Abstract
Background
Shigella species are an important cause of acute diarrheal disease worldwide. This study describes the prevalence of Shigella spp. serotypes and their resistance profile in Vellore, South India from 2014 to 2015.
Methods
From 2014 to 2015, 338 Shigella strains were isolated from stool samples at Christian Medical College, Vellore, India. Identification and serotyping was carried out using standard protocols. Antimicrobial susceptibility testing was done against commonly used antibiotics. Multidrug resistance was detected in 157 isolates. A subset of 73 isolates was randomly characterized further for acquired antimicrobial resistance genes in this study.
Results
The resistance profile of the study isolates varied by species and year. S. sonnei isolates were 100% resistant to all tested antibiotics in 2014, whereas in 2015, resistance was found for AMP-NAL-TAX-SXT-FIX. The resistance phenotypes among S. flexneri isolates for the year 2014 and 2015 were AMP-SXT-NAL-NOR-FIX-TAX and AMP-NAL-SXT-TAX-NOR-FIX respectively. Screening for antimicrobial resistance genes in S. flexneri found dhfr1A, sulII, blaOXA, blaTEM, blaCTX-M-1, qnrB, qnrS and AmpC genes while S. sonnei were found to have only dhfr1A, sulII, blaCTX-M-1 and qnrS genes respectively. Antimicrobial resistance genes were predominantly seen in AMP-SXT-NAL and AMP-SXT-NAL-NOR resistance phenotypes.
Conclusion
Shigella prevalence of 4.8% to 4.6% was documented between the years 2014 to 2015 in this study. We show evidence that resistance to commonly used antibiotics continues to increase among Shigella spp. in South India. The presence of qnrS and blaCTX-M-15 in the study isolates further indicates the threat of spreading resistance to quinolones and third-generation cephalosporins.
Keywords: Shigella prevalence, antimicrobial resistance, qnrS, blaCTX-M-1, blaTEM, non-agglutinable Shigella
Introduction
Shigella species are an important cause of acute diarrheal disease worldwide. The majority of cases occur among children less than five years of age.1,2 Shigella has been generally characterized by serological typing for epidemiological and diagnostic purposes. Shigella species include four serogroups: S. dysenteriae, S. flexneri, S. sonnei, and S. boydii. Each serogroup contains several serotypes differentiated based on the O-antigen structure.1 All four Shigella spp. may cause disease, the most common being S. sonnei in developed countries but this species has shown a propensity for intercontinental spread to developing countries as a single and rapidly evolving lineage. In contrast S. flexneri is predominant in developing countries, while S. dysenteriae and S. boydii are rarely isolated.3 Although shigellosis is primarily self-limiting, antibiotics are recommended for reducing illness duration and for preventing transmission; the current drugs of choice are fluoroquinolones and third generation cephalosporins.4
However, antimicrobial resistance (AMR) is an emerging problem among Shigella spp., particularly in Asia and Africa.5 Resistance to first line drugs such as ampicillin and trimethoprim-sulfamethoxazole has become high.6 Reports on the emergence of ciprofloxacin resistance further narrows the list of effective antimicrobials. Infection due to resistant bacteria can persist longer than infections with susceptible bacteria.7
Currently there are no vaccines available for shigellosis due to their large dependence on the prevailing species and serotypes, as only serotype specific immunity has been demonstrated in humans.8,9 The distribution of Shigella serogroups/serotypes and antimicrobial resistance differs over time and place. Due to the scarcity of data especially from developing countries like India, a comprehensive understanding of the epidemiology of Shigella spp. is essential to control shigellosis. The present study describes the trends in Shigella species, serotypes and their resistance mechanisms, in isolates from patients with diarrhea over the years 2014 and 2015.
Methods
Bacterial strains
Isolation and identification of the organism was carried out using conventional biochemical reactions (urea, citrate, triple sugar iron, indole, motility).10 All the isolates were serotyped using commercial antisera as per the manufacturer’s instructions (Denka Seiken, Tokyo, Japan).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing of Shigella isolates against ampicillin 10 μg (AMP), trimethoprim/sulfamethoxazole 1.25/23.75 μg (SXT), nalidixic acid 30 μg (NAL), norfloxacin 10 μg (NOR), cefotaxime 30 μg (TAX) and cefixime 5 μg (FIX) was performed using Kirby-Bauer disk diffusion method and results were interpreted using breakpoints recommended by the Clinical Laboratory Standards Institute (CLSI) guidelines 2014 and 2015.
Molecular analysis of antibiotic resistance genes
Genomic DNA was extracted with QIAamp DNA mini kit (Qiagen, Hilden, Germany). The presence of β-lactamase genes (blaOXA, blaTEM), ESC genes blaCTX-M-1, AmpC (blaMOX, blaCIT, blaDHA, blaACC, blaEBC and blaFOX), trimethoprim resistance gene (dhfr1a), sulphonamide resistance gene (sulII) and plasmid mediated quinolone resistance (PMQR) (qnrA, qnrB, qnrS) genes were assessed by PCR as described earlier.11
Statistical analysis
Comparisons of variables were derived using two-tailed Chi-squared test. A p-value of less than 0.05 was considered significant.
Results
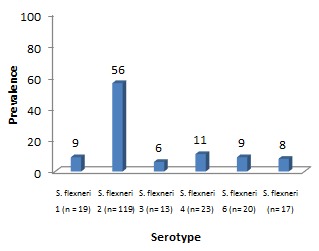
Between 2014 and 2015, a total of 7212 stool specimens were processed, of which 338 were confirmed as Shigella spp. and serotyped as follows: S. flexneri (n=211), S. sonnei (n=90), S. dysenteriae (n=12), S. boydii (n=8) and non-agglutinable Shigella (n=17). Multidrug resistance (MDR) (resistance to 3 or more classes of antibiotics) was detected in 157 isolates. The species distribution over time is shown in Figure 1. S. flexneri was the prevalent serogroup found, followed by S. sonnei, S. dysenteriae, and S. boydii. The isolation rate of S. sonnei has gradually increased from one year to the next (19.8% to 33.9%). The trends in S. flexneri serotypes are described in Figure 2, with S. flexneri 2 being the most commonly isolated serotype followed by other serotypes. In addition, the rate of non-typeable Shigella was found to be higher in the year 2014 (8.5%) and decreased to 1.2% in 2015 (p=0.002). Of the Shigella positive isolates, 60% (202/338) were from children less than or equal to five years of age. Shigella infection was found more frequently among males (57%) than females (43%). Seasonal variation of Shigella infection was observed during the study period with a peak isolation rate seen between the months May-July.
Figure 1. Prevalence of Shigella species isolated between the years 2014 and 2015.
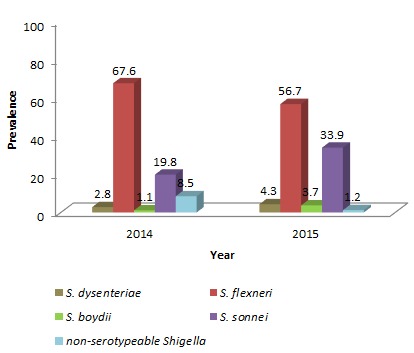
Figure 2. Distribution of S. flexneri serotypes isolated over the study period.
Overall, multidrug resistance was more frequently detected in S. flexneri and S. sonnei than in S. dysenteriae and S. boydii. The antimicrobial resistance trend change was calculated between the species and years. A significant decrease was seen in the norfloxacin resistance profile of Shigella spp. during the study period (p<0.001) and a notable change was observed for ampicillin resistance between the species S. flexneri and S. sonnei (p<0.001). The resistance profile of other antibiotics remains unchanged between the years and the species (Tables 1 and 2). The resistance rate was found to be higher in patients with less than five years and followed by those greater than fifteen years of age (Figure 3).
Table 1. Comparison of antimicrobial resistance profiles of S. flexneri and S. sonnei between 2014 and 2015.
| S. flexneri | |||||||
|---|---|---|---|---|---|---|---|
| Year (n) | Phenotype | AMP | SXT | NAL | NOR | TAX | FIX |
| 2014 (119) | R | 84 | 113 | 111 | 80 | 9 | 12 |
| S | 35 | 6 | 8 | 39 | 110 | 107 | |
| 2015 (92) | R | 63 | 88 | 85 | 20 | 12 | 9 |
| S | 29 | 4 | 7 | 72 | 80 | 83 | |
| p value | 0.741 | 0.810 | 0.802 | <0.005 | 0.187 | 0.942 | |
| Chi (df) | 0.1 (210) = 21 | 0.05 (210) = 10.5 | 0.06 (210) = 12.6 | 43 (210) = 9030 | 1.73 (210) = 363.3 | 0.005 (210) = 1.05 | |
| S. sonnei | |||||||
|---|---|---|---|---|---|---|---|
| Year (n) | Phenotype | AMP | SXT | NAL | NOR | TAX | FIX |
| 2014 (35) | R | 4 | 32 | 35 | 23 | 1 | 3 |
| S | 31 | 3 | 0 | 12 | 34 | 32 | |
| 2015 (55) | R | 13 | 54 | 54 | 10 | 3 | 4 |
| S | 42 | 1 | 1 | 45 | 52 | 51 | |
| p value | 0.149 | 0.128 | 0.423 | <0.005 | 0.561 | 0.825 | |
| Chi (df) | 2.08 (89) = 185.1 | 2.29 (89) = 203.8 | 0.64 (89) = 56.9 | 20.8 (89) = 1851 | 0.33 (89) = 29.3 | 0.05 (89) = 4.45 | |
R – resistant, S – susceptible, AMP – ampicillin, SXT – trimethoprim/sulfamethoxazole, NAL – nalidixic acid, NOR – norfloxacin, TAX – cefotaxime, FIX – cefixime.
Table 2. Comparison of antimicrobial resistance profiles between species: S. flexneri vs. S. sonnei (2014-2015).
| Year | Serotype (n) | Phenotype | AMP | SXT | NAL | NOR | TAX | FIX |
|---|---|---|---|---|---|---|---|---|
| 2014 | S. flexneri | R | 84 | 113 | 111 | 80 | 9 | 12 |
| (119) | S | 35 | 6 | 8 | 39 | 110 | 107 | |
| S. sonnei | R | 4 | 32 | 35 | 23 | 1 | 3 | |
| (35) | S | 31 | 3 | 0 | 12 | 34 | 32 | |
| p value | <0.005 | 0.435 | 0.114 | 0.865 | 0.322 | 0.787 | ||
| Chi (df) | 38.6 (153) = 5905 | 0.61 (153) = 93.3 | 2.48 (153) = 379.4 | 0.02 (153) = 3.0 | 0.98 (153) = 150 | 0.07 (153) = 11 | ||
| 2015 | S. flexneri | R | 63 | 88 | 85 | 20 | 12 | 9 |
| (92) | S | 29 | 4 | 7 | 72 | 80 | 83 | |
| S. sonnei | R | 13 | 54 | 54 | 10 | 3 | 4 | |
| (55) | S | 42 | 1 | 1 | 45 | 52 | 51 | |
| p value | <0.005 | 0.412 | 0.133 | 0.603 | 0.141 | 0.603 | ||
| Chi (df) | 27.7 (146) = 4044 | 0.67 (146) = 98 | 2.24 (146) = 327 | 0.26 (146) = 38 | 2.16 (146) = 315 | 0.26 (146) = 38 |
R – resistant, S – susceptible, AMP – ampicillin, SXT – trimethoprim/sulfamethoxazole, NAL – nalidixic acid, NOR – norfloxacin, TAX – cefotaxime, FIX – cefixime.
Figure 3. Antimicrobial resistance profile of Shigella spp. by age group.
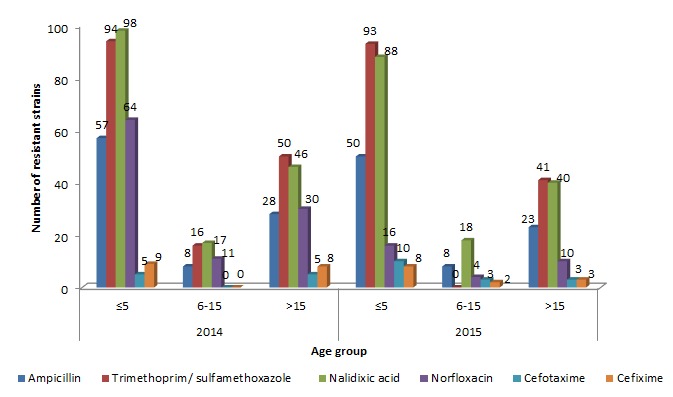
A subset of 73 MDR (2014 – S. flexneri (n=28), S. sonnei (n=1); 2015 – S. flexneri (n=35), S. sonnei (n=9)) isolates was randomly selected and characterized for the antimicrobial resistance mechanism in this study. Among the selected isolates (n=73), S. sonnei from the year 2014 were 100% resistant to all tested antibiotics, whereas in 2015, S. sonnei was found to be resistant to AMP (100%), SXT (55%), NAL (100%), TAX (67%) and FIX (33%) and none were found to be resistant to norfloxacin. The resistance rate among S. flexneri isolates for the year 2014 and 2015 were AMP (100 and 100%), SXT (100 and 77%), NAL (96 and 97%), NOR (78 and 26%), TAX (18 and 43%) and FIX (28 and 20%) respectively. Further molecular characterization of the isolates showed varied antimicrobial resistance genes. Screening of resistance by PCR in S. flexneri found dhfr1A, sulII, blaOXA, blaTEM, blaCTX-M-1, qnrB,S and AmpC genes while S. sonnei were found to have only dhfr1A, sulII, blaCTX-M-1 and qnrS genes respectively. None of the isolates were positive for qnrA gene. The results are illustrated in Table 3. Notably a higher number of resistance genes was found in individuals less than five years of age. AMR genes were predominantly seen in the AMP-SXT-NAL and AMP-SXT-NAL-NOR resistance phenotype (data not shown).
Table 3. Molecular characterization of multidrug resistant Shigella spp.
| Antibiotic | Year | % resistance (n=73) | Genes | AMR gene prevalence (n=73) | ||
|---|---|---|---|---|---|---|
| S. flexneri | S. sonnei | S. flexneri % (n=63) | S. sonnei % (n=10) | |||
| AMP | 2014 | 100 | 100 | blaOXA | 43 (12) | 0 (0) |
| AMP | 2015 | 100 | 100 | blaOXA | 51 (18) | 0 (0) |
| TMP-SXT | 2014 | 100 | 100 | dhfr1A | 100 (28) | 100 (1) |
| sulII | 43 (12) | 100 (1) | ||||
| TMP-SXT | 2015 | 77 | 55 | dhfr1A | 80 (28) | 100 (9) |
| sulII | 77 (27) | 56 (5) | ||||
| NAL | 2014 | 96 | 100 | qnrS | 14 (4) | 100 (1) |
| NOR | 78 | 100 | qnrB&S | 3 (1) | 0 | |
| NAL | 2015 | 97 | 100 | qnrS | 23 (8) | 0 (0) |
| NOR | 26 | 0 | qnrB&S | 0 (0) | 0 (0) | |
| TAX | 18 | 100 | blaTEM | 14 (4) | 0 (0) | |
| 2014 | AmpC | 7 (2) | 0 (0) | |||
| FIX | 28 | 100 | blaCTX-M-15 | 7 (2) | 100 (1) | |
| TAX | 43 | 67 | blaTEM | 34 (12) | 0 (0) | |
| 2015 | AmpC | 20 (7) | 0 (0) | |||
| FIX | 20 | 33 | blaCTX-M-15 | 3 (1) | 11 (1) | |
2014 – S. flexneri (n=28), S. sonnei (n=1); 2015 – S. flexneri (n=35), S. sonnei (n=9)
AMR – antimicrobial resistance, AMP – ampicillin, TMP-SXT – trimethoprim/sulfamethoxazole, NAL – nalidixic acid, NOR – norfloxacin, TAX – cefotaxime, FIX – cefixime.
Discussion
According to the World Health Organization (WHO), diarrheal disease was considered to be the second leading cause of mortality in children less than five years of age worldwide.12 Shigellosis represents a major cause of gastroenteritis, especially in low income countries. The isolation rate of Shigella varies from 2% to 6% in different studies from India.11
The present study documents a prevalence of 4.8% to 4.6% between the years 2014 to 2015.
S. flexneri was found to be the predominant serogroup and subtype 2 was the most common. Similar results were reported from Manipal and Puducherry in South India, where >90 percent of the isolates were S. flexneri.4,13 In an earlier study conducted in Vellore, South India (1997-2004) Shigella accounted for 5.4%, whereas S. flexneri was the most common serogroup (57.6%) followed by S. sonnei (31%).14
In recent years, an epidemiological shift in Shigella serogroups was reported. S. sonnei is now emerging in regions that used to be dominated by S. flexneri.13,15-17 This shift has been documented in many regions in Asia, Latin America, and the Middle East.17,18 S. flexneri was the most common serotype in the United States in the early 1960s but was replaced by S. sonnei during 1964-1968.19 Similarly, the present study documents a significant increase in the prevalence of S. sonnei from 2014 to 2015 with the simultaneous fall of S. flexneri. The exact reason behind this phenomenon is unclear.
However, the growing prevalence of S. sonnei in developing countries might be due to improvement in the quality of water and sanitation, which prevents the passive immunization derived from P. shigelloides, normally found in contaminated water. In addition, the amoeba A. castellani acts as a reservoir of S. sonnei, which allows their persistence in highly chlorinated environments while S. flexneri is not able to grow. Finally, S. sonnei has a greater ability to acquire resistance than S. flexneri, which provides an added advantage over S. flexneri, especially in areas with low antimicrobial usage.17
In addition, the incidence of non-typeable Shigella spp. seems to be increasing over years worldwide.13 About 5% of the isolates were found to be non-typeable during the study period. AMR was occasionally reported in the provisional/non-typeable serovars of Shigella.20 In this study 41% of the isolates were found to be MDR (data not shown). A similar trend of surfacing increase in non-typeable Shigella strains (13%) was reported in Kolkata.21
Worldwide, the majority of Shigella infections were found to occur in young children.13 A multicentric study from six Asian countries demonstrated the incidence of shigellosis with higher rates in patients younger than five years and patients above 40 years of age.8 The present study also evidences the similar trend in accordance with earlier reports.
In India, AMR was more common in Shigella than in other enteric bacteria.22 Although WHO recommends ceftriaxone, pivmecillinam and azithromycin for shigellosis due to the increase in fluoroquinolone resistant shigellae, the species was found to be resistant to one of the third generation cephalosporins and azithromycin. These have also been reported from other parts of south India.13 Ceftriaxone resistance was also observed by a multicentre study in eight Asian countries, among Shigella isolates.6
Generally, ampicillin resistance was encoded by OXA type β-lactamases. Overall, in the current study, 41% of the isolates harbored blaOXA genes. Among those exhibiting co-trimoxazole resistance, dhfr1A trimethoprim resistance gene was more prevalent, being considered the most common gene in the genus, followed by the sulII gene. Navia et al.23 demonstrated evidence of the aforementioned results of the current study.
Screening of Shigella isolates for the presence of PMQR genes showed 18% and 1% positivity for qnrS and co-occurrence of qnrB & S respectively. The presence of PMQR genes is a great concern because they can promote mutations within the quinolone resistance determining region (QRDR), which results in fluoroquinolone resistance, but may also disseminate to other species of Enterobacteriaceae.3 In this study, qnr genes were frequently seen in S. flexneri isolates than in S. sonnei, which could be due to the fact that qnrS gene was first detected in a S. flexneri serotype 1a in 2002, and later reported from Japan in 2005.24 At the same time, qnrS was the commonly found allele of the qnr genes, followed by qnrB in Shigella spp.25,26
Among Shigella spp. blaTEM was the most common β-lactamase gene followed by blaCTX-M-1, which was found to be the most frequent type of cephalosporinase conferring resistance to third generation cephalosporins especially to cefotaxime.6,11,27 Among the study isolates 22% and 7% were found to carry blaTEM and blaCTX-M-1 genes respectively. Similarly Taneja et al.6 reported blaCTX-M-1 (n=10) in S. flexneri isolates, thus indicating the consistent prevalence in India. Consequently, there are reports of Shigella spp. carrying different ESBL genes from other Asian countries. In clinical isolates, CTX-M encoding genes are found on plasmids; transposons or comprised cassettes in integrons.27 In addition, cephalosporin resistance was mediated by plasmid-mediated AmpC β-lactamases. In the present study, 12% of the isolates had AmpC genes. The AMR gene prevalence by species and year are given in Table 3.
This study provides evidence to show that resistance to antibiotics such as ampicillin and trimethoprim-sulfamethoxazole is not surprising among Shigella spp. Cephalosporin and quinolones such as norfloxacin are considered to be effective in the management of shigellosis. However, the increasing prevalence of ESBL and PMQR genes may have been an important driving force for selection of resistance to these antimicrobials. Moreover, the antimicrobial resistance pattern differs from place to place and between two regions in the same place.28
Many developing countries lack proper antibiotic resistance surveillance. Often changes in the incidence and antimicrobial resistance pattern from time to time make the treatment ineffective for shigellosis. Although the present study provides data only for a small number of isolates, the available information would help regional or national surveillance programs to document the resistance rates and trends of Shigella spp. in our locality.
Conclusion
The study highlights the growing importance of S. sonnei in a developing country like India, and emphasizes that antimicrobial resistant Shigella spp. is a raising problem among enteric pathogens in India. In our study, most of the isolates were resistant to ampicillin, trimethoprim-sulfamethoxazole and nalidixic acid. The presence of ESBL producing genes such as blaCTX-M-1 in the study isolates further indicates the threat of spreading resistance to third-generation cephalosporins, as cephalosporins are used for treatment in children, where quinolones are usually not recommended. Notably, an increasing number of non-agglutinable Shigella highlights the need for effective surveillance of circulating Shigella species and serotypes. Furthermore, monitoring changing trends in antibiotic resistance over periods of time is necessary for effective treatment.
Footnotes
Authors’ contributions statement: DP analyzed, interpreted data and wrote the manuscript. SA helped write the manuscript, analyzed and interpreted data. DP and RG carried out bench work, generated data. BV, KW and VP critically revised and approved the manuscript. All authors read and approved the final version of the manuscript.
Conflicts of interest: All authors – none to declare.
Funding: The study has been funded by the Indian Council of Medical Research, New Delhi, India (ref. no: AMR/TF/55/13ECDII dated 23/10/2013).
References
- 1.Khaghani S, Shamsizadeh A, Nikfar R, Hesami A. Shigella flexneri: a three-year antimicrobial resistance monitoring of isolates in a Children Hospital, Ahvaz, Iran. Iran J Microbiol. 2014;6:225–9. [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff KL, Nataro JP, Blackwelder WC, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–22. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 3.Nüesch-Inderbinen M, Heini N, Zurfluh K, Althaus D, Hächler H, Stephan R. Shigella antimicrobial drug resistance mechanisms, 2004-2014. Emerg Infect Dis. 2016;22:1083–5. doi: 10.3201/eid2206.152088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh S, Pazhani GP, Chowdhury G, et al. Genetic characteristics and changing antimicrobial resistance among Shigella spp. isolated from hospitalized diarrhoeal patients in Kolkata, India. J Med Microbiol. 2011;60:1460–6. doi: 10.1099/jmm.0.032920-0. [DOI] [PubMed] [Google Scholar]
- 5.Pazhani GP, Niyogi SK, Singh AK, et al. Molecular characterization of multidrug-resistant Shigella species isolated from epidemic and endemic cases of shigellosis in India. J Med Microbiol. 2008;57:856–63. doi: 10.1099/jmm.0.2008/000521-0. [DOI] [PubMed] [Google Scholar]
- 6.Taneja N, Mewara A, Kumar A, Verma G, Sharma M. Cephalosporin-resistant Shigella flexneri over 9 years (2001-09) in India. J Antimicrob Chemother. 2012;67:1347–53. doi: 10.1093/jac/dks061. [DOI] [PubMed] [Google Scholar]
- 7.Mauldin PD, Salgado CD, Hansen IS, Durup DT, Bosso JA. Attributable hospital cost and length of stay associated with health care-associated infections caused by antibiotic-resistant gram-negative bacteria. Antimicrob Agents Chemother. 2010;54:109–15. doi: 10.1128/AAC.01041-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Seidlein L, Kim DR, Ali M, et al. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med. 2006;3:e353. doi: 10.1371/journal.pmed.0030353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kotloff KL, Winickoff JP, Ivanoff B, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77:651–66. [PMC free article] [PubMed] [Google Scholar]
- 10.Bopp CA, Brenner FW, Fields PL, et al. Escherichia, Shigella, and Salmonella. In: Murray PR, Baron EJ, Jorgensen J, Pfaller MA, Yolken RH, editors. Manual of clinical microbiology. (8th ed.) Washington, DC: American Society for Microbiology; 2003. pp. 654–71. [Google Scholar]
- 11.Muthuirulandi Sethuvel DP, Anandan S, Devanga Ragupathi NK, Veeraraghavan B. Identification of typical class 1 and class 2 integron gene cassettes in clinical isolates of MDR Shigella flexneri in South Indian population. Br Microbiol Res J. 2015;10:1–6. [Google Scholar]
- 12.Sambe-Ba B, Espié E, Faye ME, Timbiné LG, Sembene M, Gassama-Sow A. Community-acquired diarrhea among children and adults in urban settings in Senegal: clinical, epidemiological and microbiological aspects. BMC Infect Dis. 2013;13:580. doi: 10.1186/1471-2334-13-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taneja N, Mewara A. Shigellosis: epidemiology in India. Indian J Med Res. 2016;143:565–76. doi: 10.4103/0971-5916.187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jesudason MV. Shigella isolation in Vellore, south India (1997-2001) Indian J Med Res. 2002;115:11–3. [PubMed] [Google Scholar]
- 15.De Lappe N, O’Connor J, Garvey P, McKeown P, Cormican M. Ciprofloxacin-resistant Shigella sonnei associated with travel to India. Emerg Infect Dis. 2015;21:894–6. doi: 10.3201/eid2105.141184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das A, Natarajan M, Mandal J. The emergence of quinolone resistant Shigella sonnei, Pondicherry, India. PLoS One. 2016;11:e0160290. doi: 10.1371/journal.pone.0160290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson CN, Duy PT, Baker S. The rising dominance of Shigella sonnei: an intercontinental shift in the etiology of bacillary dysentery. PLoS Negl Trop Dis. 2015;9:e0003708. doi: 10.1371/journal.pntd.0003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qu M, Zhang X, Liu G, et al. An eight-year study of Shigella species in Beijing, China: serodiversity, virulence genes, and antimicrobial resistance. J Infect Dev Ctries. 2014;8:904–8. doi: 10.3855/jidc.3692. [DOI] [PubMed] [Google Scholar]
- 19.Khan E, Jabeen K, Ejaz M, Siddiqui J, Shezad MF, Zafar A. Trends in antimicrobial resistance in Shigella species in Karachi, Pakistan. J Infect Dev Ctries. 2009;3:798–802. doi: 10.3855/jidc.500. [DOI] [PubMed] [Google Scholar]
- 20.Dutta S, Jain P, Nandy S, Matsushita S, Yoshida S. Molecular characterization of serologically atypical provisional serovars of Shigella isolates from Kolkata, India. J Med Microbiol. 2014;63:1696–703. doi: 10.1099/jmm.0.081307-0. [DOI] [PubMed] [Google Scholar]
- 21.Nair GB, Ramamurthy T, Bhattacharya MK, et al. Emerging trends in the etiology of enteric pathogens as evidenced from an active surveillance of hospitalized diarrhoeal patients in Kolkata, India. Gut Pathog. 2010;2:4. doi: 10.1186/1757-4749-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mamatha B, Rituparna C. Decreased susceptibility to antimicrobials among Shigella flexneri isolates in Manipal, South India--a 5 year hospital based study. Southeast Asian J Trop Med Public Health. 2012;43:1447–51. [PubMed] [Google Scholar]
- 23.Navia MM, Capitano L, Ruiz J, et al. Typing and characterization of mechanisms of resistance of Shigella spp. isolated from feces of children under 5 years of age from Ifakara, Tanzania. J Clin Microbiol. 1999;37:3113–7. doi: 10.1128/jcm.37.10.3113-3117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pu XY, Pan JC, Wang HQ, Zhang W, Huang ZC, Gu YM. Characterization of fluoroquinolone-resistant Shigella flexneri in Hangzhou area of China. J Antimicrob Chemother. 2009;63:917–20. doi: 10.1093/jac/dkp087. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Hu L, Pan Y, et al. Prevalence of plasmid-mediated quinolone resistance determinants in association with β-lactamases, 16S rRNA methylase genes and integrons amongst clinical isolates of Shigella flexneri. J Med Microbiol. 2012;61:1174–6. doi: 10.1099/jmm.0.042580-0. [DOI] [PubMed] [Google Scholar]
- 26.Cui X, Yang C, Wang J, et al. Antimicrobial resistance of Shigella flexneri serotype 1b isolates in China. PLoS One. 2015;10:e0129009. doi: 10.1371/journal.pone.0129009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auda IG. Occurrence of CTX-M-I and CTX-M-III genes on plasmids of Shigella species isolated from cases of diarrhea in Baghdad. World J Pharm Res. 2014;3:1273–80. [Google Scholar]
- 28.Muthuirulandi Sethuvel DP, Devanga Ragupathi NK, Anandan S, Veeraraghavan B. Update on: Shigella new serogroups/serotypes and their antimicrobial resistance. Lett Appl Microbiol. 2017;64:8–18. doi: 10.1111/lam.12690. [DOI] [PubMed] [Google Scholar]


