Abstract
GPR151 is a G‐protein coupled receptor for which the endogenous ligand remains unknown. In the nervous system of vertebrates, its expression is enriched in specific diencephalic structures, where the highest levels are observed in the habenular area. The habenula has been implicated in a range of different functions including behavioral flexibility, decision making, inhibitory control, and pain processing, which makes it a promising target for treating psychiatric and neurological disease. This study aimed to further characterize neurons expressing the Gpr151 gene, by tracing the afferent connectivity of this diencephalic cell population. Using pseudotyped rabies virus in a transgenic Gpr151‐Cre mouse line, monosynaptic afferents of habenular and thalamic Gpr151‐expressing neuronal populations could be visualized. The habenular and thalamic Gpr151 systems displayed both shared and distinct connectivity patterns. The habenular neurons primarily received input from basal forebrain structures, the bed nucleus of stria terminalis, the lateral preoptic area, the entopeduncular nucleus, and the lateral hypothalamic area. The Gpr151‐expressing neurons in the paraventricular nucleus of the thalamus was primarily contacted by medial hypothalamic areas as well as the zona incerta and projected to specific forebrain areas such as the prelimbic cortex and the accumbens nucleus. Gpr151 mRNA was also detected at low levels in the lateral posterior thalamic nucleus which received input from areas associated with visual processing, including the superior colliculus, zona incerta, and the visual and retrosplenial cortices. Knowledge about the connectivity of Gpr151‐expressing neurons will facilitate the interpretation of future functional studies of this receptor.
Keywords: habenula, rabies, thalamus, RRID: IMSR_JAX:000664, RRID: IMSR_JAX:024109, RRID: AB_10743815, RRID: AB_2571870, RRID: SCR_003070
1. INTRODUCTION
The habenula is a paired epithalamic brain region whose structure and connectivity are largely conserved throughout the vertebrate subphylum (Bianco & Wilson, 2009; Díaz, Bravo, Rojas, & Concha, 2011; Stephenson‐Jones, Floros, Robertson, & Grillner, 2011). In mammals, it can be further divided into two major subnuclei designated the medial and lateral habenula, homologous to the dorsal and ventral habenula in fish (Amo et al., 2010). In the mouse brain, the medial habenula receives input from the medial septal nucleus, the triangular nucleus of septum, the nucleus of the diagonal band, the bed nucleus of the anterior commissure, and the septofimbrial nucleus (Qin & Luo, 2009). The efferent axons from the medial habenula form the core of the fasciculus retroflexus fiber bundle and terminate in the interpeduncular nucleus. The afferent projections to the lateral habenula have not been investigated in detail in the mouse brain. In rats however, the most prominent afferent connections arise in the lateral preoptic area, the entopeduncular nucleus and the lateral hypothalamic area, and to a minor extent in the lateral septal nucleus, the nucleus of the diagonal band, the prefrontal cortex, the median raphe, and the ventral tegmental area (Greatrex & Phillipson, 1982; Herkenham & Nauta, 1977; Vertes, Fortin, & Crane, 1999; Warden et al., 2012). Fibers from the lateral habenular neurons travel in the outer layer of fasciculus retroflexus and terminate primarily in the hypothalamus, the ventral tegmental area, the rostromedial tegmental nucleus, the rhabdoid nucleus, the median and dorsal raphe nuclei, and the pontine central gray (Broms, Antolin‐Fontes, Tingström, & Ibañez‐Tallon, 2015; Quina et al., 2014).
The habenula has been implicated in several apparently distinct brain functions, such as maternal behavior (Corodimas, Rosenblatt, & Morrell, 1992), aggressive behavior (Chou et al., 2016; Golden et al., 2016), social play (van Kerkhof, Damsteegt, Trezza, Voorn, & Vanderschuren, 2013), drug seeking (Jhou et al., 2013), noxious substance aversion (Donovick, Burright, & Zuromski, 1970; Fowler & Kenny, 2013), decision making (Stopper & Floresco, 2013), behavioral flexibility (Baker, Raynor, Francis, & Mizumori, 2016), and pain modulation (Shelton, Becerra, & Borsook, 2012). Subdivision of the habenular complex into smaller regions, defined by the expression of certain molecules or neuronal connectivity patterns, is a necessary strategy for deepening our understanding of its role in physiology and disease. This approach has been useful to target neuronal subpopulations and investigate their anatomical and functional features, especially in the medial habenula (Chou et al., 2016; Gardon et al., 2014; Hsu, Morton, Guy, Wang, & Turner, 2016; Hsu et al., 2014; Kobayashi et al., 2013).
G‐protein coupled receptor 151 (GPR151; also known as PGR7, GALR4, GPCR‐2037, and galanin‐receptor like) is an orphan receptor phylogenetically related to the galanin receptor family (Berthold, Collin, Sejlitz, Meister, & Lind, 2003; Ignatov, Hermans‐Borgmeyer, & Schaller, 2004; Vassilatis et al., 2003). In this work, we use capital letters to refer to GPR151 protein while the term Gpr151 is used for mRNA, promoter or gene. In a previous study using immunohistochemistry, we found that the protein expression of GPR151 was limited to habenular neurons targeting the interpeduncular nucleus, the rostromedial tegmental nucleus, the rhabdoid nucleus, the median raphe, the caudal dorsal raphe, and the dorsal tegmentum in rats and mice (Broms et al., 2015). The protein expression pattern was similar in zebrafish, with strong expression in habenular projections to the interpeduncular nucleus and ventral median raphe, and this distinct and phylogenetically conserved expression pattern suggests an important role of GPR151 in modulating habenular function.
Weak Gpr151 mRNA expression can also be detected in various thalamic areas such as the paraventricular, the reunions, the rhomboid, the central lateral, and the parafascicular nuclei (Ignatov et al., 2004; Lein et al., 2006; Wagner, French, & Veh, 2014). Surprisingly, these structures display no or very weak immunohistochemical staining of GPR151 protein (Broms et al., 2015). In a recent study, specific ablation of adult neurons exhibiting Gpr151 promoter activity resulted in several behavioral alterations, such as increased anxiety, inability to habituate to repeated exposure to a novel environment, increased impulsivity, impaired pre‐pulse inhibition, and spatial memory deficits (Kobayashi et al., 2013). Decision making was also affected, where lesioned mice preferred effortless and immediate small rewards over larger rewards that required the animals to either wait longer for the reward or to climb an obstacle. The observed behavioral changes were accompanied by structural and neurochemical remodeling of the habenulo‐interpeduncular system. The authors concluded that the Gpr151‐expressing neurons may play an important role in inhibitory control and cognition‐dependent executive functioning (Kobayashi et al., 2013).
In addition to the expression in the above mentioned juxtaposed diencephalic brain regions of the brain, Gpr151 mRNA is also detected in certain sensory ganglia and cranial nerve nuclei (Ignatov et al., 2004). In the spinal nerve ligation model of chronic neuropathic pain and in a model of heat induced pain, the expression of Gpr151 mRNA was strongly upregulated in dorsal root ganglia (Reinhold et al., 2015; Yin, Deuis, Lewis, & Vetter, 2016). Gpr151 mRNA expression is also increased in the facial nucleus of the brainstem in response to damage of the facial nerve (Gey et al., 2016). These findings suggest that GPR151, besides having a possible role in modulation of habenulo‐thalamic functions, might also be important for pain modulation in cranial and peripheral nerves. However, a recent study utilizing mice carrying a Gpr151 loss‐of‐function mutation could not confirm this hypothesis (Holmes et al., 2017).
In a previous investigation (Broms et al., 2015), we characterized the efferent connectivity of habenular neurons expressing the GPR151 protein using immunohistochemistry. The aim of the current study was to identify the afferent regions projecting to Gpr151‐expressing neurons using monosynaptic pseudotyped rabies virus tracing, to gain further knowledge about this diencephalic neuronal subpopulation. Using pseudotyped rabies virus, Gpr151‐expressing neurons can be targeted specifically, avoiding some of the problems associated with conventional tracers, such as uptake in fibers of passage and diffusion of tracer to neighboring structures (Watabe‐Uchida, Zhu, Ogawa, Vamanrao, & Uchida, 2012; Wickersham et al., 2007). A detailed anatomical characterization of the input and output of Gpr151‐expressing neurons could facilitate future functional studies of this orphan receptor.
2. MATERIALS AND METHODS
2.1. Nomenclature
In this article, we will primarily use the nomenclature and area delineations (Table 3) of Franklin and Paxinos (2008). We use the nomenclature of Wagner, Stroh, and Veh, (2014) for the subnuclear organization of the habenula. The categorization of brain areas was done according to the hierarchy of the Allen Mouse Brain Atlas (Lein et al., 2006).
Table 3.
Abbreviations
| Abbreviation | Area | Abbreviation | Area |
|---|---|---|---|
| 3V | 3rd ventricle | Me | medial amygdaloid nucleus |
| aca | anterior commissure, anterior part | MG | medial geniculate nucleus |
| Acb | accumbens nucleus | MHb | medial habenular nucleus |
| AD | anterodorsal thalamic nucleus | MHbD | dorsal subnucleus of the MHb |
| AH | anterior hypothalamic area | MHbS | superior subnucleus of the MHb |
| AI | agranular insular cortex | MHbV | ventral part of the MHb |
| Arc | arcuate hypothalamic nucleus | MHbVc | central subnucleus of the MHbV |
| Au | auditory cortex | MHbVl | lateral subnucleus of the MHbV |
| AV | anteroventral thalamic nucleus | MHbVm | medial subnucleus of the MHbV |
| BAC | bed nucleus of the anterior commissure | MnPO | median preoptic nucleus |
| BL | basolateral amygdaloid nucleus | MnR | median raphe nucleus |
| BM | basomedial amygdaloid nucleus | MPA | medial preoptic area |
| CG | cingulate cortex | MPT | medial pretectal nucleus |
| Cg1 | cingulate cortex, area 1 | MPtA | medial parietal association cortex |
| Cg2 | cingulate cortex, area 2 | mRt | mesencephalic reticular formation |
| CL | centrolateral thalamic nucleus | MS | medial septal nucleus |
| CI | claustrum | MTu | medial tuberal nucleus |
| CPu | caudate putamen (striatum) | Pa | paraventricular hypothalamic nucleus |
| DB | nucleus of the diagonal band | PAG | periaqueductal gray |
| DG | dentate gyrus | Pe | periventricular hypothalamic nucleus |
| DLG | dorsal lateral geniculate nucleus | PF | parafascicular thalamic nucleus |
| DM | dorsomedial hypothalamic nucleus | PG | pregeniculate nucleus |
| DR | dorsal raphe nucleus | PH | posterior hypothalamic nucleus |
| EP | entopeduncular nucleus | PLH | peduncular part of lateral hypothalamus |
| f | fornix | PnO | pontine reticular nucleus, oral part |
| fmi | forceps minor of the corpus callosum | Po | posterior thalamic nuclear group |
| fr | fasciculus retroflexus | PrC | precommissural nucleus |
| IC | inferior colliculus | PrL | prelimbic cortex |
| IF | interfascicular nucleus | PT | paratenial thalamic nucleus |
| IL | infralimbic cortex | PV | paraventricular thalamic nucleus |
| PVA | paraventricular thalamic nucleus, anterior part | ||
| IP | interpeduncular nucleus | PVP | paraventricular thalamic nucleus, posterior part |
| LD | laterodorsal thalamic nucleus | RCh | retrochiasmatic area |
| LDTg | laterodorsal tegmental nucleus | RLi | rostral linear nucleus (midbrain) |
| LHb | lateral habenular nucleus | RMg | raphe magnus nucleus |
| LHbA | anterior division of the LHb | RSD | retrosplenial dysgranular cortex |
| LHbL | lateral division of the LHb | Rt | reticular nucleus (prethalamus) |
| LHbLB | basal subnucleus of the LHbL | S1 | primary somatosensory cortex |
| LHbLMc | magnocellular subnucleus of the LHbL | S2 | secondary somatosensory cortex |
| LHbLMg | marginal subnucleus of the LHbL | SC | superior colliculus |
| LHbLO | oval subnucleus of the LHbL | SCh | suprachiasmatic nucleus |
| LHbLPc | parvocellular subnucleus of the LHbL | SFi | septofimbrial nucleus |
| LHbM | medial division of the LHb | SHy | septohypothalamic nucleus |
| LHbMC | central subnucleus of the LHbM | sm | stria medullaris |
| LHbMMg | marginal subnucleus of the LHbM | ST | bed nucleus of the stria terminalis |
| LHbMPc | parvocellular subnucleus of the LHbM | TS | triangular septal nucleus |
| LHbMS | superior subnucleus of the LHbM | V1 | primary visual cortex |
| LHbP | posterior division of the LHb | V2 | secondary visual cortex |
| LP | lateral posterior thalamic nucleus | VMH | ventromedial hypothalamic nucleus |
| LPO | lateral preoptic area | VP | ventral pallidum |
| LS | lateral septal nucleus | VTA | ventral tegmental area |
| M | medial mammillary nucleus | ZI | zona incerta |
| ME | median eminence | ZIR | zona incerta, rostral part |
Abbreviations of mouse brain regions according to Franklin and Paxinos (2008) and Wagner, Stroh, et al. (2014).
2.2. Animals
All procedures described in the current investigation been approved by the Malmö‐Lund Ethical Committee for the use and care of laboratory animals (permit no. M37‐16). The Tg(Gpr151‐Cre)#Ito transgenic mouse line containing random insertion of bacterial artificial chromosome (BAC) clone MSMg01‐81G4 (Kobayashi et al., 2013) was kindly provided by Dr. Itohara (Laboratory for Behavioral Genetics, RIKEN Brain Science Institute, Saitama, Japan) and maintained on a C57BL/6J background (RRID:IMSR_JAX:000664). To identify individual mice carrying the Cre recombinase allele, polymerase chain reaction genotyping was performed (Transnetyx, Cordova, TN) using the proprietary “CRE” probe set.
A total number of 46 adult (11–14 weeks old) mice was used in the experiment. Wild type litter mates (Cre‐negative) were used as negative controls. The mice were housed in groups of two to six animals with free access to standard lab chow and water, in an air conditioned room at 22–23°C with a standard 12‐hr light/dark cycle (lights on 07:00 a.m., off 07:00 p.m.). The cages (1284L Eurostandard Type II L, Techniplast, Italy) were enriched with aspen wooden bedding and nesting material.
2.3. Retrograde Cre‐dependent tracing using pseudotyped rabies virus
Monosynaptic tracing of the afferents of Cre‐expressing neurons (Callaway & Luo, 2015; Watabe‐Uchida et al., 2012) was carried out in two steps. First, Cre‐dependent expression of an avian sarcoma‐leukosis virus receptor (TVA) fused to the fluorescent protein mCherry (TVA‐mCherry) and rabies glycoprotein (RG) was achieved by an intracerebral injection of adeno‐associated virus (AAV) vectors. Second, SADΔG‐eGFP(EnvA), a glycoprotein‐deleted (ΔG) rabies vector pseudotyped with avian sarcoma‐leukosis virus envelope protein (EnvA) was injected. In Cre‐expressing neurons, TVA‐mCherry then becomes expressed and acts as an entry receptor for the EnvA coated rabies vector, while infection should not be possible in TVA‐negative neurons. RG allows budding of the viral particle and transsynaptic transport to afferent neurons. The rabies vector also carried the gene for enhanced green fluorescent protein (eGFP), allowing detection of infected neurons. We will hereafter refer to Cre‐positive and TVA‐mCherry/RG‐expressing neurons that were infected by SADΔG‐eGFP(EnvA) as starter neurons. The eGFP‐positive/TVA‐mCherry‐negative afferent neuronal populations lack expression of RG which prohibits further propagation of the rabies virus (Figure 1).
Figure 1.
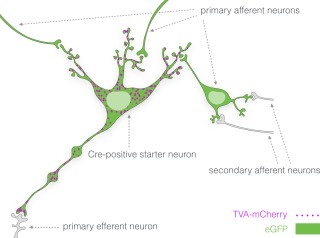
Distribution of fluorescent reporters in the Cre‐dependent rabies virus retrograde tracing system. Cre‐expressing neurons infected by AAV8‐Ef1a‐FLEX‐TVA‐mCherry express the sarcoma‐leukosis virus receptor (TVA) fused to the fluorescent protein mCherry (TVA‐mCherry; magenta). TVA enables entry of EnvA‐coated pseudotyped glycoprotein‐deleted rabies virus particles (SADΔG‐eGFP(EnvA)) into Cre‐positive neurons. Coinfection of these neurons with AAV8‐CA‐FLEX‐RG leads to expression of rabies glycoprotein (RG) which enables transsynaptic retrograde transport of the rabies virus particles. Both the Cre‐positive starter neurons and the primary afferent neurons will thus express the enhanced green fluorescent protein (eGFP; green) while TVA‐mCherry expression will be limited to Cre‐positive neurons
The three AAV vectors, AAV8‐Ef1a‐FLEX‐TVA‐mCherry, AAV8‐CA‐FLEX‐RG, and AAV8‐hSyn1‐FLEX‐mCherry, were obtained from the UNC Vector Core (University of North Carolina at Chapel Hill, NC). The FLEX‐switch makes these three vectors Cre‐dependent by allowing one stable Cre‐mediated inversion of the reversed transgene sequence (Atasoy, Aponte, Su, & Sternson, 2008). SADΔG‐eGFP(EnvA) was obtained from the Gene Transfer, Targeting and Therapeutics Core (GT3) at Salk Institute (La Jolla, CA) and from Dr. Meletis at Karolinska Institute (Stockholm, Sweden).
Gpr151‐Cre mice (n = 38) and wild type controls (n = 5) were anesthetized with isoflurane (induction at 5% and maintenance at 1–2%) and placed in a stereotaxic frame. The scalp was locally anesthetized with bupivacaine (0.25%) and the skull exposed. After adjusting the skull to a horizontal position, a hole was drilled through the skull, exposing the brain surface. Using a thin glass pipette, 0.6 µl of a 1:1 mixture of AAV8‐Ef1a‐FLEX‐TVA‐mCherry (5.4 × 1012 viral genomes/ml) and AAV8‐CA‐FLEX‐RG (2.4 × 1012 viral genomes/ml) was injected unilaterally into the habenula at two different anterior‐posterior levels (coordinates anteroposterior: −1.75, mediolateral: +0.35 or −0.35, dorsoventral: −2.7 and anteroposterior: −1.4, mediolateral: +0.35 or −0.35, dorsoventral: −2.7 relative to Bregma), the paraventricular thalamic nucleus (anteroposterior: −0.46, mediolateral: +0.1 or −0.1, dorsoventral: −3.25 relative to Bregma), or the lateral posterior thalamic nucleus (anteroposterior: −1.7, mediolateral: +1.3 or −1.3, dorsoventral: −2.7 relative to Bregma). In three of the animals injected in the habenula, AAV8‐CA‐FLEX‐RG was omitted to verify that eGFP expression in neurons outside the injected area is a result of transsynaptic transport of rabies virus particles rather than direct infection of afferent terminals. After the injection, the pipette was left in the brain for 4 min to minimize back flow of the vector into the needle tract. The skin was sutured with absorbable suture, and the mouse was kept on a heat pad until recovered from the anesthesia, before returned to its home cage.
After 21 days, the stereotaxic procedure was repeated in order to inject the animals with 0.5 µl of the SADΔG‐eGFP(EnvA) vector (1.65–3.0 × 107 transforming units/ml). Seven days after the last vector injection, the mice were deeply anesthetized with an overdose of sodium pentobarbital and transcardially perfused with isotonic sodium chloride solution (0.9%) followed by cold phosphate buffered saline (PBS; 0.1M sodium phosphate buffer with 0.15M sodium chloride) containing 4% paraformaldehyde for 8 min (∼100 ml per animal).
2.4. Immunohistochemistry
The brains were dissected and postfixed in 4% paraformaldehyde for 18 hr at 4°C. After post‐fixation, the brains were immersed in sucrose solution (25% in PBS) at 4°C for 1–3 days until equilibration had occurred (i.e., the brains sank to the bottom of the vial). The brains were frozen on dry ice, sectioned in 30 µm sections using a sliding microtome (Thermo Scientific HM450) and cryoprotected in antifreeze solution (30% glycerol, 30% ethylene glycol, 40% 0.5M PBS) before storage at −20°C. During sectioning, the left hemisphere was marked with a needle to keep track of the orientation of the sections.
Immunohistochemistry was performed on free floating brain sections as described previously (Broms et al., 2015). The sections were washed in PBS, blocked with normal donkey serum (5%) in PBST (PBS containing 0.05% Tween‐20) for 1 hr at room temperature and subsequently incubated in blocking solution with primary antibody over night at 4°C. The primary antibodies used in the experiment are described below. After repeated washing in PBS, the sections were incubated with biotinylated or Cy5‐conjugated secondary antibodies at 3 µg/ml (Jackson ImmunoResearch Europe Ltd., Suffolk, United Kingdom) in PBST, 2 hr at room temperature. For immunofluorescent detection, sections were washed and mounted on Superfrost Plus glass slides (Menzel Gläser, Brunswick, Germany) with a polyvinyl alcohol and glycerol based mounting solution containing the anti‐fading agent 1,4‐diazabicyclo[2.2.2]octane (PVA‐DABCO).
Fluorescent images were acquired using a Nikon Eclipse Ti confocal microscope with NIS‐Elements (version 4.40) software (Nikon Instruments Europe BV, Amsterdam, The Netherlands). For automatic detection and marking of eGFP‐expressing neurons in scanned images, ImageJ (version 2.0.0‐rc‐59/1.51n, RRID:SCR_003070) was used. The eGFP channel was thresholded to create a binary mask. The positions of eGFP‐expressing cells were then obtained using the “Analyze Particles…” subroutine of ImageJ. Each location was marked with a green circle. A grayscale look‐up table was applied to the mCherry channel and the background fluorescence of this channel provided a background for each micrograph.
For light microscopy detection, biotinylated secondary antibodies were used. After washing in PBS, the sections were incubated with avidin‐biotin‐peroxidase complex (diluted 1:125 in PBS) (VECTASTAIN Elite ABC Kit, Vector Laboratories, Burlingame, CA) and developed using 3,3'‐diaminobenzidine (DAB, 0.5 mg/ml) with nickel chloride (0.5 mg/ml). Following the DAB reaction, sections were mounted, dehydrated in increasing concentrations of ethanol followed by xylene and finally coverslipped with p‐xylene‐bis‐pyridinium bromide (DPX) (Fisher Scientific, Pittsburgh, PA). Low resolution micrographs were acquired using a slide scanner (PathScan Enabler 5, Meyer Instruments Inc., Houston, TX).
2.5. Antibody characterization
Rabbit GPR151 polyclonal antibody (SAB4500418; Sigma‐Aldrich, St. Louis, MO; RRID: AB_10743815) was raised against a synthetic peptide containing amino acids 370‐419 (EKEKPSSPSSGKGKTEKAEIPILPDVEQFWHERDTVPSVQDNDPIPWEHEDQETGEGVK) of the C‐terminal of human GPR151. It recognizes a single band of 46 kDa on western blots of lysate of GPR151‐expressing K562 cells. The specificity was confirmed by pre‐adsorption with the immunogen peptide (according to the manufacturer's technical information). In homozygous Gpr151‐knockout mice, the immunoreactivity was completely abolished, further proving the specificity of the antibody (Broms et al., 2015). In the present experiment, a working concentration of 0.125 µg/ml was used.
Rabbit mCherry polyclonal antibody (ab167453; Abcam, Cambridge, MA; RRID:AB_2571870) was raised against recombinant full length mCherry protein (MVSKGEEDNMAIIKEFMRFKVHMEGSVN GHEFEIEGEGEGRPYEGTQTAKLKVTKGGPLPFAWDILSPQFMYGSKA YVKHPADIPDYLKLSFPEGFKWERVMNFEDGGVVTVTQDSSLQDGEF IYKVKLRGTNFPSDGPVMQKKTMGWEASSERMYPEDGALKGEIKQRL KLKDGGHYDAEVKTTYKAKKPVQLPGAYNVNIKLDITSHNEDYTIVEQ YERAEGRHSTGGMDELYK), a fluorescent protein derived from the DsRed protein. According to the manufacturer's specifications, the antibody recognizes a ∼27 kDa band in lysate of HEK293 cells transfected with a pFin‐Ef1a‐mCherry vector. In our experiment, the antibody (used at a working concentration of 0.2 µg/ml) produced a specific immunohistochemical signal in brains of Gpr151‐Cre animals injected with the AAV8‐Ef1a‐FLEX‐TVA‐mCherry vector.
A different set of Gpr151‐Cre mice (n = 3) were injected bilaterally in the habenula with AAV8‐hSyn‐FLEX‐mCherry (0.6 µl, 6.1 × 1012 viral genomes/ml) to label cell bodies of Cre‐expressing neurons. Four weeks following the injections the brains were prepared for in situ hybridization. Perfusion and postfixation in paraformaldehyde was carried out as described above. After equilibration in 25% phosphate buffered sucrose solution, 14 µm thick brain sections mounted onto Superfrost Plus glass slides using a cryostat. Detection of Gpr151‐mRNA was performed using the RNAscope® Fluorescent Multiplex Assay with a probe set targeting base pairs 27–988 of mouse Gpr151‐mRNA (Cat No. 317321, Advanced Cell Diagnostics, Inc., Newark, CA) (Wang et al., 2012). Briefly, the sections were air dried for 20 min, washed in PBS and subsequently immersed in boiling 1x Target Retrieval solution for 5 min. After washing in distilled water and 100% ethanol for 2 min each, the sections were incubated with Protease IV solution for 30 min at 40°C in a humidified tray. After washing in distilled water for 2 min, the sections were then hybridized with Gpr151 target probe for 2 hr at 40°C followed by amplification solutions 1 (30 min), 2 (15 min), 3 (30 min), and 4 (15 min) at 40°C. Between each amplification step, the sections were rinsed in 1x Wash Buffer for 2 min at room temperature. Finally, the sections were counterstained with 4',6‐Diamidine‐2'‐phenylindole dihydrochloride (DAPI) for 30 s and coverslipped in PVA‐DABCO. The fluorochrome used for detection in this system was Alexa488. Images were acquired using a Nikon Eclipse Ti confocal microscope with NIS‐Elements software.
3. RESULTS
3.1. GPR151 protein and mRNA detected in Cre‐expressing neurons
As previously reported, immunofluorescent GPR151 staining was pronounced in habenular axonal projections, while often barely above background level in the neuronal somata (Broms et al., 2015). To better visualize cell bodies with expression of Cre, we injected Gpr151‐Cre mice with AAV8‐hSyn1‐FLEX‐mCherry in the habenular region (Figure 2).
Figure 2.
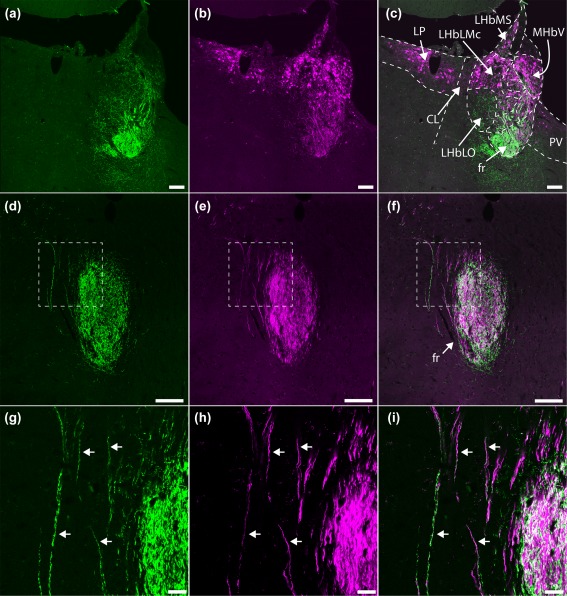
GPR151 protein expression in axons of Gpr151‐Cre neurons. Coronal sections of a Gpr151‐Cre mouse injected with AAV8‐hSyn1‐FLEX‐mCherry in the habenular region. Panels (g–i) represent magnifications of the insets in (d–f). Cell bodies positive for mCherry (magenta) were observed in the ventral medial habenula, the lateral habenula (excluding the oval subnucleus of the lateral division of lateral habenula), the lateroposterior, and paraventricular thalamic nuclei (b, c; −1.94 mm posterior to Bregma). Fibers expressing mCherry and GPR151 protein (green) exited the habenula through the fasciculus retroflexus (a–i; −2.30 mm posterior to Bregma), where colocalization between mCherry and GPR151 was observed (arrows; g–i). Scale bar (a–f) = 100 µm, (g–i) = 20 µm
Colocalization between GPR151 protein and mCherry could be observed in fibers in the fasciculus retroflexus (Figure 2d–i), which contains the efferent axons from the habenula. In the habenula, mCherry‐expressing cell bodies were localized in the ventral division of the medial habenula and in most lateral habenular subnuclei, excluding the oval subnucleus of the lateral division of lateral habenula (Figure 2c). Furthermore, mCherry‐positive neurons were also observed in the paraventricular thalamic nucleus, and in the lateral dorsal and lateral posterior thalamic nuclei. No GPR151 protein could be detected in these thalamic areas using immunohistochemistry (Figure 2c; Broms et al., 2015).
Next, we performed fluorescent in situ hybridization using a RNAscope® probe targeting mouse Gpr151 mRNA on brain sections of mice that had been injected with AAV8‐hSyn1‐FLEX‐mCherry into the habenula. In Figure 3, expression of Gpr151 mRNA in mCherry‐expressing neurons is seen in neurons located in the habenula, paraventricular thalamic nucleus and the lateral posterior thalamic nucleus. The strongest Gpr151 mRNA expression was observed in medial and lateral habenula, compared to much weaker expression in the paraventricular thalamic nucleus while only a very faint expression was observed in neurons of the lateral posterior thalamic nucleus. In the habenula (Figure 3e) and the paraventricular thalamic nucleus, mCherry colocalized with the Gpr151 mRNA signal. In the lateral posterior thalamic nucleus, Gpr151 was undetectable in some mCherry‐positive neurons (Figure 3d). This finding is further considered in the discussion.
Figure 3.
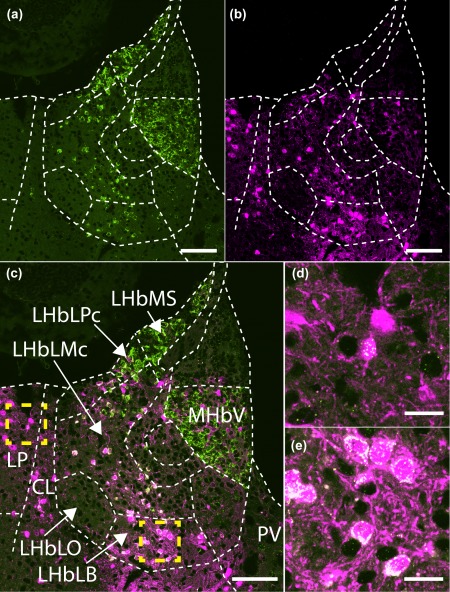
Gpr151 mRNA expression in Gpr151‐Cre neurons. Coronal section of a Gpr151‐Cre mouse injected with AAV8‐hSyn1‐FLEX‐mCherry in the habenular region (−1.82 mm posterior to Bregma). Gpr151 mRNA expression (green) can be seen in the ventral medial habenula, lateral habenula (excluding the oval subnucleus of the lateral division), as well as in the lateral posterior, central lateral, and paraventricular thalamic nuclei (panels a, c, d, e). Expression of mCherry (magenta, panels b–e) show partial overlap with Gpr151 mRNA. Transduction efficiency and injection localization could explain the lack of mCherry expression in certain Gpr151‐positive subregions like the LHbMS. Examples of coexpression of mCherry and Gpr151 mRNA in the lateral posterior thalamic nucleus and lateral habenula are shown in panel (d) and (e) (magnifications of insets in panel c). Scale bar (a–c) = 100 µm, (d, e) = 20 µm
Using a crossing between Gpr151‐Cre mice and two different ß‐galactosidase reporter strains, Kobayashi et al. (2013) identified Cre‐expressing cells in the medial and lateral habenula, paraventricular thalamic nucleus and the reuniens thalamic nucleus. This expression pattern largely agrees with our findings, except that more Cre‐expressing cells were detected in the laterodorsal and lateral posterior thalamic nucleus in the current investigation. Given that Gpr151 mRNA expression is accentuated in the habenula and paraventricular thalamic nucleus (Ignatov et al., 2004; Lein et al., 2006), no viral injections targeting the reuniens thalamic nucleus were performed in the current investigation.
3.2. Monosynaptic tracing of afferents to Cre‐expressing neurons
Using the Cre‐dependent rabies virus tracing system, Gpr151‐Cre starter neurons were labeled with TVA‐mCherry and eGFP, while monosynaptic afferents to these starter neurons were labeled with eGFP only. Not all TVA‐mCherry‐expressing neurons coexpressed eGFP. This may be explained by different transduction efficiency of the AAV and rabies vectors or that the vectors were injected at slightly different locations due to the margin of error of the stereotaxic technique. The injections targeted one of three areas, the habenula, the paraventricular thalamic nucleus, or the lateral posterior thalamic nucleus. The starter neurons at the injection sites and the resulting eGFP expression in afferent structures are summarized in Tables 1 and 2.
Table 1.
Starter neuron populations
| Habenula | Paraventricular thalamic nucleus | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M17 | M18 | M80 | M102 | M103 | M104 | M105 | M74 | M112 | M113 | M120 | M121 | ||
| Medial habenula | MHbD | ||||||||||||
| MHbS | |||||||||||||
| MHbVl | +++++ | +++++ | +++ | ++ | ++ | ++++ | |||||||
| MHbVc | +++++ | +++++ | +++ | ++ | ++ | +++ | |||||||
| MHbVm | + | + | |||||||||||
| Lateral habenula | LHbA | + | +++ | + | ++ | ++ | + | ||||||
| LHbMS | +++++ | ++ | +++++ | ++++ | ++ | ++++ | |||||||
| LHbMPc | + | +++ | ++ | ||||||||||
| LHbMC | ++ | ++ | +++ | ++ | +++ | ||||||||
| LHbMMg | +++ | + | |||||||||||
| LHbLMg | ++ | ++ | ++ | + | |||||||||
| LHbLPc | ++ | ++ | +++ | +++ | ++++ | ||||||||
| LHbLMc | + | +++++ | +++ | +++++ | +++++ | ++ | ++ | ||||||
| LHbLB | ++++ | ++++ | ++++ | ++ | |||||||||
| LHbLO | |||||||||||||
| LHbP | + | ++++ | ++++ | +++ | ++ | + | |||||||
| Paraventricular thalamic nucleus | PVA | +++ | +++++ | +++++ | +++ | +++++ | |||||||
| PV | + | + | +++ | + | + | + | ++ | ++ | +++++ | ||||
| PVP | + | ||||||||||||
| Lateral thalamic nuclei | LP | ++ | ++ | ||||||||||
| LD | + | ++ | |||||||||||
| CL | + | + | + | ||||||||||
| Posterior thalamic nuclei | PF | ||||||||||||
| PrC | + | ||||||||||||
| Po | |||||||||||||
| MPT | + | ||||||||||||
| Lateral posterior thalamic nucleus | Habenula no RG | Habenula wild type | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M114 | M115 | M116 | M119 | M106 | M117 | M118 | M113 | M16 M77 | M77 | M78 | M81 | ||
| Medial habenula | MHbD | ||||||||||||
| MHbS | |||||||||||||
| MHbV | |||||||||||||
| MHbVc | |||||||||||||
| MHbVm | |||||||||||||
| Lateral habenula | LHbA | ++ | ++ | ++ | + | ||||||||
| LHbMS | +++ | +++ | +++ | ++++ | ++ | + | |||||||
| LHbMPc | + | ++ | + | + | + | ++ | |||||||
| LHbMC | +++ | ++ | ++ | +++ | ++ | ||||||||
| LHbMMg | ++ | ||||||||||||
| LHbLMg | ++ | +++ | |||||||||||
| LHbLPc | ++ | ++ | +++ | ++++ | +++ | ||||||||
| LHbLMc | +++ | ++ | +++++ | + | |||||||||
| LHbLB | ++++ | ||||||||||||
| LHbLO | |||||||||||||
| LHbP | +++ | ++ | +++ | +++++ | +++++ | +++++ | |||||||
| Paraventricular thalamic nucleus | PVA | ||||||||||||
| PV | + | ++ | |||||||||||
| PVP | ++ | +++ | |||||||||||
| Lateral thalamic nuclei | LP | +++++ | +++++ | +++++ | +++++ | +++ | ++ | ||||||
| LD | +++++ | +++++ | +++++ | +++++ | ++++ | ||||||||
| CL | + | ++ | +++ | + | +++ | ||||||||
| Posterior thalamic nuclei | PF | + | ++ | ++++ | |||||||||
| PrC | + | + | ++ | +++ | +++ | ||||||||
| Po | ++++ | ++ | ++++ | ||||||||||
| MPT | + | ++ | ++ | + | ++ | ||||||||
Semiquantitative estimate of neurons coexpressing TVA‐mCherry and eGFP (starter neurons) in target areas and neighboring structures. Each area was graded from + to +++++ depending on the number of mCherry/eGFP coexpressing cells observed.
Table 2.
Locations of primary afferents to Gpr151‐Cre neurons.
| Habenula | Paraventricular thalamic nucleus | Lateral posterior thalamic nucleus | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M17 | M18 | M80 | M102 | M103 | M104 | M105 | M74 | M112 | M113 | M120 | M121 | M114 | M115 | M116 | M119 | ||
| Cerebral cortex | Al | +/+ | |||||||||||||||
| Au | +/‐ | +/‐ | |||||||||||||||
| BL | +/+ | ||||||||||||||||
| BM | +/‐ | ||||||||||||||||
| Cg1 | +/‐ | +/‐ | +/‐ | ++/‐ | +++/‐ | +++/‐ | ++/‐ | ||||||||||
| Cg2 | +/‐ | +/‐ | +/‐ | ‐/+ | ++/+ | +++/‐ | +/‐ | ++/+ | |||||||||
| Cl | +/‐ | +/‐ | +/+ | +/+ | +/‐ | ||||||||||||
| DG | +/‐ | +/‐ | +/‐ | ||||||||||||||
| IL | +/‐ | +/+ | +/‐ | +/‐ | ‐/+ | +/‐ | +/+ | ++/‐ | |||||||||
| MPtA | +/‐ | ||||||||||||||||
| PrL | +/+ | ++/+ | +/‐ | +/+ | +++/++ | ‐/+ | ++/+ | +/‐ | +/‐ | ++/+ | +++/+ | ||||||
| RSD | +/‐ | +/‐ | +++/‐ | ||||||||||||||
| S1 | +/‐ | ++/‐ | |||||||||||||||
| S2 | +/‐ | ||||||||||||||||
| V1 | ++/‐ | +/‐ | |||||||||||||||
| V2 | ++/‐ | +/‐ | +/‐ | ||||||||||||||
| Pallidum | BAC | +++/++ | +/‐ | +/‐ | +/‐ | ++/‐ | +/‐ | +/‐ | ++/‐ | ++/++ | |||||||
| DB | +++/++ | ++++/+ | ++++/+ | ++/+ | +/+ | +++/++ | +++/+ | ++/++ | ++/‐ | +/+ | ++/++ | ++/‐ | ++/‐ | +/‐ | +++/+ | ||
| EP | ++/‐ | +++++/+ | +++++/+ | +/‐ | ++/‐ | +/‐ | +++/+ | ||||||||||
| MS | ++ | + | ++ | ++ | + | + | + | + | |||||||||
| ST | ++++/+++ | +++/‐ | ++/‐ | +/‐ | +++/‐ | ++/++ | ++/++ | +/‐ | +/+ | ++++/++ | +++/+ | ++++/++ | +++/++ | ||||
| TS | +/‐ | +/‐ | +/+ | +++/+ | +/‐ | ||||||||||||
| VP | ++/++ | ++/‐ | ++/+ | +/‐ | ++/‐ | +++/++ | ++/+ | +/+ | +/‐ | ‐/+ | +/+ | +++/+ | ++/‐ | +/‐ | ++++/++ | ||
| Striatum | Acb | +/‐ | ++/++ | ++/++ | +/‐ | +/+ | +/‐ | ||||||||||
| CPu | ++/‐ | +/+ | +/‐ | ++/++ | +/+ | ++/‐ | +/‐ | ++/‐ | |||||||||
| LS | +/‐ | +/‐ | +/+ | +/‐ | +/‐ | +/‐ | +/+ | +/‐ | +/‐ | ++/+ | +/‐ | +/‐ | +/+ | ||||
| Me | +/‐ | +/‐ | |||||||||||||||
| Hypothalamus | AH | +/‐ | +/‐ | +/‐ | +/+ | +/‐ | +/‐ | +/+ | ++/++ | +/‐ | +/+ | +/+ | |||||
| Arc | +/‐ | ‐/+ | +/‐ | +/‐ | ++/++ | +/+ | +/‐ | ++/++ | +/‐ | +/‐ | ++/‐ | ‐/+ | |||||
| DM | ++/‐ | +/‐ | +/‐ | +/+ | ++/+ | +++/+++ | ‐/+ | +/+ | +/‐ | +/‐ | +/‐ | ||||||
| LPO | +++/++ | ++++/++ | +/‐ | ++/+ | ++/+ | +++++/++ | ++/‐ | +/+ | +/+ | ++/++ | +/+ | ++++/+ | +++/+ | +++++/+ | ++++/++ | ||
| M | +/‐ | +/+ | +/‐ | +/+ | +/‐ | ++/+ | |||||||||||
| MnPO | + | + | + | ++ | + | + | |||||||||||
| MPA | ‐/+ | +/+ | ++/‐ | +/‐ | ++/++ | ++++/+++ | +/+ | ++/+++ | +/‐ | ++/+ | ++/+ | ||||||
| MTu | +/+ | ‐/+ | |||||||||||||||
| Pa | +/‐ | +/‐ | ++/+ | +/‐ | |||||||||||||
| Pe | +/‐ | +/+ | ++/++ | +/+ | |||||||||||||
| PH | ‐/+ | +/‐ | +/‐ | +/‐ | |||||||||||||
| PLH | +++/++ | +++++/+++ | ++/+ | +++/+ | ++++/++ | +++++/++ | ++++/++ | +/‐ | ++/++ | +++/++ | +/‐ | ++/++ | +++++/++ | +++/++ | +++++/+++ | +++++/+++ | |
| RCh | +/+ | +/+ | ++/‐ | +++/++ | ‐/+ | ||||||||||||
| SCh | +/‐ | +/+ | ++/+ | +/‐ | +/+ | ||||||||||||
| SFi | ++/‐ | +++/‐ | |||||||||||||||
| SHy | +/‐ | +/+ | ++/‐ | ++/++ | |||||||||||||
| VMH | +/‐ | +/‐ | ++/‐ | +/‐ | +/+ | +++/+++ | ++/+ | +/‐ | +/‐ | ||||||||
| ZI | ++/‐ | +/‐ | +++/‐ | +/‐ | +/‐ | +++/+++ | +++++/+++++ | ++/+ | +++/+++ | ++++/‐ | ++/‐ | ++++/+ | ++++/+ | ||||
| Thalamus | AD | +/‐ | |||||||||||||||
| LD/LP (contralateral side) | + | ||||||||||||||||
| LHb (contralateral side) | + | + | + | + | |||||||||||||
| MD | +/‐ | ++/‐ | +/+ | +/‐ | |||||||||||||
| MG/PG/DLG | +/+ | +/‐ | +/‐ | ++/+ | +/‐ | ++/‐ | +/‐ | +++/+ | +++/+ | ||||||||
| MHb (contralateral side) | + | + | + | ||||||||||||||
| PF | +/‐ | +/+ | +/‐ | +/‐ | ++/+ | +/‐ | +/‐ | +/+ | ++/+ | ++/‐ | +/‐ | ||||||
| Po | +++/‐ | ||||||||||||||||
| PrC | +/‐ | +/‐ | +/‐ | ++/‐ | +/‐ | +/‐ | |||||||||||
| PVA | +/‐ | ++/‐ | +/‐ | +/+ | +/‐ | +++/++ | +/‐ | +/‐ | |||||||||
| Rt | ++/‐ | ‐/+ | ++++/+ | +++/‐ | +++++/‐ | +++++/+ | |||||||||||
| Midbrain | DR | +/‐ | +/‐ | +/‐ | ++/++ | + | +/‐ | +/+ | ++ | +/+ | ++/+ | ++/‐ | |||||
| IC | +/‐ | ||||||||||||||||
| IP | +/+ | +/‐ | ‐/+ | +/+ | ++/‐ | +/‐ | + | +/+ | |||||||||
| MnR | ++/+ | +/‐ | +/‐ | +/+ | +/+ | ++/+ | + | +/+ | +/‐ | +/+ | ++ | + | +/+ | ||||
| MPT | ++/‐ | ++/‐ | ++/+ | +++/++ | +++/++ | ||||||||||||
| mRT | ++/+ | +/‐ | ++/+ | +/+ | ‐/+ | +/+ | +/‐ | +/‐ | ++/‐ | ||||||||
| PAG | +/‐ | +/‐ | +/‐ | ++/++ | +/‐ | +/+ | ++/++ | +/‐ | +/+ | +/‐ | ++/‐ | +/+ | |||||
| RLi | + | + | + | + | + | ||||||||||||
| SC | +/‐ | +/‐ | +/‐ | ‐/+ | +/‐ | +/+ | +/+ | +++++/+ | ++++/++ | +++++/++ | +++++/++ | ||||||
| VTA | +/‐ | +/‐ | +/‐ | +/‐ | +/‐ | ++/+ | +/‐ | +/+ | +/‐ | +/‐ | +/+ | ||||||
| Hindbrain | LDTg | +/‐ | +/‐ | +/‐ | +/‐ | ++/‐ | |||||||||||
| PnO | +/+ | +/‐ | +/+ | +/‐ | +/‐ | ||||||||||||
| RMg | +/‐ | +/‐ | |||||||||||||||
Semiquantitative estimate of primary afferents to Gpr151‐Cre neurons. The entire brain was investigated and areas were graded from + to +++++ depending on the number of eGFP expressing cells found. In paired structures, the ipsilateral estimate is shown to the left and the contralateral estimate to the right (ipsilateral/contralateral).
In wild type animals, occasional neurons expressing mCherry and eGFP could be observed close to the injection tract in the habenula, which implies minor local Cre‐independent expression (Figure 4j–l; Table 1). However, no eGFP+ neurons were detected outside of the injected area, so the analysis of eGFP+ neurons far from the injected site is not impacted.
Figure 4.
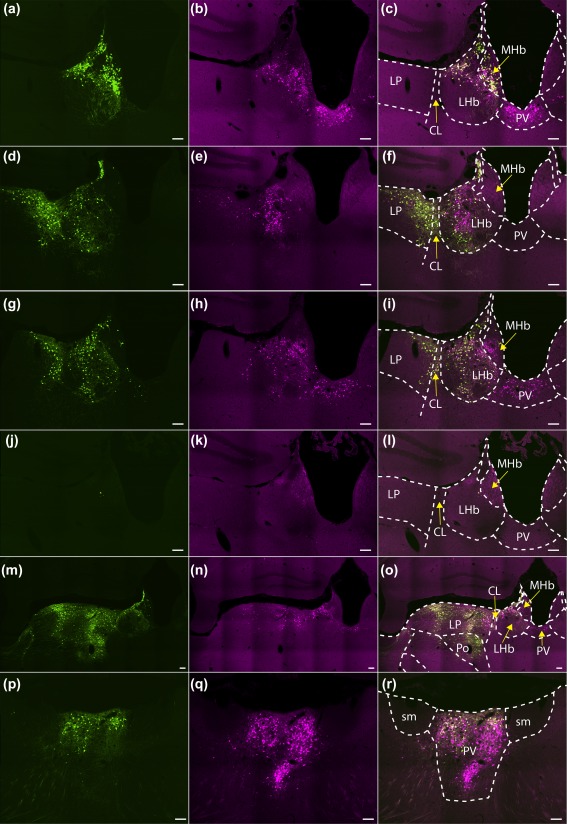
Expression of TVA‐mCherry and eGFP after injections of viral vectors at target sites. Coronal sections through the target areas of cases M80 (habenula; a–c), M18 (habenula; d–f), M118 (habenula, no RG; g–i), M77 (habenula, wild type; j–l), M119 (lateral posterior thalamic nucleus; m–o), and M113 (paraventricular thalamic nucleus; p–r) showing sarcoma‐leukosis virus receptor (TVA) fused to the fluorescent protein mCherry (TVA‐mCherry; magenta; b, e, h, k, n, q), eGFP expression (green; a, d, g, j, m, p), and an overlay between the two (c, f, i, l, o, r). Scale bar = 100 µm
SADΔG‐(EnvA) is capable of infecting neurons with minute expression of TVA (Callaway & Luo, 2015). Since it has been shown that trace amounts of TVA‐expression can result from retrograde transport of AAV‐TVA (Menegas et al., 2015), it is important to confirm that the starter neurons are indeed restricted to the injected area. By omitting AAV8‐CA‐FLEX‐RG, transsynaptic transport is inhibited and eGFP expression should be restricted to local TVA‐expressing neurons. In Gpr151‐Cre animals where AAV8‐CA‐FLEX‐RG was omitted, a strong expression of TVA‐mCherry and eGFP was only observed in the vicinity of the injection, but not anywhere else in the brain (Figures 4g–i and 5; Table 1). This confirms that any eGFP+ neurons found outside of the injected area result from RG‐dependent transsynaptic transport of rabies virus.
Figure 5.
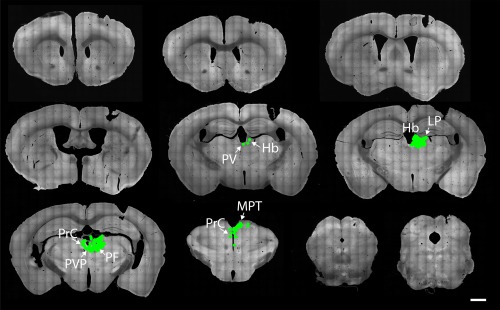
Omission of RG limits eGFP expression to the injected area. Coronal brain sections from 1.34 to −4.84 mm relative to Bregma of a Gpr151‐Cre mouse (case M118) in which AAV8‐Ef1a‐FLEX‐TVA‐mCherry and SADΔG‐eGFP(EnvA) was injected unilaterally in the habenula. Since AAV8‐CA‐FLEX‐RG was omitted, transsynaptic transport of the rabies vector is inhibited. Although many eGFP positive (green) neurons could be seen in various nuclei in the targeted area (medial and lateral habenula, lateral posterior thalamic nucleus, parafascicular thalamic nucleus, paraventricular thalamic nucleus, and precommissural nucleus), no eGFP expressing neurons were detected elsewhere in the brain. Scale bar = 1000 µm
3.3. Afferents to Gpr151‐Cre neurons in the habenula
Seven cases with good quality injections targeting the habenula were considered for further analysis (Figure 4a–f; Table 1). In all cases, a majority of starter neurons were detected in the habenula, although a varying degree of spread to neighboring thalamic structures, such as the paraventricular thalamic nucleus and the lateral posterior thalamic nucleus was also observed.
Very few eGFP+ neurons were detected in the cerebral cortex (Figure 6; Table 2). Occasional eGFP+ neurons were observed in medial prefrontal areas, most notably bilaterally in the prelimbic and infralimbic cortices.
Figure 6.
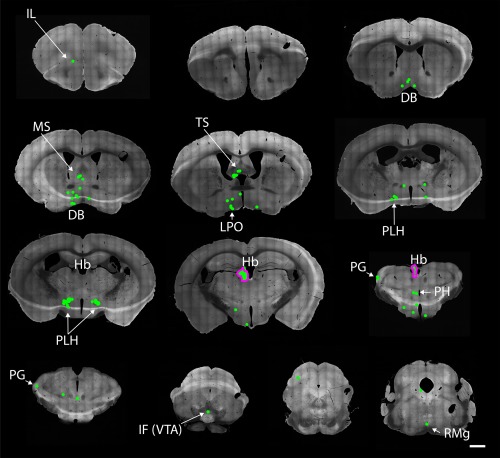
Afferents of habenular Gpr151‐Cre neurons. Coronal brain sections from 1.98 to −4.96 mm relative to Bregma of a Gpr151‐Cre mouse (case M80) where AAV8‐Ef1a‐FLEX‐TVA‐mCherry, AAV8‐CA‐FLEX‐RG and SADΔG‐eGFP(EnvA) was injected unilaterally in the habenula. Neurons coexpressing eGFP and mCherry (starter neurons; magenta outline) were almost exlusively located in the medial and lateral habenula. EGFP positive neurons (green) were found throughout the brain, most notably in the medial septal nucleus, nucleus of the diagonal band, lateral preoptic area, and the lateral hypothalamus (peduncular part). Scale bar = 1000 µm
Several structures in the pallidum contained large numbers of eGFP+ neurons (Table 2). Moderate numbers of eGFP+ neurons were detected in the medial (Figure 7a) and triangular septum (Figure 7d). The nucleus of the diagonal band contained many eGFP+ neurons, both in the vertical and horizontal limb (Figure 7a). Moderate numbers of eGFP+ neurons were also observed in the bed nucleus of the anterior commissure (Figure 7d). The bed nucleus of the stria terminalis contained great numbers of eGFP+ neurons, especially in case M17 (Figure 7c). In two cases (M18 and M104), a dense network of eGFP+ neurons was seen in the entopeduncular nucleus (Figure 7e,f). Interestingly, these specific cases were distinguished by a large number of starter neurons in lateral division of the lateral habenula as well as spread into the lateral posterior thalamic nucleus.
Figure 7.
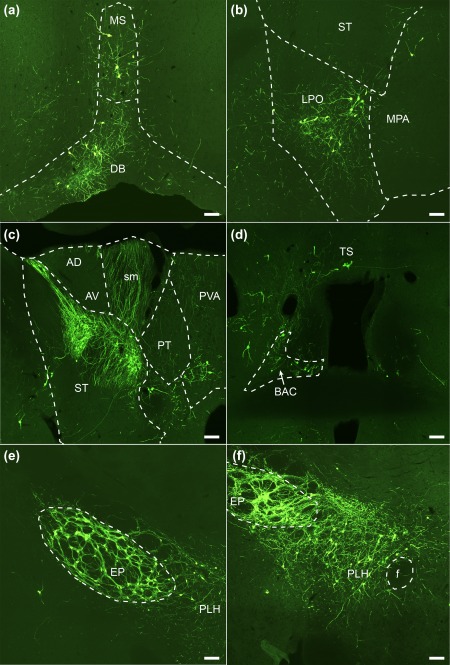
Main afferent neuronal populations of the habenular Gpr151‐Cre starter neurons. Coronal sections showing eGFP‐expressing afferent neurons (green cell bodies and fibers) in the medial septal nucleus and the nucleus of the diagonal band (a; case M17), the lateral and medial preoptic area (b; case M18), the bed nucleus of stria terminalis and the paraventricular thalamic nucleus (c; case M17), the triangular nucleus of the septum and the bed nucleus of the anterior commissure (d; case M17), the entopeduncular nucleus and the lateral hypothalamus (e, f; case M18). Scale bar = 100 µm
The hypothalamus was another major source of input to the Gpr151‐Cre neurons of the habenula (Table 2). A continuous group of eGFP‐expressing neurons stretching from the lateral preoptic area to the peduncular part of the lateral hypothalamus was observed in all cases (Figures 6 and 7b,e,f). Scattered neurons were also observed in several medial hypothalamic nuclei (Table 2). In two cases (M18 and M104), the same animals which displayed eGFP+ neurons in the entopeduncular nucleus, eGFP+ neurons could also be observed in the zona incerta.
Because undetectable expression of TVA‐mCherry may be sufficient to allow entry of the rabies vector (Callaway & Luo, 2015), it cannot be ruled out that eGFP+ neurons close to the injected area that appear to lack mCherry are indeed afferent neurons and not starter neurons. Because of the proximity of the injection target area to some Gpr151‐expressing thalamic nuclei, it makes interpretation of eGFP+ neurons in these areas uncertain. Scattered eGFP+ neurons were detected in the pregeniculate, dorsolateral geniculate, and medial geniculate nuclei which are far from the injected area. The parafascicular thalamic nucleus and the precommissural nucleus also contained a small number of eGFP+ neurons, along with occasional starter neurons. Occasionally, eGFP+ neurons were visible in the medial or lateral habenula contralateral to the injected side which suggests minor contribution from an intrahabenular projection.
In the midbrain, scattered eGFP+ neurons were observed in the pretectal area, superior colliculus, and the periaqueductal gray. A few eGFP+ cells could also be detected in the interpeduncular nucleus, which is the projection target of neurons in the medial habenula. This implies a reciprocal connection between the habenular complex and the interpeduncular nucleus. EGFP+ neurons were also observed in many monoaminergic midbrain areas such as the ventral tegmental area (primarily the interfascicular nucleus), rostral linear nucleus, median, and dorsal raphe nuclei.
Very few eGFP+ neurons were detected in hindbrain areas, including the laterodorsal tegmental nucleus, pontine reticular nucleus, and raphe magnus nucleus.
3.4. Afferents to Gpr151‐Cre neurons in the paraventricular thalamic nucleus
Five animals where starter neurons could be observed in the paraventricular thalamic nucleus (Table 1) were further analyzed regarding afferents (Figures 8 and 9; Table 2). The starter neuron population was entirely restricted to the target area except for one case (M113) where a few cells were also observed in the anterior division of the lateral habenula.
Figure 8.
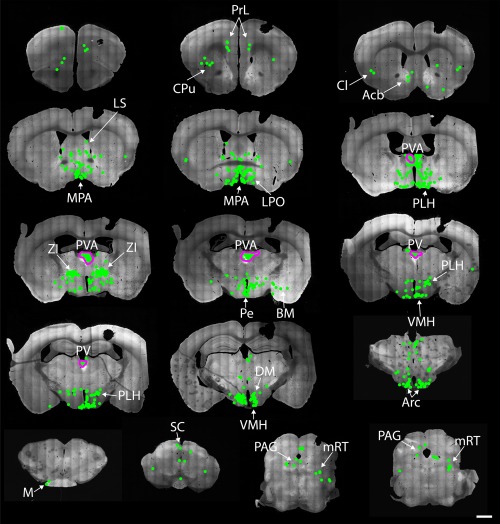
Afferents of Gpr151‐Cre neurons in the paraventricular thalamic nucleus. Coronal brain sections from 1.98 to −4.84 mm relative to Bregma of a Gpr151‐Cre mouse (case M113) where AAV8‐Ef1a‐FLEX‐TVA‐mCherry, AAV8‐CA‐FLEX‐RG, and SADΔG‐eGFP(EnvA) was injected into the anterior part of the paraventricular thalamic nucleus. Afferent eGFP expressing neurons (green) were found in great numbers in the zona incerta, medial preoptic area, medial and lateral hypothalamus and to a minor extent in the prelimbic cortex, accumbens nucleus, lateral preoptic area, bed nucleus of stria terminalis, and periaqueductal gray. Neurons coexpressing eGFP and mCherry (starter neurons; magenta outline) were located in the paraventricular thalamic nucleus. Scale bar = 1000 µm
Figure 9.
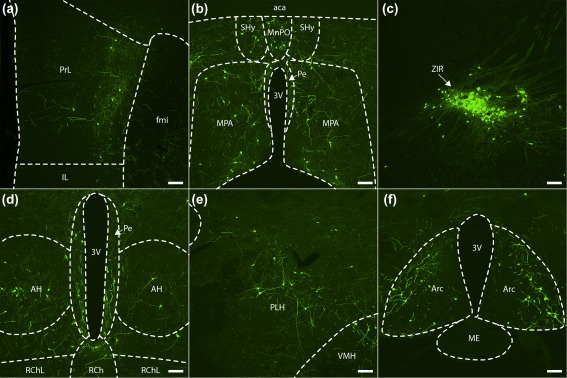
Main afferent neuronal populations of Gpr151‐Cre starter neurons in the paraventricular thalamic nucleus. Coronal sections (case M113) showing eGFP‐expressing afferent neurons (green cell bodies and fibers) in the prelimbic cortex (a, left hemisphere), the septohypothalamic nucleus (b), the median preoptic nucleus (b), the periventricular hypothalamic nucleus (b, d), the medial preoptic area (b), the zona incerta (c, right hemisphere), the anterior hypothalamic area (d), the retrochiasmatic area (d), the peduncular part of lateral hypothalamus (e, right hemisphere), the ventromedial hypothalamic nucleus (e, right hemisphere), and the arcuate hypothalamic nucleus (f). Scale bar = 100 µm
Cortical afferents were restricted to frontal cortical areas such as the prelimbic, infralimbic, and agranular insular cortices. In two cases (M113 and M121), a few eGFP+ neurons could also be observed in the claustrum.
The distribution of eGFP+ neurons in the pallidum was remarkably similar to that of the afferents to the habenular Gpr151‐Cre population (Table 2). The nucleus of the diagonal band, bed nucleus of stria terminalis, and the ventral pallidum contained many eGFP+ neurons. No eGFP+ neurons were observed in the medial septal nucleus or the entopeduncular nucleus however, in contrast to the afferents of the Gpr151‐Cre neurons in the habenula.
In the striatum, moderate numbers of eGFP+ neurons were detected in the accumbens nucleus, caudate putamen, and the lateral septum.
The main source of input to the paraventricular thalamic Gpr151‐Cre population was the hypothalamus (Table 2). Similarly to the habenular Gpr151‐Cre population, eGFP+ neurons could be detected in the lateral preoptic area and the peduncular part of the lateral hypothalamus (Figure 9e). Great numbers of eGFP+ neurons were also seen in the medial preoptic area, and to a lesser extent the median preoptic area (Figure 9b). Medial hypothalamic areas such as the anterior, ventromedial, dorsomedial, arcuate, paraventricular, and periventricular hypothalamic nuclei also contained many eGFP+ neurons (Figure 9b,d,e,f). Some afferent neurons were also seen in the retrochiasmatic (Figure 9d) and suprachiasmatic nuclei. The strongest projection arose from a cluster of cells in the rostral part of the zona incerta (Figure 9c).
Some eGFP+ neurons were also detected in thalamic areas such as the parafascicular and mediodorsal thalamic nuclei. In the midbrain, there were also a small number of eGFP+ neurons in the periaqueductal gray and the mesencephalic reticular formation.
3.5. Efferent projections of the Cre‐expressing population in the paraventricular thalamic nucleus
Since the paraventricular thalamic Gpr151‐Cre population could be targeted specifically without spread into other Gpr151‐expressing populations, we decided to also analyze the efferent connectivity of this population. Immunohistochemistry using mCherry antiserum was performed to visualize the efferent projections of the TVA‐mCherry‐expressing neurons (Figure 10). The cell bodies of these neurons were confined to the paraventricular nucleus, while dense fields of mCherry‐immunoreactivity could be observed in the prelimbic area, the shell and core regions of accumbens nucleus, the basolateral amygdala, and the zona incerta. It should be noted that while no GPR151 protein could be detected by immunohistochemistry in any of these areas (Broms et al., 2015), Gpr151 mRNA was observed in the paraventricular thalamic nucleus in the current study (Figure 3c) and has also been reported previously (Ignatov et al., 2004; Wagner, French, et al., 2014).
Figure 10.
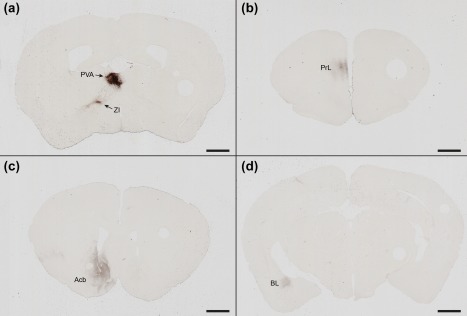
Efferent projections of Gpr151‐Cre neurons in the paraventricular thalamic nucleus. Coronal sections of a Gpr151‐Cre mouse injected with AAV8‐hSyn1‐FLEX‐mCherry in the paraventricular nucleus of the thalamus (case M74) showing 3,3'‐diaminobenzidine enhanced mCherry immunostaining in cell bodies in the injected area and in efferent projections to the zona incerta (a), the prelimbic area (b), the shell and core of accumbens nucleus (c), and the basolateral amygdala (d). Scale bar = 1000 µm
3.6. Afferents to Gpr151‐Cre neurons in the lateral posterior thalamic nucleus
Four animals with injections targeting the lateral posterior thalamic nucleus were analyzed (Figure 11; Table 2). Large quantities of starter neurons were observed in a group of neurons in the lateral dorsal and lateral posterior thalamic nuclei. In two cases, the labeled population also extended into the posterior thalamic group (M114 and M119). Unfortunately, all cases also contained a varying number of starter neurons in the habenula which impose a limit on the analysis of the afferents to the lateral thalamic group. Some striking differences between the cases targeting the lateral posterior thalamic Gpr151‐Cre population and the habenular Gpr151‐Cre population could nonetheless be observed. The cingulate cortical area 1 and area 2, as well as the prelimbic cortex, contained many eGFP+ neurons in animals with starter neurons in the lateral posterior thalamic nucleus, but were completely devoid of eGFP+ cells when the injections were restricted to the habenula. Similarly, other cortical areas such as the retrosplenial dysgranular cortex as well as the primary and secondary visual cortex and primary and secondary somatosensory cortex, only contained eGFP+ neurons in cases with lateral posterior thalamic starter neurons.
Figure 11.
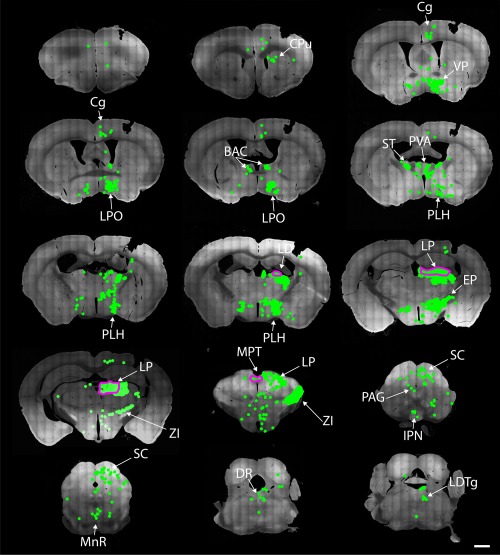
Afferents of Gpr151‐Cre neurons in lateral thalamic nuclei. Coronal brain sections from 1.98 to −5.02 mm relative to Bregma of a Gpr151‐Cre mouse (case M119) where AAV8‐Ef1a‐FLEX‐TVA‐mCherry, AAV8‐CA‐FLEX‐RG, and SADΔG‐eGFP(EnvA) was injected into the lateral posterior thalamic nucleus. Starter neurons expressing both mCherry and eGFP (outlined in magenta) were not restricted to the target area, but were also found in the laterodorsal thalamic nucleus, central lateral thalamic nucleus, parafascicular thalamic nucleus, posterior thalamic group, lateral habenula, precommissural nucleus, and medial pretectal nucleus. Afferent eGFP expressing neurons (green) were detected in many areas throughout the brain including the cingulate and prelimbic cortices, bed nucleus of stria terminalis, ventral pallidum, reticular thalamic nucleus, lateral preoptic area, lateral hypothalamic area, zona incerta, and superior colliculus. Scale bar = 1000 µm
Similar to what was observed when the habenula was targeted, many eGFP+ neurons could be observed in the bed nucleus of the anterior commissure, nucleus of the diagonal band, ventral pallidum, bed nucleus of stria terminalis, lateral preoptic area, lateral hypothalamic area, and the entopeduncular nucleus. The caudate putamen also contained eGFP+ positive neurons in contrast to habenula‐targeting injections. Large number of eGFP+ neurons was observed in the zona incerta, mainly in the caudal part.
In the thalamus, dense populations of eGFP+ neurons were observed in pregeniculate, dorsolateral geniculate, and medial geniculate nuclei as well as the reticulate and paraventricular thalamic nuclei. In the midbrain, great numbers of eGFP+ neurons were detected in the superior colliculus as well as in the pretectal region. In the hindbrain, only a few eGFP+ neurons were detected in the laterodorsal tegmental nucleus.
4. DISCUSSION
In this study, monosynaptic tracing using pseudotyped rabies virus was used to identify primary afferents to populations of Gpr151‐Cre neurons in the mouse diencephalon.
In the medial habenula, TVA‐mCherry+/eGFP+ starter neurons were observed in the ventral part of the medial habenula (Table 1). This distribution is consistent with the pattern of Gpr151 mRNA and protein expression (Figures 2 and 3). Several nuclei in the pallidum, including the triangular septal nucleus, septofimbrial nucleus, bed nucleus of the anterior commissure, medial septal nucleus, and the nucleus of the diagonal band have previously been identified as afferents to the medial habenula (Herkenham & Nauta, 1977; Qin & Luo, 2009; Yamaguchi, Danjo, Pastan, Hikida, & Nakanishi, 2013). All of these areas contained eGFP+ neurons in cases where starter neurons were found in the medial habenula, although the most prominent and consistent projection appeared to arise in the nucleus of the diagonal band and the medial septal nucleus. This projection is reported to be GABAergic (Contestabile & Fonnum, 1983; Qin & Luo, 2009), although we did not test whether the neurons that project onto Gpr151‐expressing cells have this phenotype.
In the lateral habenula, Cre expression could be detected in all subnuclei except for the oval subnucleus of the lateral division (Figures 2 and 3; Table 1). Afferent neurons were observed in many areas (Figure 7; Table 2) that have previously been reported to contain afferents to the lateral habenula, including the nucleus of the diagonal band, bed nucleus of stria terminalis, ventral pallidum, lateral preoptic area, lateral hypothalamic area and entopeduncular nucleus and the median raphe nucleus (Dong & Swanson, 2004; Herkenham & Nauta, 1977; Wallace et al., 2017).
In a recent study, the nucleus of the diagonal band was shown to provide a possible source of input to the lateral habenula, which allowed the habenular neurons to phase lock with hippocampal theta oscillations during anesthesia and rapid‐eye‐movement (REM) sleep (Aizawa et al., 2013). Lesions of the lateral habenula and the fasciculus retroflexus reduce theta power in the hippocampus and substantially decrease REM sleep duration, while non‐REM sleep remains unaffected (Aizawa et al., 2013; Valjakka et al., 1998). Since Gpr151‐Cre neurons receive input from the nucleus of the diagonal band and in turn project to the interpeduncular nucleus and median raphe, structures that has been implicated in theta oscillations and REM sleep (Funato et al., 2010; Vertes, Kinney, Kocsis, & Fortin, 1994), it is possible that this neuronal population (or a subset of it) plays a role in controlling these functions.
A fruitful approach for determining the valence of experiencing activation or inhibition of specific neurons is by optogenetic stimulation, combined with the real‐time place preference test. In this test, the animals may freely choose to enter or avoid an area where the laser light is activated. It has been shown that activation of projections from the lateral hypothalamic area and entopeduncular nucleus to the lateral habenula is aversive, that is, the animals avoid stimulation (Shabel, Proulx, Trias, Murphy, & Malinow, 2012; Stamatakis et al., 2016). In line with this finding, inhibition of the projection from the lateral hypothalamic area to the lateral habenula using an inhibitory light‐sensitive chloride channel, or activation of inhibitory afferents from the ventral tegmental area, was appetitive (Stamatakis et al., 2013). Stimulation of lateral habenular efferents to the rostromedial tegmental nucleus also produced an aversive response (Stamatakis & Stuber, 2012). It was recently demonstrated that activation of a projection from the paraventricular thalamic nucleus to accumbens nucleus evoked the similar avoidance behavior (Zhu, Wienecke, Nachtrab, & Chen, 2016). Given that the aforementioned projections were observed in our study, it is tempting to speculate that modulation of GPR151‐expressing neurons could alter the affective component of an experience. Indeed, selective ablation of Gpr151‐Cre neurons resulted in increased anxiety in the elevated plus maze and open field tests (Kobayashi et al., 2013).
Two distinct populations of entopeduncular neurons project to the lateral habenula (Wallace et al., 2017). Glutamatergic parvalbumin‐expressing neurons preferentially targets the oval subnucleus of the lateral division of the lateral habenula, while a GABA/glutamate coreleasing somatostatin‐expressing population display a more distributed terminal pattern within the lateral habenula. Given that no Gpr151‐Cre was observed in the oval subnucleus, the entopeduncular afferents may belong to this population of somatostatin‐expressing neurons.
4.1. Similarities and differences between habenular and thalamic Gpr151‐Cre populations
It is clear from the efferent connectivity pattern that Gpr151‐Cre neurons of the habenula and the paraventricular thalamic nucleus are distinct populations. The Gpr151‐Cre neurons in paraventricular nucleus project to the accumbens nucleus, zona incerta, basolateral amygdala, and prelimbic area, a pattern that is distinct from the mesencephalic projection of the habenular GPR151‐expressing neurons (Figure 10; Broms et al., 2015). On the other hand, many similarities in afferent connectivity could be noted between the habenular and paraventricular thalamic Gpr151‐Cre populations. Both populations were targeted by neurons in the bed nucleus of stria terminalis, ventral pallidum, nucleus of the diagonal band, lateral preoptic area, and lateral hypothalamic area. However, some striking differences in afferent connectivity were observed. The zona incerta produced a heavy projection onto paraventricular thalamic Gpr151‐Cre neurons, while habenular afferents from this area were sparse. The same held true for the medial preoptic area. In contrast to the habenular population, afferents to the paraventricular thalamic population could not be detected in the bed nucleus of the anterior commissure, entopeduncular nucleus, or in the triangular septum.
The paraventricular nucleus has been implicated in food and drug reward seeking (reviewed in Matzeu, Zamora‐Martinez, & Martin‐Fardon, 2014). In a recent study, it was demonstrated that inhibition of a projection from the paraventricular nucleus to the accumbens nucleus ameliorates symptoms of opiate withdrawal (Zhu et al., 2016). The lateral habenula has been implicated in cocaine‐induced avoidance behavior (Jhou et al., 2013) and the medial habenula contributes to nicotine aversion (Fowler, Lu, Johnson, Marks, & Kenny, 2011). It is therefore an interesting possibility that, despite differences in connectivity patterns, the diencephalic GPR151‐expressing neurons could be functionally related by involvement in the modulation of drug seeking behavior.
4.2. Afferent connectivity of the lateral thalamic Gpr151‐Cre population, involvement in visual processing?
Many neurons in the lateral posterior nucleus, lateral dorsal nucleus, and posterior group of the thalamus displayed Cre recombinase activity (Table 1; Figure 4m–o). In animals where the viral vector injections were targeted on the lateral posterior nucleus, there was always some degree of spread in to the habenula. This precludes clear conclusions to be drawn from these experiments. However, some characteristics of the eGFP+ neuronal distribution nonetheless provide clues regarding the connectivity and function of this thalamic population. Compared to the afferents of the habenular Gpr151‐Cre neurons, a large number of afferents to lateral thalamic Gpr151‐Cre neurons were detected in prefrontal cortical areas (primarily the cingulate cortex), the visual cortex, the caudate putamen, the reticular nucleus, and most notably in the zona incerta and the superior colliculus (Figure 11; Table 2).
Several studies have previously implicated the lateral posterior thalamic nucleus, the superior colliculus, and the zona incerta in visual information processing (Benevento & Fallon, 1975; Gale & Murphy, 2014; Krauzlis, Lovejoy, & Zénon, 2013; Legg, 1979; Power, Leamey, & Mitrofanis, 2001). Interestingly, it has been demonstrated that the lateral posterior thalamic nucleus contains multimodal neurons that are sensitive to both pain and light (Noseda et al., 2010). To clarify if GPR151 is involved in the processing of visual or somatosensory information will require further investigation.
4.3. Cre‐independent expression of viral constructs
In wild type mice injected with Cre‐dependent viral vectors, we occasionally saw TVA‐mCherry and eGFP expression in the vicinity of the injection tract. This finding suggests that there is a certain degree of leakiness in the FLEX switch, but that this ectopic expression of TVA‐mCherry (and presumably also RG) is not strong enough to enable transsynaptic transport of rabies virus particles. This problem has also been noted by other researchers using the same pseudotyped rabies tracing system (Callaway & Luo, 2015; Pollak Dorocic et al., 2014). In Cre‐expressing animals, neurons that are TVA‐mCherry‐positive/eGFP‐positive as a result of leakiness are impossible to distinguish from true starter neurons. We can therefore not conclude that all TVA‐mCherry‐positive/eGFP‐positive neurons in the vicinity of the injection tract are actual starter neurons. Nevertheless, since no neurons were observed outside of the injected area in wild type mice, the Cre‐independent expression does not affect the validity of the analysis of long‐range projections.
4.4. Different sensitivity of the methods used for Gpr151 detection
There are notable differences in expression pattern of GPR151 protein, Gpr151 mRNA, and Gpr151 promoter activity. Immunohistochemistry reveals strong GPR151 protein expression in habenular projections (Broms et al., 2015). In situ hybridization detects Gpr151 mRNA in the habenula, but also in certain thalamic areas (Figure 3; Berthold et al., 2003; Ignatov et al., 2004; Lein et al., 2006; Vassilatis et al., 2003). The expression of Cre‐dependent reporters found in the current study largely corresponds to the mRNA distribution. One exception is the lateral posterior thalamic nuclei, in which only very few neurons expressing Gpr151 mRNA has been detected using in situ hybridization (Figure 3c,d; Lein et al., 2006), but ample Cre‐dependent mCherry expression was seen in the current study (Figures 2, 3, and 4m–o). A possible explanation for this discrepancy is that there is a difference in the sensitivity of the different detection methods. The constructs for the viral reporter employ strong promoters and enhancer sequences (woodchuck hepatitis virus posttranscriptional regulatory element) for improved expression, elements that the endogenous Gpr151 gene is lacking. Furthermore, the reporter constructs are permanently activated by Cre recombination, meaning that even a transient surge in Cre expression will result in persistent tagging of that neuron. As long as a no specific GPR151 agonists or antagonists are available, there is no gold standard for assessing functional GPR151 expression. In the future when such substances have possibly been discovered, the Cre‐expressing populations described in this study should be examined electrophysiologically, in order to verify that GPR151 is expressed at physiologically relevant levels.
4.5. Caveats of using BAC transgenic Cre mice
Gpr151 mRNA and protein could be detected in many, but not all, neurons that exhibited Cre recombinase activity. One possibility is that Cre mediated activation of the reporter construct can occur even at low or transient Gpr151 promoter activation. This would result in persistent reporter expression, while GPR151 protein and Gpr151 mRNA would remain undetectable. Although the Gpr151‐Cre BAC construct is large (108 kbp) and should contain most of the regulatory elements of the endogenous Gpr151 gene, we cannot rule out that the BAC transgenic approach used in this study suffers from ectopic Cre expression independent of the Gpr151 promoter. This could be the result of inserting the BAC transgene into the vicinity of a strong promoter or enhancer (Liu, 2013). Ectopic expression could also arise if the endogenous Gpr151 promoter is epigenetically silenced, while the Gpr151 promoter of the BAC construct remains active. In future studies, knock‐in animals targeting the endogenous Gpr151 gene could be used to minimize the risk of ectopic Cre expression.
5. CONCLUSION
Here, we present a map of the afferent connectivity of a diencephalic neuronal population defined by Gpr151 expression. The connectivity of thalamic versus habenular Gpr151‐expressing neurons differed substantially, indicating a neuroanatomical heterogeneity within the Gpr151‐system. In depth knowledge of the connectivity of this, neuronal population will facilitate the design of investigations regarding the function of GPR151 as well as the interpretation of the effects of pharmacological modulators of this system.
ACKNOWLEDGMENTS
We sincerely thank Maria Ekemohn, Alice Lagebrant, Joakim Ekstrand, and Martin Lundblad for valuable input on the manuscript. This work was supported by the Swedish Research Council, the Royal Physiographic Society of Lund, Stiftelsen Professor Bror Gadelius minnesfond, Gyllenstiernska Krapperupsstiftelsen, and Stiftelsen Ellen och Henrik Sjöbrings minnesfond.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTION
All authors had full access to all the data in the study and take responsibility for the integrity and presentation of the data. Study concept and design: JB, MG, TB, and AT. Acquisition of data: JB, MG, and LH. Analysis and interpretation of data: JB, LH, and AT. Writing of the manuscript: JB and AT. Conceptualization and critical revision of the manuscript for intellectual content: AT and KM. Obtained funding: JB and AT. Study supervision: AT.
Broms J, Grahm M, Haugegaard L, Blom T, Meletis K, Tingström A. Monosynaptic retrograde tracing of neurons expressing the G‐protein coupled receptor Gpr151 in the mouse brain. J Comp Neurol. 2017;525:3227–3250. https://doi.org/10.1002/cne.24273
Funding information Swedish Research Council; Royal Physiographic Society of Lund; Vetenskapsrådet; Stiftelsen Professor Bror Gadelius minnesfond; Kungliga Fysiografiska Sällskapet i Lund; Gyllenstiernska Krapperupsstiftelsen; Stiftelsen Ellen och Henrik Sjöbrings minnesfond
REFERENCES
- Aizawa, H. , Yanagihara, S. , Kobayashi, M. , Niisato, K. , Takekawa, T. , Harukuni, R. , … Okamoto, H. (2013). The synchronous activity of lateral habenular neurons is essential for regulating hippocampal theta oscillation. Journal of Neuroscience, 33(20), 8909–8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amo, R. , Aizawa, H. , Takahoko, M. , Kobayashi, M. , Takahashi, R. , Aoki, T. , & Okamoto, H. (2010). Identification of the zebrafish ventral habenula as a homolog of the mammalian lateral habenula. Journal of Neuroscience, 30(4), 1566–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy, D. , Aponte, Y. , Su, H. H. , & Sternson, S. M. (2008). A FLEX switch targets Channelrhodopsin‐2 to multiple cell types for imaging and long‐range circuit mapping. Journal of Neuroscience, 28(28), 7025–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, P. M. , Raynor, S. A. , Francis, N. T. , & Mizumori, S. J. Y. (2016). Lateral habenula integration of proactive and retroactive information mediates behavioral flexibility. Neuroscience, 345, 89–98. [DOI] [PubMed] [Google Scholar]
- Benevento, L. A. , & Fallon, J. H. (1975). The ascending projections of the superior colliculus in the rhesus monkey (Macaca mulatta). Journal of Comparative Neurology, 160(3), 339–361. [DOI] [PubMed] [Google Scholar]
- Berthold, M. , Collin, M. , Sejlitz, T. , Meister, B. , & Lind, P. (2003). Cloning of a novel orphan G protein‐coupled receptor (GPCR‐2037): In situ hybridization reveals high mRNA expression in rat brain restricted to neurons of the habenular complex. Molecular Brain Research, 120, 22–29. [DOI] [PubMed] [Google Scholar]
- Bianco, I. H. , & Wilson, S. W. (2009). The habenular nuclei: A conserved asymmetric relay station in the vertebrate brain. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 364, 1005–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broms, J. , Antolin‐Fontes, B. , Tingström, A. , & Ibañez‐Tallon, I. (2015). Conserved expression of the GPR151 receptor in habenular axonal projections of vertebrates. Journal of Comparative Neurology, 523(3), 359–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway, E. M. , & Luo, L. (2015). Monosynaptic circuit tracing with glycoprotein‐deleted rabies viruses. Journal of Neuroscience, 35, 8979–8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, M.‐Y. , Amo, R. , Kinoshita, M. , Cherng, B.‐W. , Shimazaki, H. , Agetsuma, M. , … Okamoto, H. (2016). Social conflict resolution regulated by two dorsal habenular subregions in zebrafish. Science, 352, 87–90. [DOI] [PubMed] [Google Scholar]
- Contestabile, A. , & Fonnum, F. (1983). Cholinergic and GABAergic forebrain projections to the habenula and nucleus interpeduncularis: Surgical and kainic acid lesions. Brain Research, 275, 287–297. [DOI] [PubMed] [Google Scholar]
- Corodimas, K. P. , Rosenblatt, J. S. , & Morrell, J. I. (1992). The habenular complex mediates hormonal stimulation of maternal behavior in rats. Behavioral Neuroscience, 106, 853–865. [DOI] [PubMed] [Google Scholar]
- Díaz, E. , Bravo, D. , Rojas, X. , & Concha, M. L. (2011). Morphologic and immunohistochemical organization of the human habenular complex. Journal of Comparative Neurology, 519, 3727–3747. [DOI] [PubMed] [Google Scholar]
- Dong, H.‐W. , & Swanson, L. W. (2004). Projections from bed nuclei of the stria terminalis, posterior division: Implications for cerebral hemisphere regulation of defensive and reproductive behaviors. Journal of Comparative Neurology, 471, 396–433. [DOI] [PubMed] [Google Scholar]
- Donovick, P. J. , Burright, R. G. , & Zuromski, E. (1970). Localization of quinine aversion within the septum, habenula, and interpeduncular nucleus of the rat. Journal of Comparative and Physiological Psychology, 71(3), 376–383. [DOI] [PubMed] [Google Scholar]
- Fowler, C. D. , & Kenny, P. J. (2013). Nicotine aversion: Neurobiological mechanisms and relevance to tobacco dependence vulnerability. Neuropharmacology, 76(Pt. B), 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, C. D. , Lu, Q. , Johnson, P. M. , Marks, M. J. , & Kenny, P. J. (2011). Habenular α5 nicotinic receptor subunit signalling controls nicotine intake. Nature, 471, 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, K. , & Paxinos, G. (2008). The mouse brain in stereotaxic coordinates, compact (3rd ed.) Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- Funato, H. , Sato, M. , Sinton, C. M. , Gautron, L. , Williams, S. C. , Skach, A. , … Yanagisawa, M. (2010). Loss of Goosecoid‐like and DiGeorge syndrome critical region 14 in interpeduncular nucleus results in altered regulation of rapid eye movement sleep. Proceedings of the National Academy of Sciences of the United States of America, 107, 18155–18160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, S. D. , & Murphy, G. J. (2014). Distinct representation and distribution of visual information by specific cell types in mouse superficial superior colliculus. Journal of Neuroscience, 34(40), 13458–13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardon, O. , Faget, L. , Chu Sin Chung, P. , Matifas, A. , Massotte, D. , & Kieffer, B. L. (2014). Expression of mu opioid receptor in dorsal diencephalic conduction system: New insights for the medial habenula. Neuroscience, 277, 595–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gey, M. , Wanner, R. , Schilling, C. , Pedro, M. T. , Sinske, D. , & Knöll, B. (2016). Atf3 mutant mice show reduced axon regeneration and impaired regeneration‐associated gene induction after peripheral nerve injury. Open Biology, 6(8). pii: 160091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, S. A. , Heshmati, M. , Flanigan, M. , Christoffel, D. J. , Guise, K. , Pfau, M. L. , … Russo, S. J. (2016). Basal forebrain projections to the lateral habenula modulate aggression reward. Nature, 534, 688–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greatrex, R. M. , & Phillipson, O. T. (1982). Demonstration of synaptic input from prefrontal cortex to the habenula in the rat. Brain Research, 238, 192–197. [DOI] [PubMed] [Google Scholar]
- Herkenham, M. , & Nauta, W. J. (1977). Afferent connections of the habenular nuclei in the rat. A horseradish peroxidase study, with a note on the fiber‐of‐passage problem. Journal of Comparative Neurology, 173, 123–146. [DOI] [PubMed] [Google Scholar]
- Holmes, F. E. , Kerr, N. , Chen, Y.‐J. , Vanderplank, P. , McArdle, C. A. , & Wynick, D. (2017). Targeted disruption of the orphan receptor Gpr151 does not alter pain‐related behaviour despite a strong induction in dorsal root ganglion expression in a model of neuropathic pain. Molecular and Cellular Neuroscience, 78, 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, Y.‐W. A. , Morton, G. , Guy, E. G. , Wang, S. D. , & Turner, E. E. (2016). Dorsal medial habenula regulation of mood‐related behaviors and primary reinforcement by tachykinin‐expressing habenula neurons. eNeuro, 3. ENEURO.0109‐16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, Y.‐W. A. , Wang, S. D. , Wang, S. , Morton, G. , Zariwala, H. A. , de la Iglesia, H. O. , & Turner, E. E. (2014). Role of the dorsal medial habenula in the regulation of voluntary activity, motor function, hedonic state, and primary reinforcement. Journal of Neuroscience, 34, 11366–11384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatov, A. , Hermans‐Borgmeyer, I. , & Schaller, H. C. (2004). Cloning and characterization of a novel G‐protein‐coupled receptor with homology to galanin receptors. Neuropharmacology, 46, 1114–1120. [DOI] [PubMed] [Google Scholar]
- Jhou, T. C. , Good, C. H. , Rowley, C. S. , Xu, S.‐P. , Wang, H. , Burnham, N. W. , … Ikemoto, S. (2013). Cocaine drives aversive conditioning via delayed activation of dopamine‐responsive habenular and midbrain pathways. Journal of Neuroscience, 33, 7501–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, Y. , Sano, Y. , Vannoni, E. , Goto, H. , Suzuki, H. , Oba, A. , … Itohara, S. (2013). Genetic dissection of medial habenula‐interpeduncular nucleus pathway function in mice. Frontiers in Behavioral Neuroscience, 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauzlis, R. J. , Lovejoy, L. P. , & Zénon, A. (2013). Superior colliculus and visual spatial attention. Annual Review of Neuroscience, 36, 165–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legg, C. R. (1979). Visual discrimination impairments after lesions in zona incerta or lateral terminal nucleus of accessory optic tract. Brain Research, 177, 461–478. [DOI] [PubMed] [Google Scholar]
- Lein, E. S. , Hawrylycz, M. J. , Ao, N. , Ayres, M. , Bensinger, A. , Bernard, A. , … Jones, A. R. (2006). Genome‐wide atlas of gene expression in the adult mouse brain. Nature, 445(7124), 168–176. [DOI] [PubMed] [Google Scholar]
- Liu, C. (2013). Strategies for designing transgenic DNA constructs In Freeman Lita A. (Ed.), Lipoproteins and cardiovascular disease. Methods in molecular biology (Vol. 1027, pp. 183–201). Totowa, NJ: Humana Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzeu, A. , Zamora‐Martinez, E. R. , & Martin‐Fardon, R. (2014). The paraventricular nucleus of the thalamus is recruited by both natural rewards and drugs of abuse: Recent evidence of a pivotal role for orexin/hypocretin signaling in this thalamic nucleus in drug‐seeking behavior. Frontiers in Behavioral Neuroscience, 8, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegas, W. , Bergan, J. F. , Ogawa, S. K. , Isogai, Y. , Umadevi Venkataraju, K. , Osten, P. , … Watabe‐Uchida, M. (2015). Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. Elife, 4, e10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noseda, R. , Kainz, V. , Jakubowski, M. , Gooley, J. J. , Saper, C. B. , Digre, K. , & Burstein, R. (2010). A neural mechanism for exacerbation of headache by light. Nature Neuroscience, 13, 239–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak Dorocic, I. , Fürth, D. , Xuan, Y. , Johansson, Y. , Pozzi L., Silberberg, G. , … Meletis, K. (2014). A whole‐brain atlas of inputs to serotonergic neurons of the dorsal and median raphe nuclei. Neuron, 83, 663–678. [DOI] [PubMed] [Google Scholar]
- Power, B. D. , Leamey, C. A. , & Mitrofanis, J. (2001). Evidence for a visual subsector within the zona incerta. Visual Neuroscience, 18, 179–186. [DOI] [PubMed] [Google Scholar]
- Qin, C. , & Luo, M. (2009). Neurochemical phenotypes of the afferent and efferent projections of the mouse medial habenula. Neuroscience, 161, 827–837. [DOI] [PubMed] [Google Scholar]
- Quina, L. A. , Tempest, L. , Ng, L. , Harris, J. , Ferguson, S. , Jhou, T. , & Turner, E. E. (2014). Efferent pathways of the mouse lateral habenula. Journal of Comparative Neurology, 523(1), 32–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold, A. K. , Batti, L. , Bilbao, D. , Buness, A. , Rittner, H. L. , & Heppenstall, P. A. (2015). Differential transcriptional profiling of damaged and intact adjacent dorsal root ganglia neurons in neuropathic pain. PLoS One, 10, e0123342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabel, S. J. , Proulx, C. D. , Trias, A. , Murphy, R. T. , & Malinow, R. (2012). Input to the lateral habenula from the basal ganglia is excitatory, aversive, and suppressed by serotonin. Neuron, 74, 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton, L. , Becerra, L. , & Borsook, D. (2012). Unmasking the mysteries of the habenula in pain and analgesia. Progress in Neurobiology, 96, 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. M. , Jennings, J. H. , Ung, R. L. , Blair, G. A. , Weinberg, R. J. , Neve, R. L. , … Stuber, G. D. (2013). A unique population of ventral tegmental area neurons inhibits the lateral habenula to promote reward. Neuron, 80, 1039–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. M. , & Stuber, G. D. (2012). Activation of lateral habenula inputs to the ventral midbrain promotes behavioral avoidance. Nature Neuroscience, 15(8), 1105–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. M. , Van Swieten, M. , Basiri, M. L. , Blair, G. A. , Kantak, P. , & Stuber, G. D. (2016). Lateral hypothalamic area glutamatergic neurons and their projections to the lateral habenula regulate feeding and reward. Journal of Neuroscience, 36, 302–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson‐Jones, M. , Floros, O. , Robertson, B. , & Grillner, S. (2011). Evolutionary conservation of the habenular nuclei and their circuitry controlling the dopamine and 5‐hydroxytryptophan (5‐HT) systems. Proceedings of the National Academy of Sciences of the United States of America, 109(3), E164–E173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopper, C. M. , & Floresco, S. B. (2013). What's better for me? Fundamental role for lateral habenula in promoting subjective decision biases. Nature Neuroscience, 17(1), 33–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valjakka, A. , Vartiainen, J. , Tuomisto, L. , Tuomisto, J. T. , Olkkonen, H. , & Airaksinen, M. M. (1998). The fasciculus retroflexus controls the integrity of REM sleep by supporting the generation of hippocampal theta rhythm and rapid eye movements in rats. Brain Research Bulletin, 47, 171–184. [DOI] [PubMed] [Google Scholar]
- van Kerkhof, L. W. M. , Damsteegt, R. , Trezza, V. , Voorn, P. , & Vanderschuren, L. J. M. J. (2013). Functional integrity of the habenula is necessary for social play behaviour in rats. European Journal of Neuroscience, 38, 3465–3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilatis, D. K. , Hohmann, J. G. , Zeng, H. , Li, F. , Ranchalis, J. E. , Mortrud, M. T. , … Gaitanaris, G. A. (2003). The G protein‐coupled receptor repertoires of human and mouse. Proceedings of the National Academy of Sciences of the United States of America, 100, 4903–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertes, R. P. , Fortin, W. J. , & Crane, A. M. (1999). Projections of the median raphe nucleus in the rat. Journal of Comparative Neurology, 407(4), 555–582. [PubMed] [Google Scholar]
- Vertes, R. P. , Kinney, G. G. , Kocsis, B. , & Fortin, W. J. (1994). Pharmacological suppression of the median raphe nucleus with serotonin1a agonists, 8‐OH‐DPAT and buspirone, produces hippocampal theta rhythm in the rat. Neuroscience, 60, 441–451. [DOI] [PubMed] [Google Scholar]
- Wagner, F. , French, L. , & Veh, R. W. (2014). Transcriptomic‐anatomic analysis of the mouse habenula uncovers a high molecular heterogeneity among neurons in the lateral complex, while gene expression in the medial complex largely obeys subnuclear boundaries. Brain Structure and Function, 221(1), 39–58. [DOI] [PubMed] [Google Scholar]
- Wagner, F. , Stroh, T. , & Veh, R. W. (2014). Correlating habenular subnuclei in rat and mouse using topographical, morphological and cytochemical criteria. Journal of Comparative Neurology, 522(11), 2650–2662. [DOI] [PubMed] [Google Scholar]
- Wallace, M. L. , Saunders, A. , Huang, K. W. , Philson, A. C. , Goldman, M. , Macosko, E. Z. , … Sabatini, B. L. (2017). Genetically distinct parallel pathways in the entopeduncular nucleus for limbic and sensorimotor output of the basal ganglia. Neuron, 94, 138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Flanagan, J. , Su, N. , Wang, L.‐C. , Bui, S. , Nielson, A. , … Luo, Y. (2012). RNAscope: A novel in situ RNA analysis platform for formalin‐fixed, paraffin‐embedded tissues. The Journal of Molecular Diagnostics, 14, 22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warden, M. R. , Selimbeyoglu, A. , Mirzabekov, J. J. , Lo, M. , Thompson, K. R. , Kim, S.‐Y. , … Deisseroth, K. (2012). A prefrontal cortex‐brainstem neuronal projection that controls response to behavioural challenge. Nature, 92(7429), 428–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe‐Uchida, M. , Zhu, L. , Ogawa, S. K. , Vamanrao, A. , & Uchida, N. (2012). Whole‐brain mapping of direct inputs to midbrain dopamine neurons. Neuron, 74, 858–873. [DOI] [PubMed] [Google Scholar]
- Wickersham, I. R. , Lyon, D. C. , Barnard, R. J. O. , Mori, T. , Finke, S. , Conzelmann, K.‐K. , … Callaway, E. M. (2007). Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron, 53, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T. , Danjo, T. , Pastan, I. , Hikida, T. , & Nakanishi, S. (2013). Distinct roles of segregated transmission of the septo‐habenular pathway in anxiety and fear. Neuron, 78, 537–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, K. , Deuis, J. R. , Lewis, R. J. , & Vetter, I. (2016). Transcriptomic and behavioural characterisation of a mouse model of burn pain identify the cholecystokinin 2 receptor as an analgesic target. Molecular Pain, 28, pii: 1744806916665366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Wienecke, C. F. R. , Nachtrab, G. , & Chen, X. (2016). A thalamic input to the nucleus accumbens mediates opiate dependence. Nature, 530, 219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]


