), and all of them were CIN family members.
Figure 4.
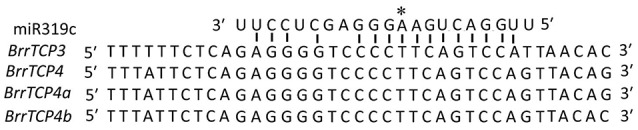
Alignment of putative target areas for miR319c (aligned in reverse). The “*” and above sequences indicate the cleavage site.
Subcellular localization
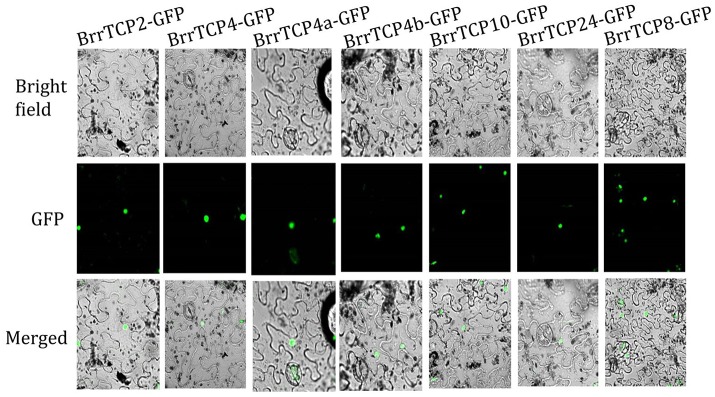
The known members of the TCP gene family function as transcription factors. The GFP gene was fused with BrrTCPs as a reporter. The GFP signals of BrrTCP-GFPs were detected in the nucleus using laser confocal microscopy (Figure 5), which suggested that BrrTCPs functioned as transcription factors.
Figure 5.
Subcellular localization of 35S:BrrTCP-GFP in Nicotiana benthamiana leaves. BrrTCP-GFP localized in the nucleus.
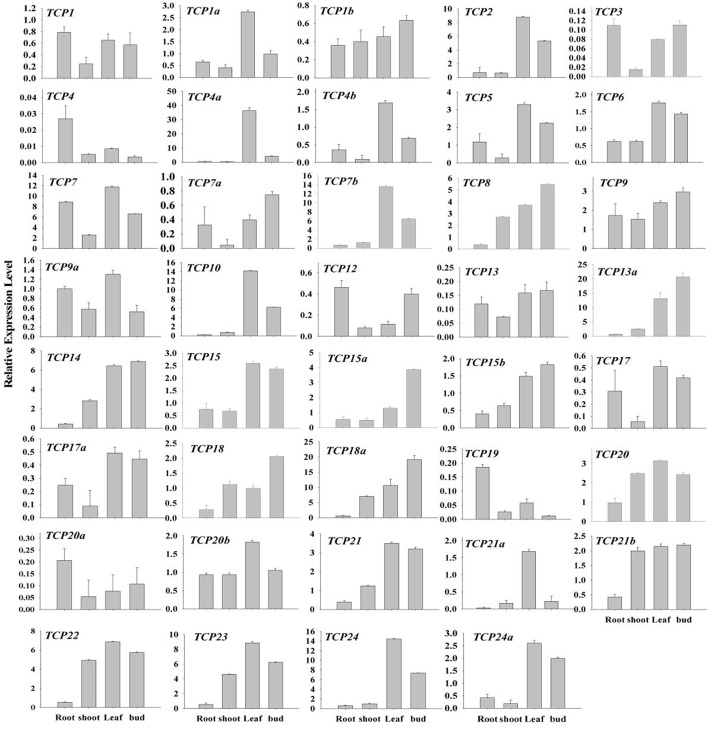
qRT-PCR analysis of the turnip TCP genes
The expression pattern of a gene is always relative to its function (Xu et al., 2015). To probe possible functions of BrrTCP genes in turnips, qRT-PCR was performed to examine the expressions of BrrTCP genes in different organs of turnips (Figure 6). Interestingly, expression levels of most CIN-type BrrTCP genes were high in leaves and weak in the roots and stems. BrrTCP18 and BrrTCP18a, the members of CYC/TB1 subclade, are highly expressed in flower buds and weakly expressed in roots. In contrast, class I clade showed more widespread but less tissue-specific expression patterns; for example, BrrTCP7 and BrrTCP9 are highly expressed in roots, stems, leaves, and flower buds; BrrTCP8, 14, 18a, 20, 21b, 22, and 23 are highly expressed in the stems, leaves, and flower buds; and BrrTCP7b, 13a, 15, and 21 are highly expressed in leaves and flower buds. These results indicated that every clade possessed a characteristic expression profile.
Figure 6.
Expression patterns of turnip TCP genes in different tissues. The expression profile data of BrrTCP genes in roots, shoots, leaves, and buds were obtained through qRT-PCR.
Interactions between turnip TCP proteins
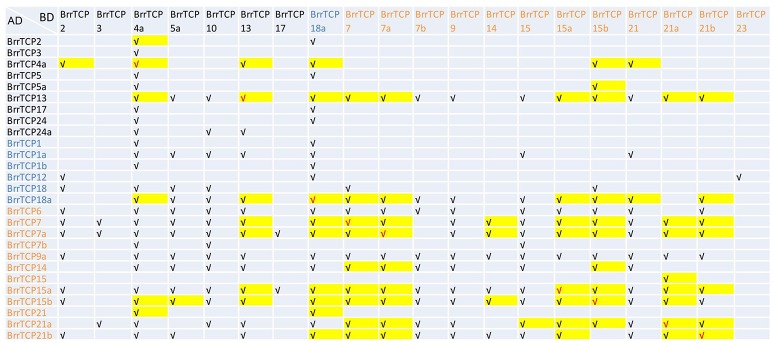
TCP proteins tend to form homodimers or heterodimers with other TCP proteins, and dimerization may be required for their DNA-binding activity and hence for their biological activity. Yeast two-hybrid screening was carried out to investigate the interactions among the BrrTCP proteins, as shown in Figure 7, where the proteins are arranged according to their subclades except their autoactivation activity. A total of 8 out of 39 AD-fusion proteins tested had autoactivation activity in yeast. Meanwhile, out of the 39 BD-fusion proteins tested, 13 had autoactivation activity in yeast. Among them, five BrrTCP proteins had autoactivation activity in AD- and BD-fusion proteins. Although we selected 219 interactions, this number may not be accurate due to autoactivation. A total of 90 (45 pairs) proteins interacted in AD- and BD-orientations, including 9 homodimer formations. The class I BrrTCP transcription factors formed 91 homo- or heterodimers within class I members. Meanwhile, class II BrrTCP transcription factors formed 42 homo- or heterodimers within class II. The members of class I proteins more preferentially formed dimerization properties in the same class than class II.
Figure 7.
Interaction of BrrTCP proteins in yeast two-hybrid assay. AD-fusion is listed in the left column. BrrTCP protein names are ordered according to their subclades (CIN subclade is represented by black, TB1 by blue, and PCF by red).
Overexpression of BrrTCP2 rescued the tcp2/4/10 phenotype
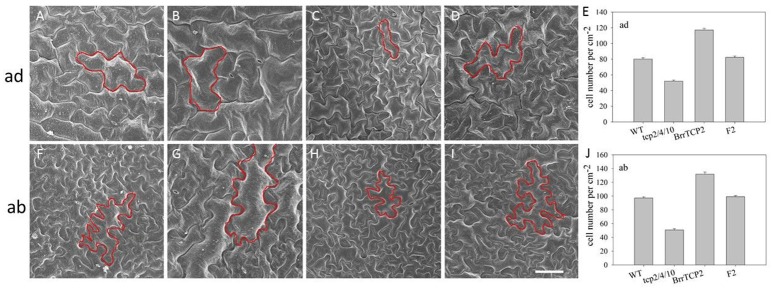
BrrTCP2 is a member of the CIN subclade of TCP in turnips. Given the unavailability of tcp-related mutant in turnips, we constructed transgenic Arabidopsis plants overexpressing BrrTCP2. As shown in Figure 8, the sixth rosette leaf of tcp2/4/10 triple mutants showed the most enhanced leaf size, with some signs of crinkling (Figures 8B,F). Meanwhile, the OXBrrTCP2 plants showed the most diminished leaf size (Figures 8C,G). OXBrrTCP2 line was crossed with tcp2/4/10, and the phenotype of the homozygote F2 plants restored the leaf phenotype of tcp2/4/10 to the phenotype of wild type (Figures 8D,H). Western blot analysis showed that OXBrrTCP2 and F2 plants had high expression levels, whereas no signal was detected in WT and tcp2/4/10 plants (Figure 8I). Arabidopsis leaf size is normally regulated by the cell size and number. To assess the cell size and number, we selected a site midway along the length of the lamina and between the margin and the midvein of the expanded sixth rosette leaf for analysis using scanning electron microscopy. The adaxial epidermal cell size of tcp2/4/10 was larger than WT (Figures 9A,B), and fewer cells were noted per unit area (Figure 9E). Meanwhile, the adaxial epidermal cell size of OXBrrTCP2 plants was smaller than WT (Figures 9A,C), and more cells were noted per unit area (Figure 9E). The F2 plant had similar cell size and cell number per unit area with WT. The abaxial epidermal cell size and cell number were similar to the adaxial epidermis. The tcp2/4/10 had larger cell size and fewer cell number per unit area than WT (Figures 9F,G,J), whereas OXBrrTCP2 plants had smaller cell size and more cell number per unit area than WT (Figures 9F,H,J). Overexpression of BrrTCP2 in tcp2/4/10 also restored the abaxial epidermal phenotype of tcp2/4/10 to WT (Figures 9D,I). BrrTCP2 may have a function in cell division and/or differentiation.
Figure 8.
Phenotype effects of constitutive expression of BrrTCP2 in transgenic Arabidopsis. (A,E) Phenotype of wild-type Arabidopsis. (B,F) Phenotype of tcp2/4/10 mutants. (C,G) Expression of BrrTCP2 in wild-type Arabidopsis. (D,H) Phenotype of the homozygote F2 plants alleviates the phenotype of the tcp2/4/10 mutants. (I) Western blot analysis of BrrTCP2 protein levels in transgenic plants. BrrTCP2 protein was analyzed through immunoblotting using an anti-GFP antibody. AtACT2 served as the control. Bar = 0.5 cm.
Figure 9.
Scanning electron micrographs of leaf epidermal cells. Adaxial epidermis in the sixth rosette leaf from WT (A), tcp2/4/10 (B), 35S:BrrTCP2 transgenic plants (C), and F2 (D). (E) Number of adaxial epidermis cells of sixth rosette leaves. Abaxial epidermis in the sixth rosette leaf from WT (F), tcp2/4/10 (G), 35S:BrrTCP2 transgenic plants (H), and F2 (I). (J) Number of abaxial epidermis cells of sixth rosette leaves. Bar = 50 μm.
Discussion
TCP proteins are plant-specific transcription factors involved in the regulation of multiple processes during plant development and growth, such as leaf morphogenesis and senescence, flower development, circadian clock, endoreduplication, branching, fiber development, and phytohormone biosynthesis (Schommer et al., 2008; Nag et al., 2009; Sugio et al., 2011; Danisman et al., 2012; Hao et al., 2012; Li Z. Y. et al., 2012; Wang et al., 2013; Ma J. et al., 2016; Zhou et al., 2016). Previous studies revealed that all Brassicaceae, including Arabidopsis and Brassica's, underwent polyploidization events, such as γ triplication (135 MYA) and β (90–100 MYA) and α (24–40 MYA) duplications (Wang and Kole, 2015). B. rapa shares this complex history, with the addition of a whole-genome triplication (WGT) thought to have occurred between 13 and 17 million years ago (MYA) making “mesohexaploidy” a characteristic of the Brassiceae tribe of the Brassicaceae (Lysak et al., 2005). The Brassica genomes diploidized after this triplication event through genome fractionation and rearrangements (Mun et al., 2009). Several studies revealed that the three subgenomes did not behave similar and that the dominat subgenome retained most genes; in addition, the genome fractionation was not a random process, as certain gene families retained more copies (Park et al., 2005; Wang et al., 2011; Chalhoub et al., 2014). The BrrTCP gene family in turnips may be caused by genome duplication processes, including multiple segmental duplications, tandem duplication, transposition events, and whole-genome duplication. Except gene duplication, differences in exon/intron organizations can also clarify the evolutionary history of the gene family. The gene structure of BrrTCPs compared with the same clade showed that TCP genes shared similar exon/intron distribution in terms of exon length and intron numbers; meanwhile, BrrTCPs with the same clade displayed similar motif distribution. Similar to tomato TCP proteins (Parapunova et al., 2014), more interactions were found for class I proteins than class II proteins (91 vs. 42), although the number of interactions for class I and class II may be underestimated because of the autoactivating members. The interactions obtained by a comprehensive yeast two-hybrid screen of turnip TCP transcription factors have not yet been reported for TCP members from other species than tomato. Expression analysis and dimerization properties may help to identify TCP protein pairs that function together and explain observed functional redundancies in case of overlapping interaction maps of turnips in the future.
In Arabidopsis, miR319-targeted AtTCP2, 3, 4, 10, and 24 regulate leaf development and petal growth (Palatnik et al., 2003; Ori et al., 2007; Nag et al., 2009). The three closest turnip TCP genes have a putative binding site for miR319c. Gene function is also related to its expression profile (Zhou et al., 2016). In this study, we detected the expression patterns of 39 BrrTCP genes in four organs using qRT-PCR. These genes vary widely among the turnip organs. Two CIN-type genes (BrrTCP4a and BrrTCP4b), which are miR319c targeted, exhibited high expression levels in leaves, particularly BrrTCP4a. Meanwhile, BrrTCP4 exhibited low expression levels in all detected organs. This phenomenon was also found in other duplicated gene pairs, such as BrrTCP7, BrrTCP7a, and BrrTCP7b. BrrTCP7 showed high expression levels in roots, leaves, and flowers, whereas BrrTCP7a showed low expression levels in all detected organs. However, BrrTCP7b exhibited high expression levels in leaves and flowers. Gene duplication plays a vital role in the process of plant genomic and organismal evolution and confers new gene functions and the evolution of gene networks (Flagel and Wendel, 2009). Gene duplication might confer new functions to the paralogous BrrTCP genes. The other CIN-type genes, such as BrrTCP2, BrrTCP10, BrrTCP13a, and BrrTCP24, exhibited high expression levels not only in leaves but also in flowers. Turnip CIN-type genes may have a function in leaf and flower development. Class I and class II have antagonistic functions based on similar putative binding sites (Danisman et al., 2012). Class I BrrTCP genes, such as BrrTCP7b and BrrTCP14, were also detected to have high expression levels in leaves and flowers. In Arabidopsis, TCP14, which is homologous to BrrTCP14, acts repeatedly with TCP15 in modulating cell proliferation in developing leaf blades and flowers (Kieffer et al., 2011). Some members of class I and class II competitively regulated the cell proliferation in leaf development.
In Arabidopsis, loss-of-function mutants tcp2, tcp4, and tcp10 caused slight phenotype defect. Meanwhile, tcp2/4 double mutants exhibited an increased phenotype defect, and tcp2/4/10 triple mutants showed the most significant phenotype defects, with increase in leaf size and signs of crinkling (Schommer et al., 2008). TCP2, 4, and 10 repeatedly regulated leaf development. BrrTCP2 overexpression in Arabidopsis exhibited as small leaves with few epidermal cells. Overexpression of BrrTCP2 in tcp2/4/10 triple mutants restored the defect leaf phenotype to mimic wild-type leaf phenotype. BrrTCP2 might function in leaf development via inhibiting cell proliferations.
Conclusion
In summary, we identified 39 TCP genes, which were distributed on 10 chromosomes with different densities, in the turnip genome. Y2H analysis showed that these transcription factors preferentially formed heterodimers. Expression analysis showed that these genes exhibited varied expression profiles. In addition, BrrTCP2 was involved in leaf development via regulating cell proliferations.
Author contributions
XS, YoY, and HS conceived and designed the study; JD, SH, QY, CW, and YuY performed the experiments and analyzed the data; XS, YoY, and JD wrote the paper; all authors have read and approved the final version.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We all would like to thank Dr. Yanxia Jia and Dr. Hongtao Li for practical assistance in the operation of the Olympus FV1000 laser confocal microscope and ABI7500.
Footnotes
Funding. This work was financially supported by the National Natural Science Foundation of China (NSFC) (31590823, 41271058, and 31400244), Natural Science foundation of Yunnan Province (2017FB050) and the Basic Research Project of the Ministry of Science and Technology of China (2012FY111400).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01588/full#supplementary-material
References
- Aguilar Martinez J. A., Sinha N. R. (2013). Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22 and AtTCP23 in leaf development. Front. Plant Sci. 4:406. 10.3389/fpls.2013.00406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Long J., He X., Yan J., Chen X., Tan Y., et al. (2016). Overexpression of spinach non-symbiotic hemoglobin in Arabidopsis resulted in decreased NO content and lowered nitrate and other abiotic stresses tolerance. Sci. Rep. 6:26400. 10.1038/srep26400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilstein M. A., Nagalingum N. S., Clements M. D., Manchester S. R., Mathews S. (2010). Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 18724–18728 10.1073/pnas.0909766107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B., Denoeud F., Liu S., Parkin I. A. P., Tang H., Wang X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. 10.1126/science.1253435 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Danisman S., van der Wal F., Dhondt S., Waites R., de Folter S., Bimbo A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. 10.1104/pp.112.200303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L. E., Wendel J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. 10.1111/j.1469-8137.2009.02923.x [DOI] [PubMed] [Google Scholar]
- Gaut B. S., Morton B. R., McCaig B. C., Clegg M. T. (1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. U.S.A. 93, 10274–10279. 10.1073/pnas.93.19.10274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao J., Tu L. L., Hu H. Y., Tan J. F., Deng F. L., Tang W. X., et al. (2012). GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63, 6267–6281. 10.1093/jxb/ers278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M., Master V., Waites R., Davies B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147–158. 10.1111/j.1365-313X.2011.04674.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619. 10.1105/tpc.9.9.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Ohashi Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. 10.1046/j.1365-313X.2002.01294.x [DOI] [PubMed] [Google Scholar]
- Li C., Potuschak T., Colon-Carmona A., Gutierrez R. A., Doerner P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. U.S.A. 102, 12978–12983. 10.1073/pnas.0504039102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Li B., Shen W. H., Huang H., Dong A. (2012). TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 71, 99–107. 10.1111/j.1365-313X.2012.04973.x [DOI] [PubMed] [Google Scholar]
- Li Z. Y., Li B., Dong A. W. (2012). The arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant. 5, 270–280. 10.1093/mp/ssr086 [DOI] [PubMed] [Google Scholar]
- Lin K., Zhang N., Severing E. I., Nijveen H., Cheng F., Visser R. G., et al. (2014). Beyond genomic variation - comparison and functional annotation of three Brassica rapagenomes: a turnip, a rapid cycling and a Chinese cabbage. BMC Genomics. 15:250. 10.1186/1471-2164-15-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M. A., Koch M. A., Pecinka A., Schubert I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15, 516–525. 10.1101/gr.3531105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Liu F., Wang Q., Wang K., Jones D. C., Zhang B. (2016). Comprehensive analysis of TCP transcription factors and their expression during cotton (Gossypium arboreum) fiber early development. Sci. Rep. 6:21535. 10.1038/srep21535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Ma J., Fan D., Li C., Jiang Y., Luo K. (2016). Genome-wide identification of TCP family transcription factors from populus euphratica and their involvement in leaf shape regulation. Sci. Rep. 6:32795. 10.1038/srep32795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Trillo M., Cubas P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. 10.1016/j.tplants.2009.11.003 [DOI] [PubMed] [Google Scholar]
- Mun J.-H., Kwon S.-J., Yang T.-J., Seol Y.-J., Jin M., Kim J.-A., et al. (2009). Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 10:R111 10.1186/gb-2009-10-10-r111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., King S., Jack T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc Natl Acad Sci, U.S.A. 106, 22534–22539. 10.1073/pnas.0908718106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaud O., Dabos P., Carnus E., Tremousaygue D., Herve C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. 10.1007/s00239-006-0174-z [DOI] [PubMed] [Google Scholar]
- O'Neill C. M., Bancroft I. (2000). Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant, J. 23, 233–243. 10.1046/j.1365-313x.2000.00781.x [DOI] [PubMed] [Google Scholar]
- Ori N., Cohen A. R., Etzioni A., Brand A., Yanai O., Shleizer, et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39, 787–791. 10.1038/ng2036 [DOI] [PubMed] [Google Scholar]
- Palatnik J. F., Allen E., Wu X., Schommer C., Schwab R., Carrington J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. 10.1038/nature01958 [DOI] [PubMed] [Google Scholar]
- Parapunova V., Busscher M., Busscher-Lange J., Lammers M., Karlova R., Bovy A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14:157. 10.1186/1471-2229-14-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. Y., Koo D. H., Hong C. P., Lee S. J., Jeon J. W., Lee S. H., et al. (2005). Physical mapping and microsynteny of Brassica rapa ssp. pekinensis genome corresponding to a 222 kbp gene-rich region of Arabidopsis chromosome 4 and partially duplicated on chromosome 5. Mol. Genet. Genomics 274, 579–588. 10.1007/s00438-005-0041-4 [DOI] [PubMed] [Google Scholar]
- Resentini F., Felipo-Benavent A., Colombo L., Blazquez M. A., Alabadi D., Masiero S. (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant. 8, 482–485. 10.1016/j.molp.2014.11.018 [DOI] [PubMed] [Google Scholar]
- Schommer C., Palatnik J. F., Aggarwal P., Chetelat A., Cubas P., Farmer E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. 10.1371/journal.pbio.0060230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz M. E., Lysak M. A., Mitchell-Olds T. (2006). The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11, 535–542. 10.1016/j.tplants.2006.09.002 [DOI] [PubMed] [Google Scholar]
- Shi P., Guy K. M., Wu W., Fang B., Yang J., Zhang M., et al. (2016). Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 16:85. 10.1186/s12870-016-0765-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A., Kingdom H. N., MacLean A. M., Grieve V. M., Hogenhout S. A. (2011). Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 108, E1254–E1263. 10.1073/pnas.1105664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. D., Feng Z. H., Meng L. S., Zhu J., Geitmann A. (2013). Arabidopsis ASL11/LBD15 is involved in shoot apical meristem development and regulates WUS expression. Planta 237, 1367–1378. 10.1007/s00425-013-1844-x [DOI] [PubMed] [Google Scholar]
- Town C. D., Cheung F., Maiti R., Crabtree J., Haas B. J., Wortman J. R., et al. (2006). Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18, 1348–1359. 10.1105/tpc.106.041665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uberti-Manassero N. G., Lucero L. E., Viola I. L., Vegetti A. C., Gonzalez D. H. (2012). The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J. Exp. Bot. 63, 809–823. 10.1093/jxb/err305 [DOI] [PubMed] [Google Scholar]
- Wang M. Y., Zhao P. M., Cheng H. Q., Han L. B., Wu X. M., Gao P., et al. (2013). The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 162, 1669–1680. 10.1104/pp.113.215673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kole C. (2015). The Brassica rapa Genome Compendium of Plant Genomes. Berlin; Heidelberg: Springer. [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 43, 1035–1039. 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- Xu Z., Sun L., Zhou Y., Yang W., Cheng T., Wang J., et al. (2015). Identification and expression analysis of the SQUAMOSA promoter-binding protein (SBP)-box gene family in Prunus mume. Mol. Genet. Genomics. 290, 1701–1715. 10.1007/s00438-015-1029-3 [DOI] [PubMed] [Google Scholar]
- Yang Y. W., Lai K. N., Tai P. Y., Li W. H. (1999). Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 48, 597–604. 10.1007/PL00006502 [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Xu Z., Zhao K., Yang W., Cheng T., Wang J., et al. (2016). Genome-wide identification, characterization and expression analysis of the TCP gene family in Prunus mume. Front. Plant Sci. 7:1301. 10.3389/fpls.2016.01301 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.