Abstract
Tyrosinase‐related protein 1 (TYRP1) is one of three tyrosinase‐like glycoenzymes in human melanocytes that are key to the production of melanin, the compound responsible for the pigmentation of skin, eye, and hair. Difficulties with producing these enzymes in pure form have hampered the understanding of their activity and the effect of mutations that cause albinism and pigmentation disorders. Herein we show that the typical tyrosinase‐like subdomain of TYRP1 contains two zinc ions in the active site instead of copper ions as found in tyrosinases, which explains why TYRP1 does not exhibit tyrosinase redox activity. In addition, the structures reveal for the first time that the Cys‐rich subdomain, which is unique to vertebrate melanogenic proteins, has an epidermal growth factor‐like fold and is tightly associated with the tyrosinase subdomain. Our structures suggest that most albinism‐related mutations of TYRP1 affect its stability or activity.
Keywords: albinism, human tyrosinase, melanogenesis, structure elucidation, zinc–copper enzymes
In humans, three enzymes are involved in the biosynthesis of melanin (Figure S1 in the Supporting Information), the primary determinant of the color of the skin, hair and iris of the eye.1 Tyrosinase (TYR) is the critical and rate‐limiting enzyme; it catalyzes the hydroxylation and subsequent oxidation of tyrosine. Tyrosinase‐related protein 2 (TYRP2) is a tautomerase,2 and tyrosinase‐related protein 1 (TYRP1) has been suggested to catalyze the oxidation of 5,6‐dihydroxyindole‐2‐carboxylic acid (DHICA) in mice,3 although this activity has been challenged in humans.4 All three enzymes are metal‐containing glycoproteins localized in melanosomes where melanin synthesis takes place. They share approximately 40 % amino acid sequence identity and contain four conserved regions: an N‐terminal signal peptide, an intramelanosomal domain, a single transmembrane α‐helix, and a small, flexible C‐terminal cytoplasmic domain (Figure 1 A). The intramelanosomal domain contains a Cys‐rich subdomain, unique to mammalian tyrosinases, and a tyrosinase‐like subdomain with a binuclear metal‐ion‐binding sequence motif. Mutations in the TYR or TYRP1 genes result in oculocutaneous albinism (OCA), a group of autosomal recessive disorders characterized by reduced production of melanin in skin, hair, and eyes, with OCA3 resulting from TYRP1 mutations. In addition, TYR and TYRP1 variants are significantly associated with risk of melanoma,5 a malignant tumor of melanocytes that, because of its aggressive nature, causes the majority of deaths related to skin cancer.6 Inhibition of melanogenesis may be an efficient therapeutic strategy, as evidenced by phase 1 clinical trials with a recombinant human monoclonal antibody against TYRP1.7
Figure 1.
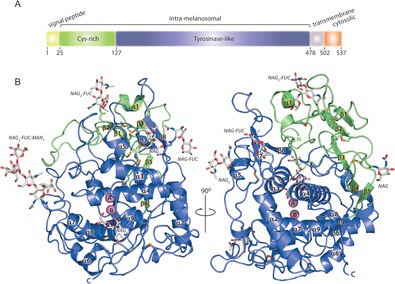
A) TYRP1 contains 4 conserved regions: the signal peptide (yellow, residues 1–24), the intramelanosomal (or luminal) domain (25–477, subdivided into the Cys‐rich (green, 25–126) and the tyrosinase‐like (blue, 127–477) subdomains), the transmembrane α‐helix (gray, 478–501), and the cytoplasmic domain (orange, 502–537). B) Overall structure of the intramelanosomal domain, same coloring as in (A). The six Asn sites with N‐linked sugars (96, 104, 181, 304, 350, and 385) are shown as sticks and the disulfide bonds are shown in orange. The two metal ions (A and B) are shown as pink spheres.
Although we recently succeeded in the recombinant production of intramelanosomal human TYR,8 its crystallization was not suitable for structure determination. Herein we report the crystal structure of intramelanosomal human TYRP1 and in complex with tyrosinase substrates and inhibitors, which reveal for the first time the structure of a mammalian tyrosinase family member. We show that the active site contains two zinc ions, rather than copper ions as in tyrosinases, which makes it unlikely that human TYRP1 is a redox enzyme, as had been assumed for the last three decades.
The 2.35 Å resolution crystal structure of intramelanosomal TYRP1 (Figure 1 B, Table S1) reveals a compact globular shape, with tight interactions between the Cys‐rich and tyrosinase subdomains. Furthermore, all six Asn N‐glycosylation motifs bear glycans (96, 104, 181, 304, 350, and 385) (Figure 1 B and Figure S2). The tyrosinase‐like subdomain (residues 127–477) has the typical tyrosinase fold, consisting of a four‐helix bundle (α3, α4, α8, and α9) connected by long loops. It is most similar to the bacterial Bacillus megaterium tyrosinase (BmTYR, PDB entry 3NM8),9 with 32 % sequence identity and an rmsd of 1.6 Å for 263 aligned amino acids. The most conspicuous differences are three long insertions that cluster together at one side of the tyrosinase‐like subdomain (Figure 2; residues 155–182, 199–204, and 291–300). These loops extend away from the entrance to the active site, and interact with the Cys‐rich subdomain. Two disulfide bonds (C258–C261 and C290–C303) stabilize the tyrosinase subdomain (disulfide bonds 6 and 7 in Figure S2).
Figure 2.
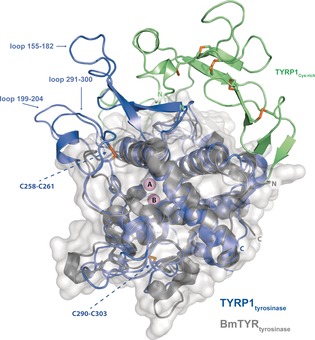
Superposition of the TYRP1 and Bacillus megaterium TYR crystal structures. BmTYR is represented as a gray structure and light gray surface (PDB 3NM8).9 The tyrosinase subdomain of TYRP1 is shown in blue and the Cys‐rich subdomain in light green. Loops with different lengths between TYRP1 and BmTYR are indicated. Disulfide bonds are shown in orange and the two metal ions (A and B) in pink. The disulfide residues stabilizing the TYRP1 tyrosinase subdomain are indicated as orange sticks.
The core of the Cys‐rich subdomain (residues 25–126) has an epidermal growth factor (EGF)‐like fold (SCOP family g.3.11.1) similar to the structure of the human epidermal growth factor (Figure S3), formed by two pairs of short antiparallel β‐strands (β1/β2 (residues 62–66/98–102) and β4/β5 (116–118/124–126; Figure 1 B) from which long loops emerge. They are stabilized by disulfide bonds that follow the [C1–C3, C2–C4, C5–C6] signature pattern of EGF‐like structures,10 corresponding to cysteine residues 42–65, 56–99, 101–110. The subdomain associates with the tyrosinase‐like subdomain via its N‐terminus and the long 67–97 loop extending from the core. The subdomain is located far from the active site, at the opposite side of the molecule (Figure 1 B), making a direct effect on the activity of TYRP1 unlikely. Although the Cys‐rich subdomain has been suggested to be involved in oligomerization,11 we did not find evidence for this, since purified intramelanosomal domains of both TYRP1 and TYR elute as monomers on size‐exclusion chromatography (Figure S4A), and co‐elution of TYRP1 and TYR did neither produce a heterodimer (Figure S4B).
TYRP1 has a binuclear metal‐binding site that resembles the classical type‐3 binuclear copper‐binding site of hemocyanins and tyrosinases (Figure 3), but the nature of the bound metal ions has been unclear.12 Using X‐ray fluorescence, we found that TYRP1 crystals contained zinc, while TYR crystals contained copper (Figure S5). The presence of zinc in TYRP1 was further confirmed by X‐ray anomalous dispersion data collected at the zinc absorption edge, which showed that the TYRP1 zinc ions occupy the positions of copper ions in tyrosinases (Figure S6 and Table S1). Both zinc ions in the active site are bound with a nearly planar trigonal geometry, slightly out of the plane defined by the Nϵ2 atoms of their ligands. The distance between the ZnA and ZnB ions (Figure 3) is 3.5±0.1 Å. A bridging water molecule or hydroxide ion can be modeled with a distance of 2.1±0.1 Å to the two zinc ions (Figure 3 A and Figure S7A). Although we carried out repeated productions of TYRP1 and TYR samples under identical cell culture conditions and in the absence of external sources of zinc or copper ions in the media, the two proteins consistently contain different ions, unambiguously proving that TYR is a copper‐binding enzyme while TYRP1 is a zinc‐binding enzyme.
Figure 3.
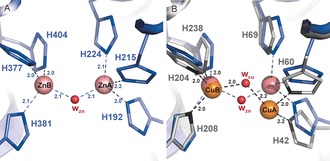
Metal‐ion binding in the TYRP1 active site. A) Close‐up of the TYRP1 active site. Zn ions are shown as pink spheres with the bridging water molecule in red (wzn). Metal coordination distances are labeled. B) Superposition of TYRP1 and BmTYR active sites. The Cu ions present in BmTYR are shown in orange, and the bridging water molecule (wcu) in red. Metal‐coordination distances are displayed in dark gray and labeled. The location of metal B is better conserved than metal A, in agreement with previous observations14 (see Figure 5). Numbering of His residues is indicated for TYRP1 (blue) and BmTYR (gray).
Generally, it has been assumed that TYRP1 is a DHICA oxidase.13 Yet, human TYRP1 was found to be devoid of DHICA activity,4 and also in our studies the intramelanosomal domain of TYRP1 did not display any DHICA activity nor tyrosine hydroxylase or l‐DOPA oxidase activity (Figure 4). The nature of the metal ions in the active site is partly responsible for these differences in activity, since exchanging the Zn2+/Cu2+ cofactors of TYRP1/TYR for Cu2+/Zn2+ conferred TYR‐level DHICA oxidase activity to TYRP1, but no significant l‐DOPA oxidase and tyrosine hydroxylase activities. Only a prolonged (up to 160 min) on‐plate tyrosinase activity assay with TYRP1‐Cu did show some low tyrosinase activity (Figure 4 D). In contrast, TYR‐Zn had almost no activity, with the residual activity likely resulting from incomplete removal of the copper ions. Amino acid substitutions in the active site affect the activity profile only to a minor extent. A triple mutant, which had the three non‐identical residues replaced by the corresponding ones of TYR (Y362F/R374S/T391V; TYRP1‐3M), had similar activities as native TYRP1, both in the Zn‐ and Cu‐substituted forms. Thus, our results suggest that it is unlikely that TYRP1 is a redox enzyme.
Figure 4.
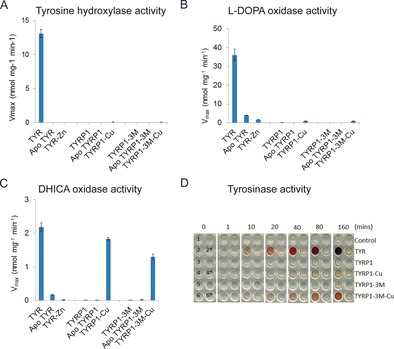
A)–C) Activity assays of TYR,8 TYRP1, and their corresponding apo‐proteins recharged with Zn or Cu ions. TYRP1‐3M is the triple Y362F/R374S/T391V mutant. For details see Table S2. D) Colorimetric activity assay. Enzymes added are indicated on the right. The pink pigment is the tyrosinase activity product dopaquinone in complex with MBTH, which appears after ca. 1 min for TYR, ca. 20 min for TYRP1‐3M‐Cu, and ca. 40 min for TYRP1‐Cu, and accumulates over time. Well 1 (W1) is a blank control. W2*, W4*, and W6* contain tyrosinase inhibitors as negative controls. W3 and W5 show no pigment, indicating that TYRP1 and TYRP1‐3M have no tyrosinase activity. W6 is darker than W4 at the same time point, indicating that TYRP1‐3M‐Cu presents higher tyrosinase activity than TYRP1‐Cu.
TYRP1 binds typical tyrosinase substrates and inhibitors (Figure 5, Table S1). Importantly, l‐tyrosine and the l‐DOPA analogue l‐mimosine do not directly interact with the zinc ions, but their aromatic hydroxy and keto‐groups are hydrogen‐bonded to the water molecule bridging the two zinc ions (Figure S7B). Additional interactions include offset aromatic stacking interactions with His381, and hydrogen bonding interactions of their carboxylate group with Arg374 and Ser394 (Figure 5 A,B). The electron density of l‐ mimosine is better defined than that of l‐tyrosine, suggesting that the interactions of the ϵ‐hydroxy group increase the binding affinity of mimosine. In contrast, the metal chelator tropolone does have direct interactions with the ZnA ion (Figure 5 C). Interestingly, ZnA has shifted by 1.5 Å towards tropolone compared to native TYRP1, indicating that the metal ion site is somewhat flexible. The skin‐whitening agent kojic acid binds with its ring hydroxy and keto groups at approximately 3 Å from the zinc ions, which is too far for good coordination (Figure 5 D). Interestingly, the amino acid differences between the active sites of TYRP1 and TYR do not affect the binding modes of these compounds, as shown by crystal structures of the TYRP1‐3M mutant (see above) with/without bound l‐mimosine, tropolone and kojic acid (Figure S8, Table S1). Despite removal of hydrogen‐bonding interactions by the mutations, the compounds bind in a similar conformation as in native TYRP1. Apparently, the stacking interaction with His381 and the hydrogen bonds of aromatic keto‐ and hydroxy groups are sufficient for binding. This finding implies that recognition of the substrate's carboxylate group is not essential and that, in addition to DHICA, TYRP1 may also be active on 5,6‐dihydroxyindole (DHI) (Figure S1).
Figure 5.
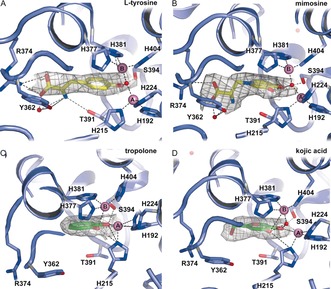
Binding of tyrosinase substrates and inhibitors. l‐tyrosine (A) and mimosine (B) bind with their carboxylate groups to R374. Tropolone (C) and kojic acid (D) bind in the same position. Each compound is shown with its 2 F o−F c electron density map (gray wire) contoured at 0.9 σ. Zinc ions are shown as pink spheres and water molecules as red spheres. Contacts of the compounds with TYRP1 residues and the Zn ions are shown as dashed lines.
Fifteen OCA3‐related mutations in the TYRP1 gene have so far been found, including 8 point mutations (C30R, R93C, H215Y, T253M, C290Y, R356Q, M452V, and P513R; Figure S9).15 Most of these residues contribute to the stability. The C30–C41 disulfide bond attaches helix α1 to the core of the Cys‐rich subdomain, the C290–C303 disulfide bond stabilizes the 290–303 loop, and R356 is part of an extensive buried hydrogen‐bonding network. H215 is a ZnA ligand, and replacement by Tyr may reduce the enzyme's zinc‐binding affinity and activity. T253M and M452V are missense mutations found in Caucasian patients that are yet to be confirmed as pathological mutations.16
T253 is located on a surface loop, far away from the active site, and the introduction of a large apolar methionine at this position may lead to local unfolding and/or aggregation. M452 is located in the interface between the Cys‐rich and tyrosinase‐like subdomains. Replacement by Val may reduce part of its hydrophobic interactions, which may have a (probably minor) effect on the stability of the enzyme. The R93C substitution causes blond hair in normally black‐haired Melanesian individuals.17 R93 is near a cluster of four disulfide bonds in the Cys‐rich subdomain (Figure S9), and replacement by a cysteine could interfere with correct disulfide bond formation or even correct folding. Finally, the P513R mutation is in the cytosolic tail domain, which is critical for targeting melanogenic proteins to the melanosomal membrane.18 The P513R replacement may thus interfere with translocation of TYRP1 to the melanosomal membrane.
In summary, the crystal structure of intramelanosomal human tyrosinase‐related protein 1 (TYRP1) has revealed for the first time the structure of a mammalian tyrosinase family member. We provide compelling evidence for the presence of two zinc ions in the active site, which makes it unlikely that human TYRP1 is a redox enzyme, in contrast to what has been assumed for the last three decades. Which reaction TYRP1 does catalyze is unclear at this moment, although the effect of mutations in the TYRP1 gene suggest the utmost importance of TYRP1 for the biosynthesis of melanin.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We thank ESRF beamline scientists D. de Sanctis (ID29), A. Popov (ID23‐1), and M. Bowler (MASSIF‐1) for assistance during data collection, and F. Garzoni (Eukaryotic Expression Platform at EMBL Grenoble) for help with protein expression in insect cells. The Structural Biology Group at the ESRF and the Groningen Biomolecular Sciences and Biotechnology Institute (University of Groningen) supported this work.
X. Lai, H. J. Wichers, M. Soler-Lopez, B. W. Dijkstra, Angew. Chem. Int. Ed. 2017, 56, 9812.
Contributor Information
Dr. Montserrat Soler‐Lopez, Email: montserrat.soler-lopez@esrf.fr.
Prof. Dr. Bauke W. Dijkstra, Email: b.w.dijkstra@rug.nl.
References
- 1. Rees J. L., Annu. Rev. Genet. 2003, 37, 67–90. [DOI] [PubMed] [Google Scholar]
- 2. Yamaguchi Y., Hearing V. J., Cold Spring Harbor Perspect. Med. 2014, 4, a017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kobayashi T., Urabe K., Winder A., Jiménez-Cervantes C., Imokawa G., Brewington T., Solano F., García-Borrón J. C., Hearing V. J., EMBO J. 1994, 13, 5818–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boissy R. E., Sakai C., Zhao H., Kobayashi T., Hearing V. J., Exp. Dermatol. 1998, 7, 198–204. [DOI] [PubMed] [Google Scholar]
- 5. Ghanem G., Fabrice J., Mol. Oncol. 2011, 5, 150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siegel R. L., Miller K. D., Jemal A., Ca-Cancer J. Clin. 2016, 66, 7–30. [DOI] [PubMed] [Google Scholar]
- 7. Khalil D. N., Postow M. A., Ibrahim N., Ludwig D. L., Cosaert J., Kambhampati S. R. P., Tang S., Grebennik D., Kauh J. S. W., Lenz H.-J., et al., Clin. Cancer Res. 2016, 22, 5204–5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lai X., Soler-Lopez M., Wichers H. J., Dijkstra B. W., PLoS One 2016, 11, e0161697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sendovski M., Kanteev M., Ben-Yosef V. S., Adir N., Fishman A., J. Mol. Biol. 2011, 405, 227–237. [DOI] [PubMed] [Google Scholar]
- 10. Lu H. S., Chai J. J., Li M., Huang B. R., He C. H., Bi R. C., J. Biol. Chem. 2001, 276, 34913–34917. [DOI] [PubMed] [Google Scholar]
- 11. Jackson I. J., Chambers D. M., Tsukamoto K., Copeland N. G., Gilbert D. J., Jenkins N. A., Hearing V., EMBO J. 1992, 11, 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Furumura M., Solano F., Matsunaga N., Sakai C., Spritz R. A., Hearing V. J., Biochem. Biophys. Res. Commun. 1998, 242, 579–585. [DOI] [PubMed] [Google Scholar]
- 13. Sarangarajan R., Boissy R. E., Pigment Cell Res. 2001, 14, 437–444. [DOI] [PubMed] [Google Scholar]
- 14. Deri B., Kanteev M., Goldfeder M., Lecina D., Guallar V., Adir N., Fishman A., Sci. Rep. 2016, 6, 34993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stenson P. D., Mort M., Ball E. V., Howells K., Phillips A. D., Thomas N. S., Cooper D. N., Genome Med. 2009, 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hutton S. M., Spritz R. A., Invest. Ophthalmol. Visual Sci. 2008, 49, 868–872. [DOI] [PubMed] [Google Scholar]
- 17. Kenny E. E., Timpson N. J., Sikora M., Yee M.-C., Moreno-Estrada A., Eng C., Huntsman S., Burchard E. G., Stoneking M., Bustamante C. D., et al., Science 2012, 336, 554–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calvo P. A., Frank D. W., Bieler B. M., Berson J. F., Marks M. S., J. Biol. Chem. 1999, 274, 12780–12789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


