Abstract
In minute virus of mice (MVM) capsids, icosahedral five-fold channels serve as portals mediating genome packaging, genome release, and the phased extrusion of viral peptides. Previous studies suggest that residues L172 and V40 are essential for channel function. The structures of MVMi wildtype, and mutant L172T and V40A virus-like particles (VLPs) were solved from cryo-EM data. Two constriction points, termed the mid-gate and inner-gate, were observed in the channels of wildtype particles, involving residues L172 and V40 respectively. While the mid-gate of V40A VLPs appeared normal, in L172T adjacent channel walls were altered, and in both mutants there was major disruption of the inner-gate, demonstrating that direct L172:V40 bonding is essential for its structural integrity. In wildtype particles, residues from the N-termini of VP2 map into claw-like densities positioned below the channel opening, which become disordered in the mutants, implicating both L172 and V40 in the organization of VP2 N-termini.
Keywords: Parvovirus, Minute virus of mice, MVM, cryo-EM image reconstruction, gatekeeper mutation, five-fold channel, mutant capsid, low resolution cryo-EM density, packaging single-stranded DNA, entry/exit portal dynamics
Background
Minute Virus of Mice (MVM) is a member of genus Protoparvovirus in the family Parvoviridae (Cotmore et al., 2014). These viruses have small (~26nm diameter) non-enveloped T=1 icosahedral protein capsids (Agbandje-McKenna et al., 1998; Llamas-Saiz, 1997), containing a single copy of a linear single-stranded DNA genome of ~5kb (Bourguignon et al., 1976). Capsids are assembled from a total of sixty VP1 (83kDa) and VP2 (63kDa) polypeptides, in an approximately 1:5 ratio (Tattersall et al., 1976), which are encoded by alternative splicing from a single structural gene, such that VP1 includes the entire sequence of VP2 plus an additional 142 amino acid N-terminal peptide (Pintel et al., 1983; Labienec-Pintel and Pintel, 1986). Atomic structures derived by X-ray crystallography of wildtype and mutant MVM particles (Llamas-Saiz et al., 1997, Agbandje et al., 1998; Kontou et al., 2005; Plevka et al., 2011) identified unique positions for the C-termini of these proteins (VP2 residues 39 to 585 in MVMi), but provided little information about the disposition of the VP N-termini. On the surface of the particle, raised cylinders or “towers” surround each icosahedral five-fold vertex (Tsao et al., 1991; Chapman & Rossmann, 1993; Llamas-Saiz et al., 1997; Agbandje-Mckenna et al., 1998). As shown in Fig. 1, these towers are formed by the juxtaposition of antiparallel β-strand hairpins contributed by the five symmetry-related structural proteins, which enclose a channel that penetrates from the outer surface of the capsid to its inner core. Previous studies indicate that these channels serve as portals, mediating the inward and outward movement of virion components at multiple stages in the viral life cycle (reviewed in Cotmore and Tattersall, 2014).
Fig 1.
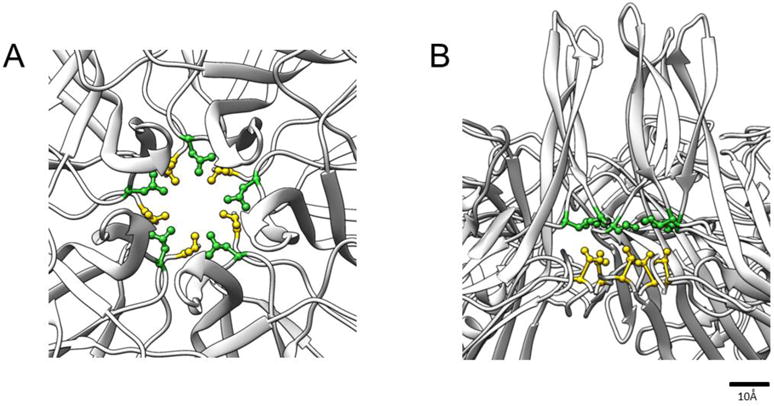
The view down the icosahedral five-fold axis (A) and a cut-away side view (B) from the crystal structure of MVMp VP2-only VLPs (grey ribbon) (PDB ID 1Z14: Kontou et al., 2005) show that the five-fold channel is constructed from symmetry-related VP2 molecules. The position of L172 (green) and V40 (yellow) are shown with their side chains rendered as ball and sticks.
One unique five-fold channel creates a packaging conduit, supporting the 3′-to-5′ encapsidation of a linear single-stranded DNA genome into a preassembled empty capsid, driven by the helicase activity of NS1 (King et al., 2001; Farr and Tattersall 2004; Cotmore and Tattersall, 2005; Plevka et al. 2011). Viral genomes are ultimately released from the intact capsid, also in a 3′-to-5′ direction, through a single five-fold channel, or “exit” portal, which initial analyses suggest is different from the packaging portal (Cotmore et al 2010; Cotmore and Tattersall, 2012). In MVM and other viruses from genus Protoparvovirus, the channels have further adapted to mediate a complex pattern of capsid protein dynamics, progressively delivering effector VP N-terminal peptide sequences to the particle surface at appropriate times during the life cycle. Thus, the five-fold channels of viruses from genus Protoparvovirus appear to have adapted to support a protracted program of nucleic acid and peptide extrusion and retraction during infectious entry, but currently we have little structural information to document how this critical process is controlled.
Here we report our use of cryo-EM to explore the structures of MVMi wildtype and mutant VLPs carrying L172T or V40A substitutions. These structures, obtained at resolutions ranging from 6.2 to 7.6 Å, allow us to re-examine the conformation of the five-fold cylinder and to identify disordered regions in the mutants. Changes occurred predominantly at constrictions we have named the mid-gate, which includes residue L172, and the inner-gate, which involves V40. In wildtype crystal structures the side chains of each symmetry-related L172 and V40 residue (shown in Fig. 1) are linked by hydrophobic bonding, and it has been suggested that this interaction might be important for the organization and stability of the base of the channel and/or the disposition of the VP2 N-terminal peptides within the virion (Plevka et al, 2011). Here we show that this suggestion is correct on both counts. First, the single L172T mutation, which cannot bond with V40, leads to complete destabilization of the inner gate even though residue 172 is physically distant from this structure and all of the normal inner gate residues are present. Second, the intermediate resolution of the maps allows us to visualize and identify VP2 N-terminal densities that have not been seen before in MVM VLPs, perhaps in part because the flexibility of this peptide causes the residues to become disordered upon further refinement or at higher contour levels. These claw and funnel-shaped densities are highly disrupted in the mutants, and by tracing the N-terminal sequence into them it is possible to predict how both of the individual mutations change the conformation of VP N-terminal extensions in the capsid interior.
Results
Cryo-EM maps showed no gross exterior differences in morphology
VLPs were purified by iodixanol gradient centrifugation, and the purity, concentration and molecular integrity of their constituent proteins verified by SDS-PAGE and western blots, using anti-peptide antibodies raised against sequences within either the extreme N-terminus or central region of VP2 (Fig. 2A). Aliquots of each of the three VLPs, wildtype, L172T, and V40A were vitrified for cryo-EM data collection and image reconstruction (Fig. 2B). The density maps derived from these particles ranged in resolution from 6.0 to 7.9 Å (Table 1). Comparison of the external capsid structures of the wildtype, L172T, and V40A MVMi VLPs showed no major structural differences, suggesting that the mutations did not affect capsid assembly (Fig. 3, top row). The characteristic raised five-fold towers and surrounding canyons appeared similar in all structures, except that the wildtype channels appear open to the external environment, while those of L172T and V40A appear closed (Fig. 3, middle row).
Fig 2.
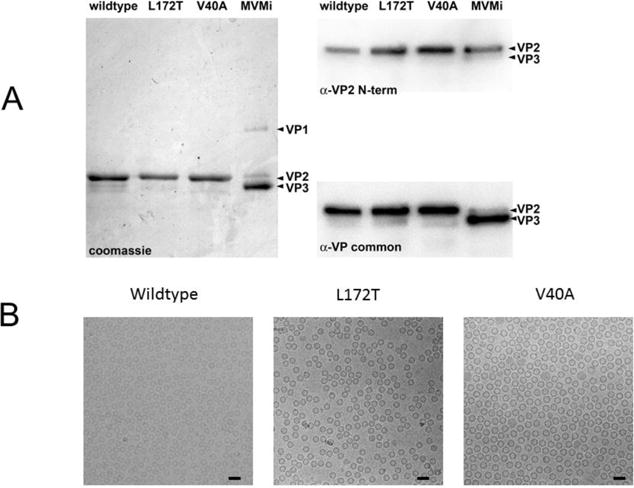
(A) The left panel shows a Coomassie Brilliant Blue-stained SDS-PAGE of total protein present in samples of purified MVMi wildtype, L172T and V40A VP2-only VLPs, run in parallel with authentic MVMi virions. The latter show the positions of VP1 and VP2, and of VP3, the proteolytic cleavage product found in DNA-containing virions, which has lost ~25 amino acids from its N-terminus. The top right panel shows a western blot of the VP2/VP3 region of a parallel SDS-PAGE probed with α-VP2 N-term, a rabbit polyclonal antibody raised against a 25mer peptide with the sequence NH2 - M S D G T S Q P D G G N A V H S A A R V E R A A - COOH, corresponding to residues 1 to 24 of the VP2 sequence. The bottom right panel shows the same western blot probed with α-VP common, a rabbit polyclonal antibody raised against an 18mer oligopeptide with the sequence NH2 - Q G S R H G A T Q M E V N W V S K - COOH, located in the MVMi VP sequence from amino acid residues 453 to 469 of VP1 and 311 to 327 of VP2. (B) Cryo-EM micrographs of vitrified MVM show (left to right) assembled wildtype, L172T, and V40A VLPs (50 nm scalebar).
Table 1.
Cryo-EM statistics of MVM Wildtype, L172T and V40A
| Capsid type | No. of Micrographs | Microscope Pixel size | Defocus | No. of particles | No. of particles in map | Resolution (Å) | Absolute pixel size |
|---|---|---|---|---|---|---|---|
| Wildtype | 38 | 1.38 | 2.85–4.48 | 31,630 | 31,614 | 6.5 | 1.28 |
| L172T | 5 | 1.38 | 2.73–4.22 | 12,311 | 5,541 | 7.9 | 1.33 |
| V40A | 49 | 1.27 | 1.10–4.79 | 83,413 | 76,820 | 6.0 | 1.27 |
Fig 3.
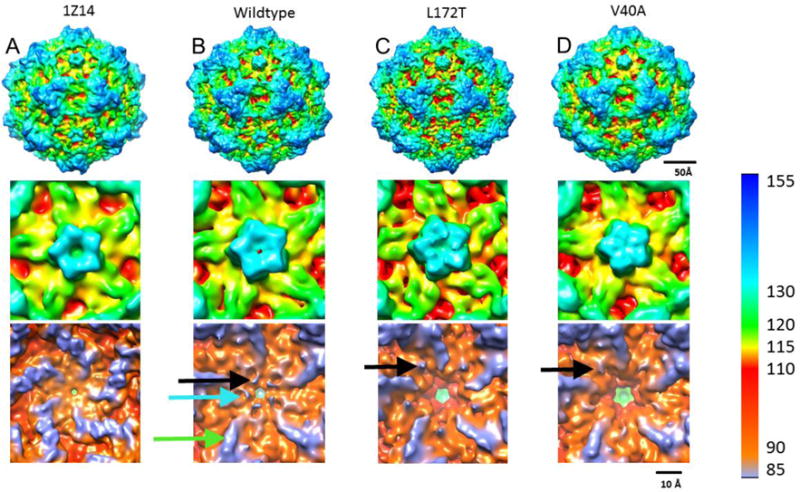
Maps calculated from the X-ray structure PDB: ID 1Z14 (A), and three cryo-EM structures of wildtype (B), L172T (C), and V40A (D) are surface rendered and displayed at a contour level of 1 sigma, with color coding according to radius (see color key). Overall exterior structures are remarkably similar between wildtype and the mutant VLP maps (upper panels). Zoomed views down the five-fold axis illustrate the vertices (middle panels). Both wildtype maps have a visible opening whereas the channel is occluded for the L172T and V40A mutants. Lower panels show the inner capsid surface of the five-fold channels, which have significantly different topologies. The calculated 1Z14 map has a smooth interior whereas the wildtype cryo-EM map has claw-like projections around the five-fold (black arrow) positioned near pit-like indentations (blue arrow) that extend out from ridges (green arrow). Rearrangements of the claw-like projections are seen in L172T and V40A (black arrows) along with the loss of the wildtype pit-like indentations.
Differences in five-fold channel morphology between wildtype and mutant VLPs
In contrast when reconstructions were viewed from the particle interior, major differences were observed in the shape and 3-D structures of the five-fold channels (Fig. 3, bottom row). As a basis for comparison, the X-ray structure of MVM VLPs (PDB ID: 1Z14; Kontou et al., 2005) was used to simulate a 6Å density map (Fig. 3A). The five-fold channel in this simulated map appears completely open and the adjacent inner surface is smooth due to the lack of any ordered N-terminal residues prior to 39. In comparison, the wildtype cryo-EM map has distinct extra density not resolved in the X-ray map. This additional density, which is as strong in magnitude as portions of the capsid, forms claw-like projections (Fig. 3B, black arrow) that encircle the base of the channel. There are also distinct pit-like indentations (Fig. 3B, cyan arrow) positioned next to the channel opening. Both mutant cryo-EM maps also exhibit claw-like projecting densities, but with distinct rearrangements compared to wildtype. The claw-like structures appear to extend from the five-fold ridges in the wildtype map (Fig. 3B, green arrow), whereas the claws appear between the ridges in L172T and V40A (Figs. 3C&D). Neither mutant has structures corresponding to the wildtype pits.
Viewed from the side, the channels of wildtype VLPs have an hourglass-like shape, with two constrictions or “gates” (Fig. 4A, panel 1). The mid-gate in the middle of the hourglass corresponds to the tightest constriction point and a second, inner-gate, encircles the base of the five-fold channel. However, while the mid-gate of the V40A mutant resembles the wildtype structure, in L172T the mid-gate exhibits stronger density and has a different topology, and in both mutants the inner-gate is largely missing and the base of the pore appears to flare open to the capsid interior (Panel 1 of Figs. 4&C).
Fig 4.
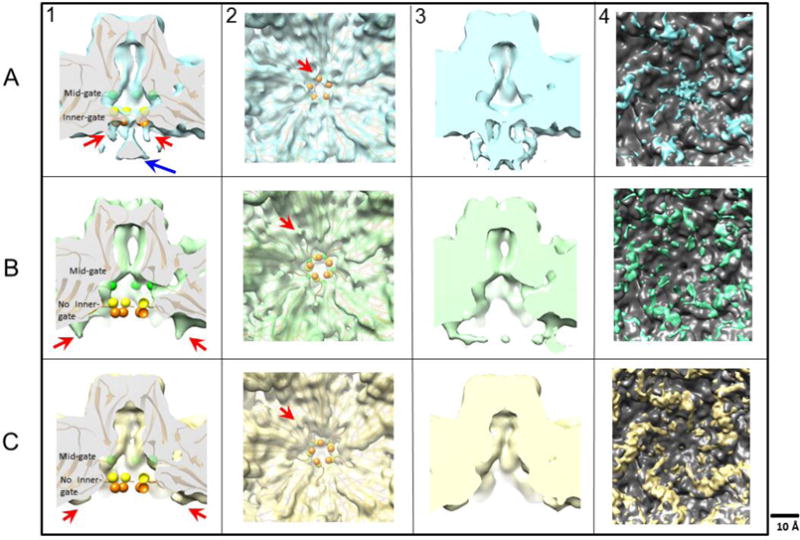
In panel 1, maps of wildtype (A-1), L172T (B-1), and V40A (C-1) VLPs were surface rendered and a central cross-section view of the five-fold channel is shown with the alpha carbon trace from the crystal structure (PDB ID 1Z14) fitted into the cryo-EM densities. The shape of the pore in L172T cuts back sharply from the enhanced mid-gate constriction, leaving the fitted L172 residue (green spheres) out of density compared to the wildtype and V40A VLPs. Below the mid-gate, the base of the pore in V40A also opens, so that the fitted residues G39 (orange spheres) and V40 (yellow spheres) are out of density, and the inner constriction of the channel is lost. In the wildtype map, the five-fold density funnel (blue arrow in panel A-1) has magnitude as strong as the capsid density, and remains strong even at higher contour levels at which some capsid densities disappear. In panel 2, the five-fold vertex is viewed along the five-fold axis from the interior of the capsid. Claw-like densities surrounding the base of the five-fold pore (red arrows) are rendered at a density contour of 1 sigma. Panel 3 compares maps at lower contour levels (sigma = 0.6), where more flexible densities can be seen. Here, the claw-like features merge with the funnel density in the wildtype map (blue). Although claw-like densities were seen for both L172T (green) and V40A (yellow), compared to wildtype they are in a different location, laterally displaced away from the five-fold axis. Panel 4 shows the difference densities between the MVM crystal structure (PDB ID: 1Z14) rendered as a density map and the cryo-EM maps of wildtype, L172T and V40A VLPs. The difference densities were segmented and the density around the five-fold were quantified. The volume of the wildtype, L172T and V40A claws were 697, 1287 and 1762 respectively (not shown).
In order to identify the residues that contribute to the gate densities, an MVMp VP2 VLP crystal structure (residues 39–587 from PDB ID: 1Z14; Kontou et al., 2005) was fitted into the cryo-EM reconstructions. Residues 1–38 of the crystal structure were not resolvable. Although residue 160 in the VP2 of the MVMp strain is leucine, while it is serine in the MVMi-derived VLPs, this difference did not appear to influence any structural changes. As seen at a contour level of 1 sigma (Figs. 4A&C, panel 1), residue 172 (green spheres) from the X-ray structure sits within reconstructed density at the base of the mid-gate constriction in the wildtype and V40A structures, but is out of density in the L172T mutant (Fig. 4B, panel 1), where it maps below the altered mid-gate constriction point, indicating that the L172T substitution changes the contour of the channel at this point. In the X-ray structure, G39 (orange spheres) and V40 (yellow spheres) are the first ordered VP2 residues. These residues fit into the arms of the wildtype inner-gate map. However, in the cryo-EM reconstructions of both L172T and V40 VLPs, the inner-gate constriction is not formed, residues G39 and V40 are out of density, and the base of the pore gapes open to the particle interior.
Significant unfilled density is present at the base of the pore in wildtype and L172T VLPs
Comparison of five-fold channel structures also revealed extra densities unaccounted for in the crystal structure extending into the particle interior from the base of the vertex (Fig. 4, panels 1 and 2; red arrows). In the wildtype particles these “claw-like” densities surrounding the five-fold axis point into the capsid interior toward a funnel-shaped density positioned directly underneath the channel (Fig. 4A). In the wildtype map, the density for the claws and the five-fold funnel (blue arrow in panel A) are equal in magnitude to capsid densities, suggesting that they are not an averaging artifact. As the contour level is lowered, these claws extend and merge with the enlarged funnel density directly beneath the channel (Fig. 4A, panel 3). In wildtype particles, residue G41 maps directly above the claw densities surrounding the five-fold pore. However, in the L172T mutant, the claw-like densities are further away from the five-fold symmetry axis (Fig. 4B), and residue S46 maps above the beginning of the claw, with N-terminal residues 39–45 extending out of density. In the V40A mutant, the claws appear closer to the five-fold compared to L172T, but further away than in the wildtype, and residue T44 maps above the claw, with N-terminal residues 39–43 extending out of density. Difference density between the MVM crystal structure (PDB ID: 1Z14) rendered as a density map and the wildtype, L172 and V40A cryo maps corresponds to the unfilled density around the 5-fold. The fittings and the difference maps therefore suggest that these densities are the N-terminal residues of VP2, which have never been resolved in X-ray structures of empty MVM particles or VLPs (Kontou et al., 2005), and which occupy two distinct patches of submolar density in crystal structures of DNA-filled particles (Agbandje-McKenna et al., 1998). As reported previously (Cifuente et al., 2013), one advantage of modest resolution cryo-EM maps is that they sometimes include densities corresponding to residues that are entirely eliminated from crystal structures because they adopt multiple, nearly similar, conformations or are flexible. We conclude that the high density, claw-like structures that can be visualized in our relatively low-resolution cryo-EM maps of wildtype and L172T MVMi are consistent with the flexible N-termini of VP2 proteins extending into the capsid interior.
To further explore the unfilled densities, the N-terminal carbon backbone of the fitted wildtype crystal structure was built into the cryo-EM claws (Fig. 5). In the alpha carbon trace for wildtype, N-terminal residues 41–43 fill the claw and, at lower contour, lead into the funnel, potentially accommodating G36, G37, and G38 although these residues could not be positioned due to the bulbous nature of the density. In the L172T mutant build, residues S46 mapped to the beginning of the claw and 39–45 extended into the capsid interior (Fig. 5B). Although there is extra density near the L172T claws, it is too noisy for reliably predicting any structure beyond residue 39 or 40, and there is no funnel of density as seen in the wildtype. For V40A, T44 mapped to the beginning of the claw, although further interpretation is not possible due to the weak density. For comparison, a recently published Human Bocavirus-1 structure was accessed from EMDB (EMDB-8598) (Mietzsch et al., 2017) and similar claw-like projections were seen around the five-fold. Though the sequence similarity between the two parvoviruses is not high, the residues of interest, i.e. G39, V40 and L172 of MVM, are conserved in HBoV-1, where their equivalent resdiues are G35, V36 and L151, respectively.
Fig 5.
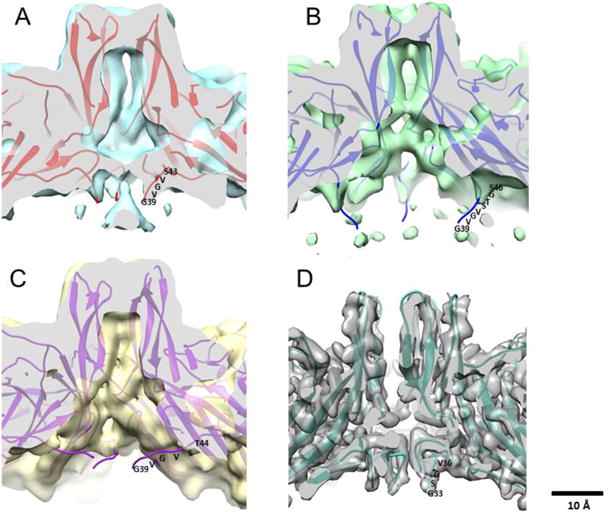
Wildtype crystal structure (PDB ID 1Z14) fitted into cryo-EM maps (surface rendered at 1 sigma) of MVMi wildtype (blue) (A), L172T (green) (B), V40A (yellow). The cut away view shows the five-fold channel of each map. N-terminal residues were built into the unfilled claw densities by tracing the alpha carbon backbone of wildtype residues 43-39 (red), L172T residues 46-39 (blue) and V40A 44-39 (purple). (D) High resolution HBoV-1 map shows claw-like features at the base of the five-fold with G33 being the first ordered residue of the N-terminus (EMDB-8598) (PDB ID: 5URF) (Mietzsch et al., 2017) for comparison to the wildtype MVM five-fold in panel A.
Compared to the wildtype crystal structure (PDB ID 1Z14), the VLP N-terminal structures for wildtype, L172T, and V40A superimposed with a root mean square deviation of 3.3, 10.2, and 6.8Å respectively, revealing a dramatic rearrangement of the VLP N-terminus, especially in the mutants. Figure 6A shows superimposition of all the available crystal structures of MVM with the first ordered residue labeled. The positions of the N-termini in the new cryo-EM builds were markedly different to those in previous crystal structures, even including the wildtype VLP N-terminus, which takes on a conformation in the cryo-EM map different from any reported previously. Fig. 6C shows the superimposition of capsid structures of other parvoviruses such as canine parvovirus (CPV)(pink), feline panleukopenia virus (FPV)(green), human erythrovirus B19 (blue), and bovine parvovirus (BPV)(orange), and illustrates that the VP2 N-termini described here have not previously been visualized.
Fig 6.
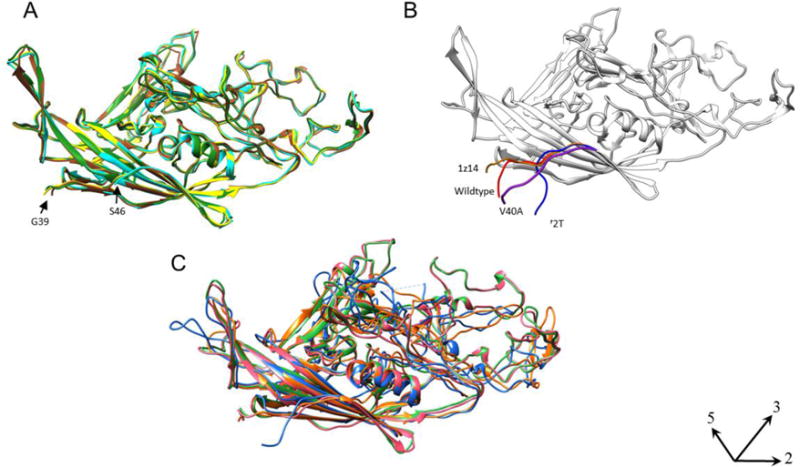
(A) The known crystal structures of MVMp (PDB ID 1Z14; Kontou et al., 2005) (Brown), MVMi with DNA (PDB ID 1MVM; Llamaz-saiz et al., 1997) (Green), MVMi VLP (PDB ID 1Z1C; Kontou et al., 2005) (Yellow), MVMi L172W (PDB ID 2XGK; Plevka et al., 2011) (Cyan) were superimposed to compare the structures of the N-termini. The first ordered N-terminal residue ranges from G39 (MVMp, MVMi, MVMi VLP) to S46 for MVMi L172W. (B) The wildtype crystal structure of MVMp (PDB ID 1Z14) (Brown) is superimposed with the N-terminal residues that were built into the cryo-EM maps of wildtype (red), L172T (blue) and V40A (purple) VLPs. Both panels show the structure oriented with five-fold axis positioned at the left (axes indicated, lower right). (C) The known crystal structures of other related parvoviruses CPV (PDB ID 1C8D; Simpson et al., 2000) (Pink), FPV (PDB ID 1C8E; Simpson et al., 2000) (Green), B19 (PDB ID 1S58; Kaufmann et al., 2004) (Blue), and BPV (PDB ID: 4QC8; Kailasan et al., 2015) (Orange) were superimposed to show that the VP2 N-termini of parvoviruses have not been visualized before.
Discussion
Although the map resolutions we obtained are modest, they allow us to compare density features, fit the crystal structure, and build the carbon chain into unfilled cryo-EM density. These maps reveal the impact of specific mutations lining the MVM five-fold cylinders, and allows us to link the structural changes to previously described defects in portal function. Indeed, the moderate resolution we obtained may have facilitated the visualization of potentially flexible features, such as the disordered N-termini of VP2, since we were able to build up to seven residues from this peptide into claw- and funnel-shaped internal densities that have not been resolved in previous MVM VLP structures. An alternative explanation for this disparity might be that capsids crystallize more efficiently, or they give more desirable resolution, when their N-termini adopt a specific conformation that is unlike the wildtype structure captured by cryo-EM. This scenario might also explain why the first five ordered VP2 N-terminal residues in the available crystal structures did not superimpose with the alpha carbon trace built into the observed wildtype cryo-EM map.
The L172T and V40A mutations were designed to explore the effects of disrupting two linked, but potentially separable, aspects of cylinder architecture, namely 1) the significance of L172:V40 hydrogen bonds that normally link the mid- and inner-gate constrictions, which are disrupted by both mutations, and 2) the role of the L172 mid-gate constriction point, which would only be apparent in the L172T mutant. Cryo-EM maps show that the V40A substitution leads to major disruption of the inner gate constriction, but so does the L172T mutation, even though this residue is physically distant from the channel entrance and all of the normal inner gate amino acids are present. Specifically, in both mutants, residue 40 is not anchored into the cylinder structure, permitting the displacement of VP2 residues 45–39 from their crystallographic positions in the cylinder wall. This allows the less-ordered N-terminal residues of VP2, which in wildtype VLPs occupy both claw- and funnel-shaped densities beneath the gate, to move progressively away from the five-fold vertex, presumably further into the particle interior. These new observations therefore provide strong physical support for the hypothesis that the L172:V40 interaction is critical for the structural integrity of the five-fold channel and the retention of flexible VP2 N-termini immediately around this entryway (Plevka et al. 2011).
At the mid-gate, five symmetry related L172 side chains encircle the portal at the base of its narrowest point, leading to the proposition that this constriction might serve a ratchet-like function that ensures the unidirectional transition of the genome into the capsid through the packaging portal. While previous studies showed that “reversing-out” of the genome was common among particles carrying the L172T mutation, virions bearing the V40A mutation underwent premature genome uncoating, ejecting their genomes in a 3′-to-5′ direction. V40A mutant virions clearly did not eject their genomes by reversing, in a 5′-to-3′ direction, the original packaging process, indicating that the ratchet mechanism remained intact (Cotmore and Tattersall, 2012). The cryo-EM structures further support the ratchet hypothesis, showing that in the L172T mutant the upper section of the mid-gate appears essentially intact, and only the region immediately below the basal 172T constriction point shows evidence of disruption not seen in V40A VLPs, mapping the altered phenotype to this part of the cylinder.
Combining the new structural observations with previous infectivity data gives additional insight into cylinder mechanics. For example, MVM virions bearing either the V40A or L172T mutation were found to package their genomes efficiently (Farr and Tattersall, 2004; Reguera et al., 2004; Farr et al., 2006; Cotmore and Tattersall, 2012), which indicates that the packaging mechanism is not significantly affected by major structural changes that occur below the cylinder mid-gate. Similarly, infectivity studies indicate that neither mutation impairs the progressive extrusion of VP2 N-termini from full virions, which can temporarily stabilize the cylinder. However, once the exposed VP2 N-termini are cleaved to VP3 during cell entry, neither mutant can block the premature externalization of VP1 N-termini from the intact particle at physiological temperatures or the 3′-to-5′ uncoating of the genome within the endosomal compartment. Thus eliminating hydrophobic interactions between L172 and V40 ultimately destroys the virion’s ability to modulate a progressive series of conformational shifts that normally keep the genome sequestered while the particle is trafficked into the cell nucleus. Whether the five-fold cylinders of parvoviruses from other genera show evidence of similar mechanics remains to be determined.
Materials and Methods
Generation of Virus-Like Particles
A pFast-Bac 1 clone (Invitrogen) containing the full length cDNA encoding wildtype MVMi VP1 capsid protein, pFB-MVMi-VP1, was digested with BamHI and BssHII, then religated to create pFB-MVMi-VP2, in which the BamHI site just downstream of the polyhedron promoter was recreated 161 basepairs upstream of the VP2 initiation codon, yielding a construct only capable of expressing VP2.
Clone pFB-L172T-VP2, expressing the L172T mutant of MVMi VP2, was generated by subcloning the SpeI-EcoRI fragment from the infectious clone pL172T (Farr & Tattersall, 2004) into SpeI-EcoRI digested pFB-MVMi(−)VP1.
Cloning of the V40A mutation into pFB-MVMi-VP2 was achieved using sequential PCR reactions. The forward primer for the first reaction, 5′- CCATCGGGCGCGGATCCGCGCTTTTGCACCTAAGCTTGCTACTGACTCTGACCTGGAACTTCTGGTGTAAGCAG -3′, included the BamHI site (underlined) upstream of the start of VP2. This primer was used with a mutagenic reverse primer, 5′- CCCAAGAATCTATAATGCGTCTGATTATCATAAGACCCAGTAGAAACACCAgCCCCACCCCCGCCAGAGCCCCCAC -3′, to amplify a 345 basepair region from the MVMi genome, changing valine to alanine at codon 40 of VP2 (underlined). This PCR product was purified and used as the forward primer in the second PCR reaction with a reverse primer, 5′- GGCATGTTTAAATGTACTAGTCTAGTTGCTAGTGCAGTAATTTCTACCCAGCCGTCACCCAAGAATCTATAATGCG -3′, located downstream of V40A, that included the SpeI site in VP2 (underlined). This 402 basepair PCR product was cloned into pCR2.1-TOPO, sequenced to confirm the V40A change, then digested with BamHI and SpeI and inserted into BamHI-SpeI digested pFB-MVMi-VP2 to create pFB-V40A-VP2.
The presence of each mutation was confirmed by DNA sequencing, and the DNA transformed into DH10Bac cells (Invitrogen). The resulting bacmid DNAs were transfected into SF9 cells to make a P1 virus stock. P2 and P3 stocks were made from infecting with the P1 and P2 stocks, respectively. The titer of the P3 stock was determined by plaque assay, and used to infect SF9 cells at 5 plaque-forming units per cell for 72 hours, following which cells were harvested, resuspended in TE8.7, frozen and thawed three times and cleared by centrifugation at 12,000 rpm for 45 minutes at 4°C. VLPs were twice purified by iodixanol gradient centrifugation as described previously (Plevka et al., 2011), and the purity and quantity of VLPs in each fraction determined by SDS-PAGE followed by staining with Coomassie Blue.
Cryo-EM Reconstructions
For each MVM sample, particle concentration was adjusted appropriately before applying 3.5 μl aliquots to cryo-EM grids for vitrification. Grids were imaged using low-dose techniques on an FEI Tecnai TF-20 FEG electron microscope operating at 200 kV, and micrographs were recorded on Kodak SO-163 film that was subsequently developed with full-strength Kodak D19 developer for 12 mins. Negatives were digitized using a Nikon Super Coolscan 9000 scanner with a scan step size of 6.35 μm/pixels to generate a pixel size of 1.27–1.33 Å/pixel at the sample, according to the magnification used for imaging. Particles were selected using the program EMAN2 and defocus values for the micrographs were measured using the ctffind3 program (Vargas et al., 2013). The selected particles were normalized, linearized, and apodized prior to image reconstruction. The “gold standard” approach was used in AUTO3DEM to split the dataset to generate a random model, assess particle orientations, calculate the final reconstruction maps, and determine the Fourier shell correlation (FSC) cutoff at 0.143 to estimate resolution (Yan et al., 2007). A temperature factor of 200 was applied during refinement to sharpen the maps. Handedness of the maps was assigned by assessing the correlation coefficient of the fit of the MVM crystal structure (PDB ID 1Z14) (Kontou et al., 2005) into unflipped maps and maps flipped along the x-axis. During these routines, the scaling allowed assignment of absolute pixel size.
Fitting and figures
Figures were generated using the program Chimera (Pettersen 2004). Icosahedral symmetry operators were applied to the MVM VP2 crystal structure (PDB ID 1Z14) (Kontou et al., 2005) using the program Multiscale Model. The resulting calculated capsid structure was fitted into the WT, L172T, and V40A cryo-EM density maps using the program “Fit in Map”, which produced correlation coefficients of 0.9019, 0.7933, and 0.8859, respectively. The final resolution of each map was estimated using the “bresolve” program of Bsoft (Heymann 2001). The map of MVM crystal structure (PDB ID: 1Z14) was generated at 6Å with a Gaussian smoothing kernel using pdb2vol from the Situs package (Chacon and Wriggers 2002). The N-termini of VP2 were built into the wildtype MVMi and L172T density maps using the program COOT (Emsley 2010). The alpha carbon backbone of the MVMp VP2-only VLP crystal structure (PDB ID 1Z14) (Kontou et al., 2005) was relocated using translation and rigid body fitting procedures into the corresponding density.
Difference maps
The MVMp VP2-only VLP crystal structure (PDB ID 1Z14) (Kontou et al., 2005) was rendered as a density map with Gaussian smoothing kernel using pdb2vol from the Situs package (Chacon and Wriggers 2002). The appropriate pixel size and resolution for the respective maps was used to generate the density map in order for it to be comparable to the wildtype, L172T and V40A cryo-EM maps. Difference densities were calculated using the Situs program voldiff. The difference densitites were then segmented using the Chimera ‘segmentation’ program and quantified using the ‘Measure Volume and Area’ program (Pettersen 2004).
Highlights.
Parvovirus VP2 N-terminal density mapped to the five fold pore.
Seven previously disordered residues of were resolved.
Differences between wild-type and mutant viruses mapped to the base of the pore.
Conformation linked to virus function published by Cotmore et al 2012.
Acknowledgments
This work was supported in part by Pennsylvania Department of Health CURE funds. Research reported in this publication was also supported by the Office of the Director, National Institutes of Health, under Award Number S10OD011986 (SH) and S10OD019995 (JFC), as well as NIH grants R01AI107121 (SH), R01CA29303 (PT), and T32CA060395 (LJO). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agbandje-McKenna M, Llamas-Saiz AL, Wang F, Tattersall P, Rossmann MG. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure. 1998;6:1369–1381. doi: 10.1016/s0969-2126(98)00137-3. [DOI] [PubMed] [Google Scholar]
- Bourguignon GJ, Tattersall PJ, Ward DC. DNA of minute virus of mice: self-priming, nonpermuted, single-stranded genome with a 5′-terminal hairpin duplex. Journal of Virology. 1976;20:290–306. doi: 10.1128/jvi.20.1.290-306.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures1. Journal of Molecular Biology. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- Chapman MS, Rossmann MG. Structure, Sequence, and Function Correlations among Parvoviruses. Virology. 1993;194:491–508. doi: 10.1006/viro.1993.1288. [DOI] [PubMed] [Google Scholar]
- Cifuente JO, Lee H, Yoder JD, Shingler KL, Carnegie MS, Yoder JL, Ashley RE, Makhov AM, Conway JF, Hafenstein S. Structures of the Procapsid and Mature Virion of Enterovirus 71 Strain 1095. Journal of Virology. 2013;87:7637–7645. doi: 10.1128/JVI.03519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Soderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. The family Parvoviridae. Archives of virology. 2014;159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, Hafenstein S, Tattersall P. Depletion of Virion-Associated Divalent Cations Induces Parvovirus Minute Virus of Mice To Eject Its Genome in a 3′-to-5′ Direction from an Otherwise Intact Viral Particle. Journal of Virology. 2010;84:1945–1956. doi: 10.1128/JVI.01563-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. A genome-linked copy of the NS-1 polypeptide is located on the outside of infectious parvovirus particles. Journal of Virology. 1989;63:3902–3911. doi: 10.1128/jvi.63.9.3902-3911.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. Encapsidation of minute virus of mice DNA: aspects of the translocation mechanism revealed by the structure of partially packaged genomes. Virology. 2005;336:100–112. doi: 10.1016/j.virol.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. Mutations at the Base of the Icosahedral Five-Fold Cylinders of Minute Virus of Mice Induce 3′-to-5′ Genome Uncoating and Critically Impair Entry Functions. Journal of Virology. 2012;86:69–80. doi: 10.1128/JVI.06119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotmore SF, Tattersall P. Parvoviruses: Small Does Not Mean Simple. Annual Review of Virology. 2014;1:517–537. doi: 10.1146/annurev-virology-031413-085444. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D: Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr GA, Cotmore SF, Tattersall P. VP2 Cleavage and the Leucine Ring at the Base of the Five-fold Cylinder Control pH-Dependent Externalization of both the VP1 N Terminus and the Genome of Minute Virus of Mice. Journal of Virology. 2006;80:161–171. doi: 10.1128/JVI.80.1.161-171.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr GA, Tattersall P. A conserved leucine that constricts the pore through the capsid five-fold cylinder plays a central role in parvoviral infection. Virology. 2004;323:243–256. doi: 10.1016/j.virol.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Heymann JB. Bsoft: Image and Molecular Processing in Electron Microscopy. Journal of Structural Biology. 2001;133:156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- Kailasan S, Halder S, Gurda B, Bladek H, Chipman PR, McKenna R, Brown K, Agbandje-McKenna M. Structure of an enteric pathogen, bovine parvovirus. Journal of Virology. 2015;89:2603–2614. doi: 10.1128/JVI.03157-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Simpson AA, Rossmann MG. The structure of human parvovirus B19. Proc Natl Acad Sci U S A. 2004;101:11628–11633. doi: 10.1073/pnas.0402992101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JA, Dubielzig R, Grimm D, Kleinschmidt JA. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. The EMBO Journal. 2001;20:3282–3291. doi: 10.1093/emboj/20.12.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontou M, Govindasamy L, Nam HJ, Bryant N, Llamas-Saiz AL, Foces-Foces C, Hernando E, Rubio MP, McKenna R, Almendral JM, Agbandje-McKenna M. Structural Determinants of Tissue Tropism and In Vivo Pathogenicity for the Parvovirus Minute Virus of Mice. Journal of Virology. 2005;79:10931–10943. doi: 10.1128/JVI.79.17.10931-10943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labieniec-Pintel L, Pintel D. The minute virus of mice P39 transcription unit can encode both capsid proteins. Journal of Virology. 1986;57:1163–1167. doi: 10.1128/jvi.57.3.1163-1167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas-Saiz AL, Agbandje-McKenna M, Wikoff WR, Bratton J, Tattersall P, Rossmann MG. Structure Determination of Minute Virus of Mice. Acta Crystallographica Section D. 1997;53:93–102. doi: 10.1107/S0907444996010566. [DOI] [PubMed] [Google Scholar]
- Mietzsch M, Kailasan S, Garrison J, Ilyas M, Chipman P, Kantola K, Janssen ME, Spear J, Sousa D, McKenna R, Brown K, Söderlund-Venermo M, Baker T, Agbandje-McKenna M. Structural insights into emerging pediatric pathogens human bocaparvoviruses. Journal of Virology. 2017 doi: 10.1128/JVI.00261-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pintel D, Dadachanji D, Astell CR, Ward DC. The genome of minute virus of mice, an autonomous parvovirus, encodes two overlapping transcription units. Nucleic Acids Research. 1983;11:1019–1038. doi: 10.1093/nar/11.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plevka P, Hafenstein S, Li L, D’Abramo A, Jr, Cotmore SF, Rossmann MG, Tattersall P. Structure of a packaging-defective mutant of minute virus of mice indicates that the genome is packaged via a pore at a 5-fold axis. Journal of Virology. 2011;85:4822–4827. doi: 10.1128/JVI.02598-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera J, Carreira A, Riolobos L, Almendral JM, Mateu MG. Role of interfacial amino acid residues in assembly, stability, and conformation of a spherical virus capsid. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2724–2729. doi: 10.1073/pnas.0307748101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AA, Chandrasekar V, Hébert Bt, Sullivan GM, Rossmann MG, Parrish CR. Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses 1. Journal of Molecular Biology. 2000;300:597–610. doi: 10.1006/jmbi.2000.3868. [DOI] [PubMed] [Google Scholar]
- Tattersall P, Cawte PJ, Shatkin AJ, Ward DC. Three structural polypeptides coded for by minite virus of mice, a parvovirus. Journal of Virology. 1976;20:273–289. doi: 10.1128/jvi.20.1.273-289.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao J, Chapman M, Agbandje M, Keller W, Smith K, Wu H, Luo M, Smith T, Rossmann M, Compans R, et al. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- Vargas J, Otón J, Marabini R, Jonic S, de la Rosa-Trevín JM, Carazo JM, Sorzano COS. FASTDEF: Fast defocus and astigmatism estimation for high-throughput transmission electron microscopy. Journal of Structural Biology. 2013;181:136–148. doi: 10.1016/j.jsb.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Yan X, Sinkovits RS, Baker TS. AUTO3DEM – an automated and high throughput program for image reconstruction of icosahedral particles. Journal of structural biology. 2007;157:73–82. doi: 10.1016/j.jsb.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]


