ABSTRACT
Bifidobacteria are important gut commensals of mammals, including humans, of any age. However, the molecular mechanisms by which these microorganisms establish themselves in the mammalian gut and persist in this environment are largely unknown. Here, we analyzed the genetic diversity of the predicted arsenal of sortase-dependent pili of known and sequenced members of the Bifidobacterium genus and constructed a bifidobacterial sortase-dependent fimbriome database. Our analyses revealed considerable genetic variability of the sortase-dependent fimbriome among bifidobacterial (sub)species, which appears to have been due to horizontal gene transfer events and for which we were able to perform evolutionary mapping. Functional assessment by transcriptome analysis and binding assays involving different substrates demonstrates how bifidobacterial pili are pivotal in promoting various abilities for adhesion to glycans and extracellular matrix proteins, thereby supporting the ecological success of bifidobacteria in the mammalian gut.
IMPORTANCE Adhesion of bifidobacterial cells to the mucosa of the large intestine is considered a hallmark for the persistence and colonization of these bacteria in the human gut. In this context, we analyzed the genetic diversity of the predicted arsenal of sortase-dependent pili of known and sequenced members of the Bifidobacterium genus, and constructed a bifidobacterial sortase-dependent fimbriome database. Our analyses revealed considerable genetic variability of the sortase-dependent fimbriome among bifidobacterial (sub)species, which appears to have been due to horizontal gene transfer events. In addition, functional assessment by transcriptome analysis and binding assays involving different substrates demonstrates how bifidobacterial pili are crucial in promoting various abilities for adhesion to glycans and extracellular matrix proteins, thereby supporting the ecological success of bifidobacteria in the mammalian gut. This study represents a complete genomic study regarding the presence of fimbriae in the genus Bifidobacterium.
KEYWORDS: bifidobacteria, gut microbiota, bacterial interactions, genomics, metagenomics
INTRODUCTION
Bifidobacteria are microorganisms that are known to colonize the gut of various mammals, including humans, birds, and social insects (1–3). In this gut ecosystem, bifidobacteria interact with their host as well as with other members of the microbiota through different strategies (4–7). Bacterial surface appendages, in particular pili or fimbriae, are considered to be important bacterial structures involved in host-microbe interactions (8). Pili were observed in Gram-positive and Gram-negative bacteria, and they are found to elicit functions in addition to adherence, such as conjugation, motility, immunomodulation, biofilm formation and electron transfer (9, 10). Structurally, Gram-positive pili contain multiple copies of one or more different pilin proteins. The pilus shaft is composed of a multimer of the major pilin and is in turn associated with a smaller number of ancillary pilins (11). Various types of pili have been characterized, including sortase-dependent (SD) pili (types I and II) and type IV pili (12). In SD pili, the pilins that constitute the pilus fiber are covalently linked by sortases (13). The genes encoding an SD pilus structure, including the corresponding sortase, are generally located in the same locus. Pilus anchoring takes place on the cell wall following Sec-dependent secretion of pilus protein components.
The existence of SD pili in bifidobacteria has only recently been appreciated (14). Bifidobacterial SD pilus loci are typically composed of (i) an fimA or fimP gene encoding the major pilin protein, (ii) an fimB or fimQ gene that specifies an ancillary or minor pilin and that commonly acts as an adhesin at the tip of the pilus (note: some clusters encode more than one such minor pilin), and (iii) a gene encoding the pilus-specific sortase (14). The deduced amino acid sequences of fimA/P and fimB/Q contain particular consensus motifs and domains characteristic of a pilin primary structure, including a Sec-dependent secretion signal, the sortase recognition site (cell wall sorting signal motif), the pilin-like motif (TVXXK), and the E box (14, 15).
Bifidobacteria have also been shown to encode type IV pili, which are similar to the tight adherence (Tad) pili (16–18) and which, since they are essential for gut colonization, are believed to mediate adhesion to the host's surfaces (19, 20).
The role of pili in modulating adhesion to human gut mucosa, while at the same time having an impact on host-microbe dialogue, has only been studied for a small number of gut-associated bifidobacterial species (19, 21). Furthermore, these extracellular appendages are also believed to mediate aggregation/interaction events involving other members of the gut microbiota (22). The accumulation of genomic data for this bacterial genus (23–25) justifies a reevaluation of the number, diversity, and distribution of and role(s) elicited by the sortase-dependent (SD) fimbriome of the genus Bifidobacterium. Here, we analyze the features of the bifidobacterial SD fimbriome (i.e., the collection of predicted SD pili of the genus Bifidobacterium) and evaluate how these extracellular structures are crucial in the interaction with glycans that are abundant in the ecological niche of bifidobacteria. Furthermore, comparison of the SD fimbriome identified here with microbiome data sets allowed us to reconstruct the contribution of bifidobacterial sortase-dependent pili to the overall predicted microbial interactome of the human gut.
RESULTS AND DISCUSSION
Identification of bifidobacterial pilus loci.
We surveyed 158 genome sequences representing all 48 currently recognized (sub)species of the Bifidobacterium genus for loci encompassing genes predicted to encode SD pilus structures, henceforth designated pilus-encoding loci (PEL). This bifidobacterial genomic data set included all of the genomes belonging to the Bifidobacterium genus that were available in NCBI at the time of writing. This analysis led to the identification of 294 PEL that collectively represent the SD fimbriome of the genus Bifidobacterium. The genome of Bifidobacterium dentium LMG11045 contains seven PEL, which is the largest number of PEL so far identified in a given genome of a member of the genus Bifidobacterium. In contrast, the chromosomes of Bifidobacterium animalis subsp. lactis, Bifidobacterium asteroides, Bifidobacterium bombi, Bifidobacterium callitrichos, Bifidobacterium kashiwanohense, Bifidobacterium mongoliense, Bifidobacterium psychraerophilum, Bifidobacterium subtile, Bifidobacterium thermacidophilum subsp. thermacidophilum, and Bifidobacterium longum subsp. suis were shown to contain just a single PEL (see Table S1 in the supplemental material), while 10 strains belonging to Bifidobacterium actinocoloniforme, Bifidobacterium longum subsp. longum, and B. longum subsp. infantis species do not contain any PEL in their genomes (Table S1). Notably, we identified a commonly occurring gene constellation, observed for 269 PEL, that consists of two genes specifying predicted pilus subunits and an associated sortase-encoding gene (Fig. 1). In addition, we identified 19 pilus loci that consist of just a single pilus subunit-encoding gene flanked by a sortase-encoding gene and 6 pilus loci consisting of two pilin-encoding genes without an associated sortase-encoding gene. Within the predicted SD fimbriome of the genus Bifidobacterium (including typical and atypical pilus loci), we identified 156 PEL that are shared by different bifidobacterial (sub)species and that therefore constitute the clusters of orthologous (sortase-dependent) pilus-encoding genes (COPGs) (described below). PEL belonging to a given COPG were defined as displaying 50% identity on sortase protein sequences. We used sortase-encoding genes because these are highly conserved compared to the pilin subunit-encoding genes (described below). Notably, cross-alignment of all fimA/P and fimB/Q genes included in the 15 COPGs revealed, in addition to high intra-COPG similarity and identity, high homology between pilus subunits of different COPGs, which is indicative of horizontal gene transfer (HGT) events (see Data Set S1 in the supplemental material) (described below). In addition, the bifidobacterial SD fimbriome includes 138 PEL that are uniquely identified in the chromosome of a single bifidobacterial (sub)species and thus represent an accessory PEL arsenal. Of these 138 unique pilus loci, 89 are species-specific clusters (i.e., species-specific PEL), and for the remaining 49, 5 are unique for a specific strain representing strain-specific PEL, while for 44, it was not possible to make such a classification since only one genome sequence of the particular species was available. Altogether, our findings highlight the existence of a very substantial arsenal of SD pilus-encoding genes within the pangenome of bifidobacteria. Furthermore, comparative analysis suggests that, in addition to species-specific genetic strategies, there are highly variable, strain-dependent mechanisms employed by these bacteria to interact with their environment.
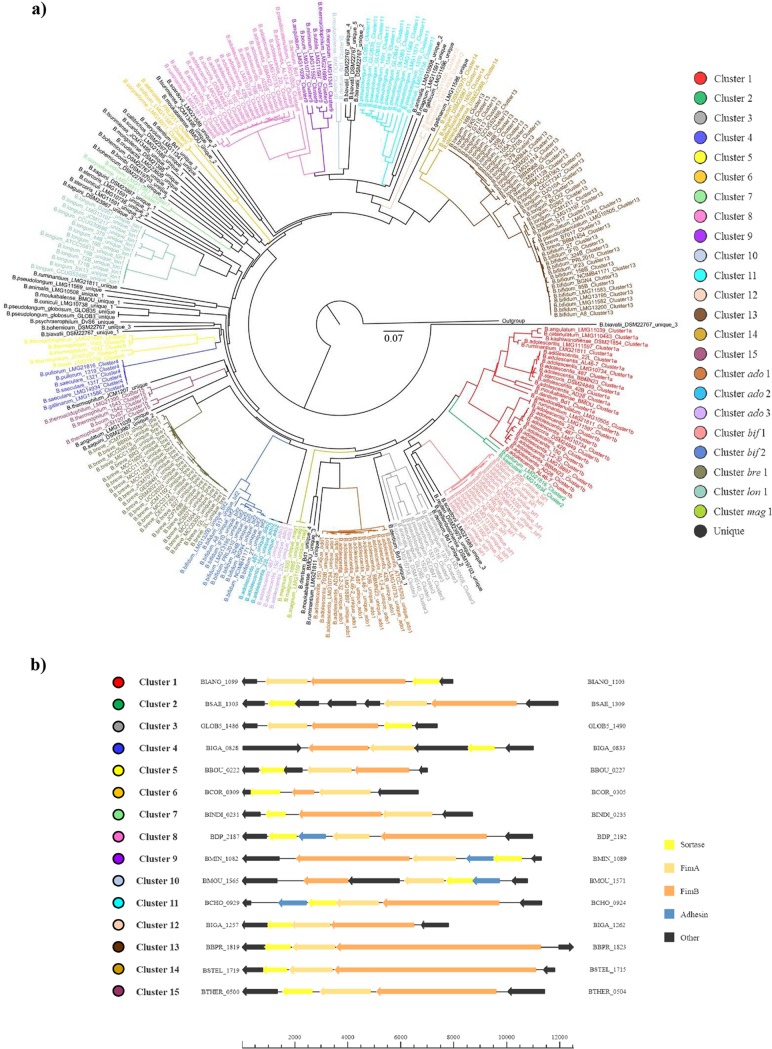
FIG 1.
Phylogenetic analyses of the SD fimbriome of bifidobacteria. Panel a represents a tree generated from hierarchical clustering based on the identified bifidobacterial sortases. Panel b depicts physical maps of a representative strain for the 15 COPGs. Genes were categorized according to their functions with differently colored arrows.
Classification of the pilus-encoding loci in the Bifidobacterium SD fimbriome.
Based on the finding that sortase proteins of Gram-positive bacteria are cell-wall-anchored transpeptidases, used by bacteria to assemble pilus subunits, a phylogenetic tree was built by comparing the deduced PEL-associated sortase proteins from all available Bifidobacterium members. Note that sortase homologs in the bifidobacterial genomes outside PEL were not included in our analyses.
The resulting phylogenetic tree was based on 287 sortases encoded by genes present in the 294 identified PEL (described above), identified not only based on the genomes of the reference strains for each (sub)species of the genus Bifidobacterium but also in all analyzed 158 bifidobacterial genomes (Fig. 1). In seven PEL, no sortase-encoding gene was identified, suggesting the involvement of a sortase that is encoded by a gene outside the identified PEL, similar to what was previously described for Streptococcus pyogenes (26). Such analyses revealed the presence of 15 main interspecies groups of PEL, named COPGs, that were shown to possess >50% sequence identity and were located in the same genomic region (see Fig. S1 in the supplemental material). Furthermore, eight COPGs were identified that are present only in multiple strains of the same species. These clusters were found in Bifidobacterium adolescentis (and named ado1, ado2, and ado3), Bifidobacterium bifidum (designated bif1 and bif2), Bifidobacterium breve (bre1), Bifidobacterium longum (lon1), and Bifidobacterium magnum (mag1).
In a small number of PEL (i.e., 39), we identified a gene encoding a third pilus subunit predicted to encode an adhesin protein displaying >98% similarity to homologous proteins involved in bacterial attachment to host tissue surfaces (12, 27). Interestingly, a phylogenetic tree based on these 39 adhesin proteins described above showed the same clusters as those retrieved by sortase analyses (see Fig. S2 in the supplemental material).
In silico characterization of the pilus-encoding loci of the Bifidobacterium SD fimbriome.
Major and minor subunits of the Bifidobacterium SD fimbriome were identified based on similarity at the amino acid level (28, 29) as well as detailed in silico analysis of domains typically found in the pilin subunits. In this context, in silico identification of domains and secondary structures of the deduced protein products of the predicted fimA/P and fimB/Q homologs revealed the typical consensus motifs and domains characteristic of a pilin primary structure, including a Sec-dependent secretion signal, the sortase recognition site (CWSS motif), the pilin-like motif (TVXXK), and the E box (14) (see Tables S1 and S2 in the supplemental material). Interestingly, identification of the latter motifs of bifidobacterial fimA/P confirms their widespread distribution in the major subunits of sortase-dependent pili in other common human gut commensals, such as members of the genera Bacteroides, Blautia, Collinsella, Enterococcus, Faecalibacterium, and Lactobacillus (Table S2).
Notably, in silico inspection of the predicted secondary structures and domains of the predicted FimA/P major subunits confirmed a similar topology associated with a backbone functionality. Analysis of putative FimB/Q minor subunits encoded by bifidobacteria revealed the presence of domains that seem to drive the adhesion of pili to different substrates (11). These include a putative collagen-binding adhesin (CNA) domain identified in the minor subunit of 11 COPGs, which was previously demonstrated to be a primary determinant of the ability to bind collagen in Staphylococcus aureus (30, 31). Notably the product of fimB/Q small subunits of COPGs 1, 3, and 5 appears to be evolutionary linked to the minor pili subunit-encoding gene rrgA of Streptococcus pneumoniae, as indicated by a similarity of >40%. This domain has been shown to be pivotal for immune evasion by S. pneumoniae as well as for mediating adhesion to the extracellular matrix proteins of the host (32). Altogether, these findings suggest a common functional role exploited by the FimB/Q small subunits of COPGs 1, 3 and 5. Moreover, in Bifidobacterium saeculare, this CNA domain is also present in the FimB/Q-like member of COPG 2. In contrast, the product of the fimB/Q gene of B. pullorum classified as COPG 2 encompasses B repeat units of the collagen-binding surface protein (CNA) of S. aureus. This domain is involved in “stalk” functions improving the projection of the binding domain (collagen-binding A region) from the bacterial surface (33).
Interestingly, the small subunit of five COPGs also contains a carbohydrate binding module (CBM), described to sustain the interaction of pili with carbohydrates (34). Notably, these FimB/Q paralogs also encompass a CNA domain, suggesting that these pili promote adhesion of bifidobacterial cells to host tissue, such as the gut mucosa, as well as adhesion to carbohydrates present in the gut, including those present in mucin or introduced with the diet (35, 36). More specifically, in addition to a CNA domain homologous to the product of the rrgC gene of S. pneumoniae (37), the products of the fimB/Q homologs that make up COPGs 8, 9, 10, 11, and 15 also include domains predicted to be involved in sugar binding. These encompass homologs of domains 4 and 5 of the S-layer sugar-binding protein SbsC of Geobacillus stearothermophilus (38), as well as, except for COPG 9, homologs of the CBM 25 of Bacillus halodurans, responsible for starch binding (39). Therefore, analysis of the 15 identified COPGs revealed that a small number contain only a CNA domain, while others possess both a CNA domain as well as a CBM domain (Fig. 2).
FIG 2.
Genetic comparison of COPGs within the genus Bifidobacterium. Panels a and b show the similarities between amino acid sequences belonging to the different COPGs reported for each COPG separately or with clustered genes and species, respectively. Column titles and colors represent different COPGs. The same colors in the heat map represent a similarity of >50% to the COPG. Panel c depicts the presence (red) or absence (black) of CNA and CBM domains in the COPGs. ND, not detected.
Genetic comparison of COPGs within the genus Bifidobacterium.
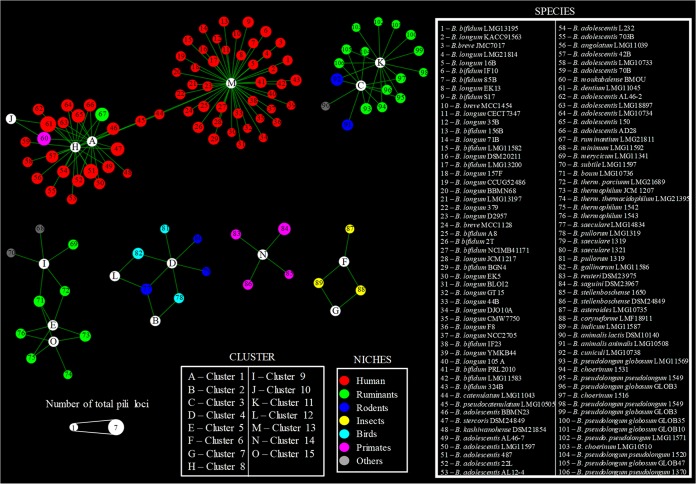
Network analyses of the predicted SD fimbriome of the genus Bifidobacterium highlighted that among the identified 15 COPGs, cluster 13 as well as clusters 1 and 8 are placed in the central nodes within the reconstructed bifidobacterial pilus-based network (Fig. 3). Other SD fimbriome clusters that were identified in different bifidobacterial (sub)species included clusters 4 and 12. Notably, these latter clusters are identified in the genomes of bifidobacterial taxa residing in the gut of birds, whereas the largest part of the network consisting of clusters 1, 8, and 13 includes the SD pilus arsenal of bifidobacterial (sub)species present in the human gut, with the exception of B. moukabalense and B. ruminantium (Fig. 3). Interestingly, the PEL from nonhuman primates cluster to form COPG 14, which is not connected with any other COPG (Fig. 3), suggesting a divergent evolution followed by COPG 14 with respect to COPG 13. In contrast, COPGs 6 and 7 were found in the genomes of bifidobacterial species present in the gut of social insects (Fig. 3), indicating that the SD fimbriome arsenal of bifidobacteria may reflect the ecological niche in which the various bifidobacterial (sub)species are commonly encountered.
FIG 3.
Network analyses of COPGs within the genus Bifidobacterium. Network analyses based on the presence/absence of COPGs in each Bifidobacterium strain are indicated. White circles represent the 15 identified COPGs, each color denotes a different ecological niche, and the diameter is proportional to the number of total pilus loci (i.e., loci belonging to COPGs as well as unique pilus loci).
Extensive acquisition and loss of COPGs during Bifidobacterium evolution.
In silico analyses of the predicted major and minor pilus subunits, as well as the sortases, demonstrated similarities between amino acid sequences of different COPGs (Fig. 2). Remarkably, 30 of the PEL classified in the 15 bifidobacterial COPGs were shown to be flanked by transposon elements, which suggests acquisition by HGT. Other findings supporting possible HGT transfer of PEL are the deviating percentages of G+C content between the sequence of the major pilus subunit-encoding gene and its corresponding genome (which is, for example, 11% lower in the case of B. asteroides LMG10735 or 8% higher in the case of B. catenulatum LMG11043) (see Table S3 in the supplemental material). A similar situation was found for the gene encoding the minor pilus subunit: the percentage of G+C content of this gene ranged from being 12% lower (in the case of B. choerinum 1516) to being 12% higher (in the case of B. saguini DSM23967) than that of the corresponding genome. In addition, the predicted FimA/P and FimB/Q proteins also have a different codon usage bias (Table S3). Furthermore, search for homologs of bifidobacterial fimA/P and fimB/Q genes in the NCBI nr database revealed that a wide range of Gram-positive gut colonizers carry genes with a BLASTp E value of <1e−30, such as members of the genera Fusicatenibacter, Ruminococcus, Faecalibacterium, Blautia, Clostridium, and Streptococcus (see Fig. S3 and S4 in the supplemental material). The presence of homologs to bifidobacterial fimA/P and fimB/Q genes in other genera may indicate that the latter may have acted as putative donors of PEL to bifidobacterial genomes. These findings are further corroborated by inspection of the phylogenetic tree based on the SD fimbriome of bifidobacteria plus the various PEL homologs from other Gram-positive microorganisms, which clearly points out that bifidobacterial FimA/P and FimB/Q proteins do not form a monophyletic group with those of other high G+C Gram-positive bacteria (Fig. S3 and S4). Altogether, the phylogenetic inconsistencies, GC content deviation, and distinctive codon usage indicate that elements of the bifidobacterial SD fimbriome, similar to the SD fimbriome of other actinobacteria (40), were acquired through HGT events (Table S3).
Transcriptional analysis of bifidobacterial pilus-encoding loci.
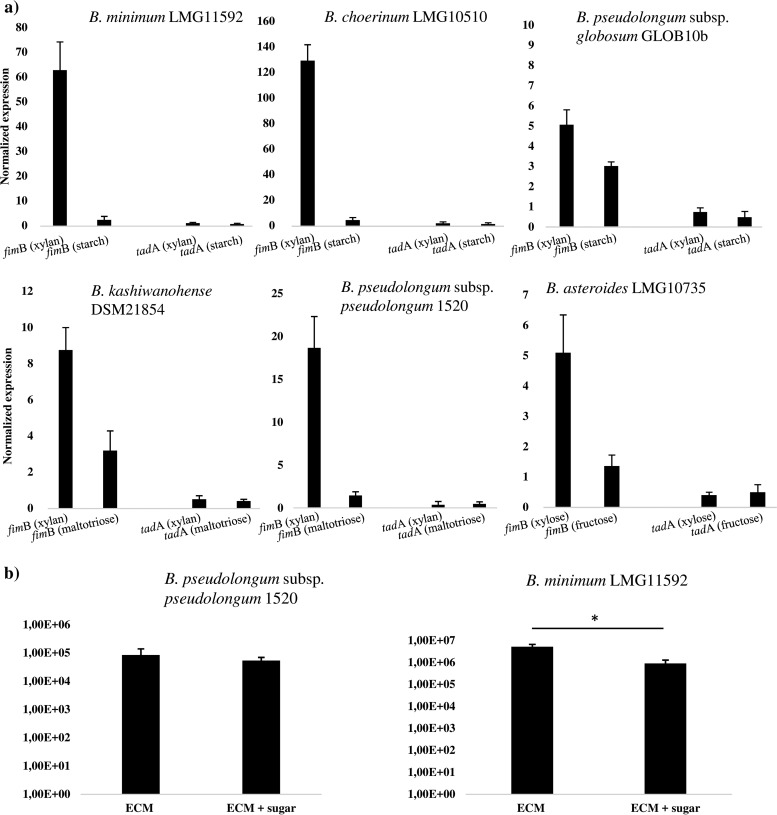
In order to investigate if the bifidobacterial PEL are differentially transcribed in response to the environment to which bifidobacterial cells are exposed, the levels of fimB homolog-specific mRNA were determined by quantitative real-time PCR (qRT-PCR) assays by in vitro experiments. We decided to investigate six bifidobacterial strains (i.e., B. minimum LMG11592, B. choerinum LMG10510, B. pseudolongum subsp. globosum GLOB10b, B. pseudolongum subsp. pseudolongum 1520, B. kashiwanohense DSM21854, and B. asteroides LMG10735). These strains encompass a unique PEL in their genome consisting of pilin-encoding genes that encode proteins with either a single CNA or a combined CNA-CBM domain (Fig. 4). The experiments were performed using mRNA samples extracted from cultures of the above-mentioned bifidobacterial strains (Fig. 4), which had been exponentially grown in de Man-Rogosa-Sharpe (MRS) medium containing, as the unique carbon source, a particular glycan found in the ecological niche where these strains are naturally residing (41): for example, starch or xylan in the case of B. minimum LMG11592, B. choerinum LMG10510, B. pseudolongum subsp. globosum GLOB10b, or B. pseudolongum subsp. pseudolongum 1520b, maltotriose or xylan in the case of B. kashiwanohense DSM21854, or xylose or fructose in the case of B. asteroides LMG10735. The observed transcription levels of the putative subunit-encoding pilin genes in response to these growth substrates were shown to be variable between the different bifidobacterial strains and highly variable between different pilus gene clusters harbored by the same organism (Fig. 4).
FIG 4.
Relative transcription level of bifidobacterial PEL and binding to human receptor fibronectin. Panel a shows transcription levels of fimB and tadA by qRT-PCR for bifidobacterial strains when grown on xylan or starch (i.e., B. minimum LMG11592, B. choerinum LMG10510, B. pseudolongum subsp. globosum GLOB10b, or B. pseudolongum subsp. pseudolongum 1520b [all isolates from the mammalian gut]), xylan or maltotriose (B. kashiwanohense DSM21854 [an isolate from infant gut]), or fructose or xylose (B. asteroides LMG10735 [an isolate from the bee hindgut]). Panel b displays the adhesion of B. pseudolongum subsp. pseudolongum 1520b and B. minimum LMG11592 to fibronectin alone or to fibronectin with the presence of the sugar xylan. The y axis represents the CFU per milliliter.
Identification of pilus-like structures on the cell envelope in bifidobacteria.
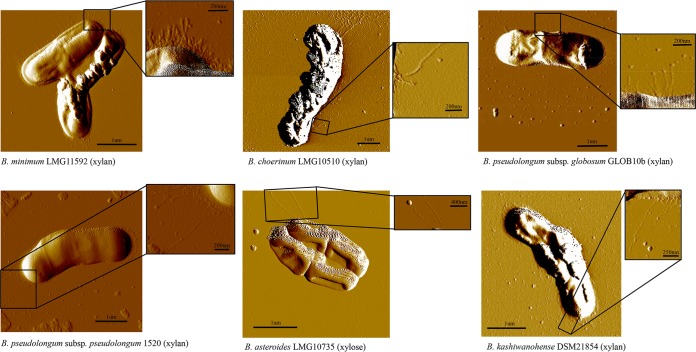
We decided to investigate the occurrence of pilus-like structures in the genus Bifidobacterium by AFM. However, in order to avoid possible misidentification of these extracellular appendages due to the potential presence of multiple different pilus-like structures for the same bifidobacterial cell as a consequence of the occurrence of different PELs, we decided to assay the cell surface of the six strains possessing a unique PEL in their genomes, which were those strains assayed by qPCR experiments (described above). Prior to AFM visualization, bacterial cells were cultivated in the presence of (partially hydrolyzed) xylan (42), or xylose only for B. asteroides LMG10735, as these conditions were shown to induce the expression of pilin subunits (described above), representing glycans that are expected to be present in their natural ecological niches (43). When we assayed the production of pilus-like structures by bifidobacteria cultivated on these different carbon sources by atomic force microscopy (AFM), we noticed the presence of pilus-like structures decorating the cell surfaces (Fig. 5). Notably, these pilus-like structures occupied different positions of the cell surface of the investigated strains, being located at the poles of the cell or uniformly distributed along the cells, while they also varied in pilus density and length (Fig. 5).
FIG 5.
Presence and morphology of pilus-like structures in various bifidobacterial species. Images were captured by atomic force microscope and show pilus-like structures of B. minimum LMG11592, B. choerinum LMG10510, B. pseudolongum subsp. globosum GLOB10b, B. pseudolongum subsp. pseudolongum 1520b, and B. kashiwanohense DSM21854 grown on xylan and B. asteroides LMG10735 grown on xylose. The scale bar is shown in each image.
It is known that bifidobacteria possess other pilus-like encoding loci—i.e., the type IV pili, which are highly conserved in the bifidobacterial genomes (4). In order to verify if the pili observed by AFM analyses were those encoded by the PEL described above or by the type IV pilus-encoded loci (Tad pili), we assayed the transcription of the tadA gene, which encodes an ATPase that is believed to be crucial for Tad pilus assembly (18), by qRT-PCR. Interestingly, transcription of the tadA gene was not detectable under conditions that allowed transcription of the fimB gene. This finding is consistent with what has previously been described for B. breve UCC2003, whose tad locus is only transcribed when this bacterium is present in the (murine) gut (19).
Binding to human receptor fibronectin.
In order to evaluate if the in silico-identified CNA motifs present in the minor pilin subunit of bifidobacteria are involved in adhesion to extracellular matrix (ECM) proteins, we performed fibronectin binding assays, following the protocol described in the Materials and Methods section, involving two strains, B. minimum LMG11592 and B. pseudolongum subsp. pseudolongum 1520b, each of which possesses a unique PEL in its genome. We used fibronectin since this ECM protein may act as a possible epithelial cell receptor involved in the recognition of bacteria (44). As a negative control, we employed bovine serum albumin (BSA). Notably, both B. pseudolongum subsp. pseudolongum 1520 and B. minimum LMG11592 cells were shown to adhere to fibronectin compared to the BSA control, consisting of 8.15E−4 CFU/ml (corresponding to 0.37% recovery of adhesion) and 5.18E−6 CFU/ml (corresponding to 7.32% recovery of adhesion), respectively (Fig. 4). Interestingly, our experiments with B. pseudolongum subsp. pseudolongum 1520 cells showed that adhesion to fibronectin is not influenced by the presence of xylan (7.00E−4 CFU/ml, corresponding to 0.23% recovery of adhesion), which may be correlated to the fact that the fimBQ homolog of this strain encodes a CNA domain only (and not a CBM domain). In contrast, B. minimum LMG11592 cells, whose minor pilin subunit is predicted to possess both CNA and CBM domains, showed a significant reduction (5.11E−5 CFU/ml [P < 0.04], corresponding to 1.29% recovery of adhesion) in adhesion efficacy to fibronectin when the ECM binding trials were performed in the presence of xylan (Fig. 4). These findings suggest that both carbohydrate residues and glycoproteins are modulating fimbrial binding of B. minimum LMG11592 cells.
Bifidobacterial SD fimbriome across the infant gut microbiota.
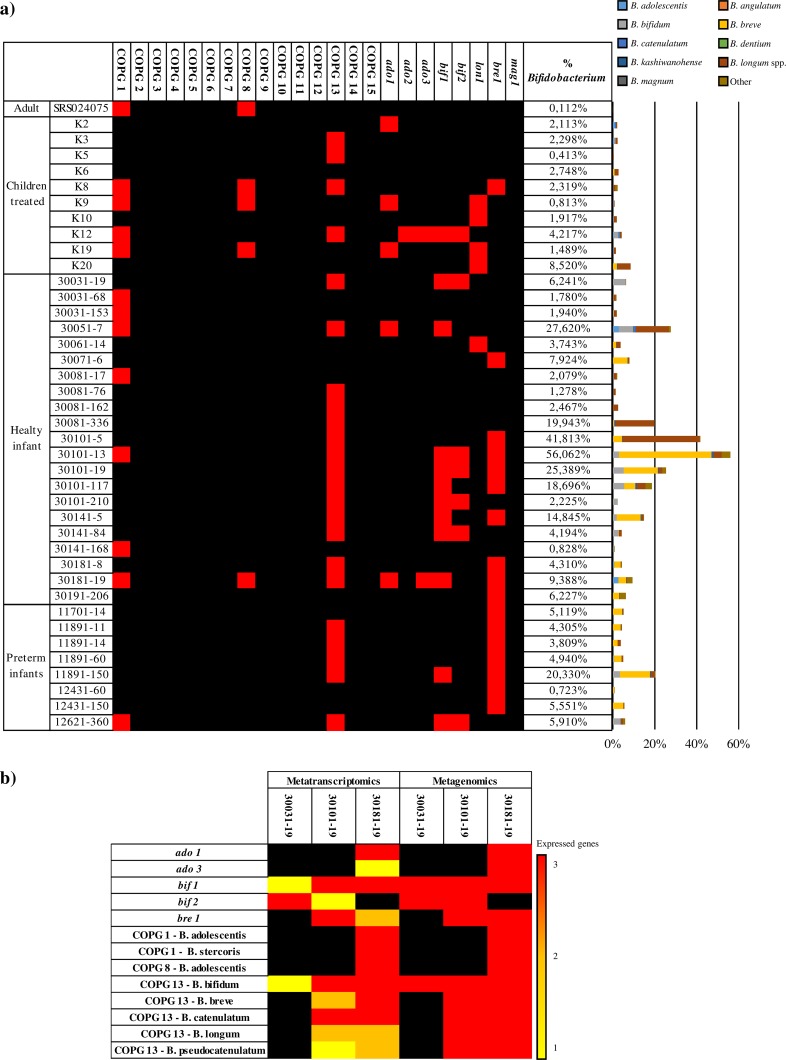
The functional contribution of bifidobacteria in terms of metabolism and resistome has recently been investigated within several gut microbiomes (42, 45). However, their contribution to the SD fimbriome of the mammalian gut has not been explored. In order to investigate the presence of bifidobacterial PEL and COPGs within the gut of humans, we employed metagenomic data sets from three different projects, encompassing healthy and preterm infants (46), children who had received an intensive antibiotic treatment (47), and healthy adults (https://www.broadinstitute.org/hmp/human-microbiome-project).
The presence of bifidobacterial sequences within these samples has previously been evaluated, highlighting a higher abundance of bifidobacterial genes in healthy children compared to children treated with antibiotics and preterm infants (45, 48). Our analyses demonstrated the presence of both bifidobacterial COPGs and PEL across these different metagenomic data sets (Fig. 6). Analyzing the overall bifidobacterial read distribution, we found that the species with the higher occurrence within the metagenomic samples reflects the bifidobacterial species of the PEL and COPG reads identified: e.g., B. breve, B. bifidum, B. longum spp., B. adolescentis, and B. catenulatum (Fig. 6). Interestingly, the COPGs that were identified in the examined microbiomes are represented by COPG 1, COPG 8, and COPG 13, while the unique PEL are scattered between the samples, the only exception being mag1, which was not identified in any of the metagenomic samples, confirming its nonhuman ecological origin (Fig. 6). These data highlight the presence of COPGs and PEL related to bifidobacterial species that typically colonize the gut of humans, such as B. breve, B. bifidum, B. longum spp., B. adolescentis, and B. catenulatum. Furthermore, COPG 8, which corresponds to the B. adolescentis group (49), has been identified in noninfant samples, suggesting a role in the colonization of a more complex ecological niche (i.e., adults and children that had received antibiotic treatment), probably due to a more varied diet.
FIG 6.
Occurrence of bifidobacterial COPGs and PEL in fecal metagenome and metatranscriptome data sets. Panel a displays the presence of bifidobacterial COPGs and PEL in the metagenomic samples from adults, healthy infants, preterm infants, and children treated with antibiotics. Within the heat map, red squares indicate the presence of the relative COPGs and PEL in each metagenomic sample. Only those samples that possess a high abundance of bifidobacterial reads are reported in the heat map. The bar plot on the right shows the relative abundance of bifidobacterial species in the analyzed metagenomic samples, while the numbers at the top of the heat map correspond to the related COPGs. Panel b displays the presence of specific PEL and COPGs in metagenomic samples and their relative expression in the corresponding metatranscriptomic samples. Within the heat map, colored squares indicate the expression of one, two, or three genes of the locus, shown in yellow, orange, and red, respectively.
In order to evaluate if the identified PEL and COPGs were expressed within the gut of infants, we employed the metatranscriptomic data set of healthy infants retrieved from the project mentioned above (46). Selecting the PEL and COPGs that were identified within healthy infants, we found a positive correlation between the presence of reads corresponding to certain PEL in metagenomic and metatranscriptomic data sets (Fig. 6). Thus, the genes encoding sortase-dependent pili of bifidobacteria were shown to be expressed when these microorganisms reside within the gut of humans. These results suggest that SD pilus-like structures of bifidobacteria are involved in the interaction with the host and perhaps also with other members of the microbiota.
Conclusions.
Bifidobacteria have been shown to be dominant microorganisms of the human infant gut microbiota (1, 50–54), where they are considered to play a pivotal role in promoting gut health of infants through the priming of the immune system as well as acting on the physiology of the gut. Adhesion of bifidobacterial cells to the mucosa of the large intestine is considered a hallmark for the persistence and colonization of these bacteria in the human gut. SD pili produced by the infant gut isolate B. bifidum PRL2010 are involved in adhesion to human cell lines through extracellular matrix proteins as well as in aggregation with other members of the human gut microbiota (21, 22). However, current knowledge on the SD fimbriome of the genus Bifidobacterium is rather limited (14, 21, 22). In this study, we provide a detailed genetic survey of the SD fimbriome in the genus Bifidobacterium, which shows that these structures are widely distributed in this group of microorganisms, suggesting an important role in facilitating environmental interactions. However, in contrast to other pilus types, like type IV fimbriae, which are much more conserved and genetically homogeneous among members of the Bifidobacterium genus, the SD pili display substantial genetic diversity in both sequence and number of PEL per strain. Notably, many of these extracellular structures are predicted to be acquired by HGT and show a patchwork genetic organization. Such genetic features of the reconstructed SD fimbriome of the genus Bifidobacterium suggest variable and exchangeable interaction capabilities with different environments. In silico analyses coupled with transcriptome analyses and ECM binding experiments revealed how the different members of the bifidobacterial SD fimbriome may interact with ECM proteins, such as fibronectin as well as with glycans or specifically only one of these substrates. These findings suggest an intriguing mechanism of genetic adaptation of bifidobacteria to the gut of mammals, birds, and insects, allowing their successful colonization and persistence in these ecological niches. More specifically, our data are in line with two possible different ecological activities for those bifidobacteria that colonize the mammalian gut. In this context, those bifidobacteria that due to their SD fimbriome adhere to diet-derived glycans (e.g., starch, xylan, and pectin) thus exhibit a luminal behavior, which may not support a long persistence in the gut. In contrast, other bifidobacterial species possess an SD fimbriome that allows adherence to the mucosa layer through the ECM proteins and consequently long-term colonization in the gut. Nevertheless, such different adhesive roles played by the bifidobacterial SD fimbriome might be further influenced by the SD fimbriome or the extracellular structures produced by other members of the gut microbiota. Nevertheless, since many bifidobacteria are practically genetically inaccessible, with just a few exceptions (55, 56), we are currently not in a position to test our predictions under in vivo circumstances. Future gene inactivation PEL experiments, when such procedures will become available, will therefore be important to confirm our in silico results.
MATERIALS AND METHODS
Identification of PEL and COPGs.
SD pilus-encoding loci (PEL), which represent type I and type II pili (12), were identified through manual inspection based on homology analyses for all 140 bifidobacterial genomes (publicly) available at the time of this study using a custom database encompassing all known sortase-dependent pilus genes reported in the NCBI RefSeq database. A further 18 novel bifidobacterial strains were selected for PEL inspection, resulting in 81 genes that were deposited in GenBank. Clusters of orthologous pilus genes (COPGs) were identified through pangenome analysis performed using the PGAP pipeline (57) as outlined below. The open reading frame (ORF) contents from all PEL identified in this study were organized in functional clusters using the gene family (GF) method involving comparison of each deduced protein to all other proteins by BLAST analysis (cutoff E value of 1 × 10−5 and 50% identity across at least 50% of both protein sequences), followed by clustering into protein families using MCL (a graph-theory-based Markov clustering algorithm) (58). Data regarding protein family distribution among PEL were implied for manual reconstruction of the COPGs. Information on the distribution of individual COPGs in bifidobacterial taxa was used to produce a network representation with Gephi software (59). Phylogenetic trees of the minor and major pilus subunits were constructed using ClustalX for sequence alignment (60) and FigTree for visualization.
Growth conditions.
Bifidobacterium cells were cultivated in an anaerobic atmosphere (2.99% H2, 17.01% CO2, and 80% N2) in a growth chamber (Concept 400; Ruskin) at 37°C for 24 h in de Man-Rogosa-Sharpe medium (MRS) (Scharlau Chemie, Barcelona, Spain), supplemented with 0.05% (wt/vol) l-cysteine hydrochloride. Bifidobacterium cells were inoculated in 10 ml of MRS without carbohydrates (Scharlau Chemie, Barcelona, Spain) supplemented with 0.5% starch, maltotriose, xylan, xylose, or fructose (Sigma-Aldrich) as unique carbon sources. Carbohydrates were purchased from Sigma (Milan, Italy) or Carbosynth (Berkshire, United Kingdom). Carbohydrates were dissolved in water and then sterilized by filtration using a 0.2-μm-pore filter and then added to autoclaved MRS, with the exception of xylan (Sigma, Aldrich), which was autoclaved with MRS. Cell suspensions were mixed and incubated at 37°C for 24 h under anaerobic conditions.
RNA isolation.
Total RNA was isolated by a previously described method (61). Briefly, cell pellets/tissue materials were resuspended in 1 ml of QIAzol (Qiagen, United Kingdom) and placed in a tube containing 0.8 g of glass beads (diameter, 106 μm; Sigma). Cells were lysed by shaking the mixture on a BioSpec homogenizer at 4°C for 2 min (maximum setting). The mixture was then centrifuged at 12,000 rpm for 15 min, and the upper phase containing the RNA-containing sample was recovered. The RNA sample was further purified by phenol extraction and ethanol precipitation according to an established method (62). The quality of the RNA was checked by analyzing the integrity of rRNA molecules by Tape Station analysis (Agilent).
Reverse transcription and qRT-PCR analyses.
Reverse transcription to cDNA was performed with the iScript Select cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA) using the following thermal cycle regimen: 5 min at 25°C, 30 min at 42°C, and 5 min at 85°C. The mRNA expression levels of cytokines were analyzed with SYBR green technology by RT-qPCR using SsoFast EvaGreen supermix (Bio-Rad Italia, Segrate, Italy) on a Bio-Rad CFX96 system according to the manufacturer's instructions (Bio-Rad Italia). The primers used are indicated in Table S4 in the supplemental material. qPCR for housekeeping genes was carried out according to the following amplification cycle protocol: initial hold at 95°C for 2 min and then 45 cycles at 95°C for 5 s and 60°C for 30 s. The same PCR cycle conditions were used for all qRT-PCR experiments performed, although the annealing temperature was adjusted according to the primers employed. Gene expression was normalized to the housekeeping gene coding for the 16S rRNA as described previously (63, 64). The amount of template cDNA used for each sample was 12.5 ng.
Sample preparation and AFM imaging.
Bacteria from an overnight culture were harvested by centrifugation, washed with phosphate-buffered saline (PBS) by resuspension, and collected by centrifugation (4,000 rpm). The washed pellet was then resuspended in 200 μl of PBS and kept on ice until AFM imaging. Mica was then rinsed with Milli-Q water (Millipore) and dried with nitrogen. After this, 20 μl of bacterial suspension was deposited onto mica for 10 min. The mica disk was then rinsed with Milli-Q water and dried under a weak gas flow of nitrogen. The quality of the sample and density of surface-bound bacteria were verified with an optical microscope.
AFM imaging was performed on dried samples with an XE-100 microscope (Park System) in tapping mode. Commercial diving board silicon cantilevers (MikroMasch) were used. The best image quality was obtained with amplitude 1 V and a low scan rate (0.5 to 0.7 Hz). Filamentous structures at the periphery of bacteria were visible in images of 256 by 256 pixels, representing a scan size of 10 μm or less. While imaging, both height and amplitude signals were collected. Height images were flattened using XEI software.
Quantification of bacterial binding to ECM proteins.
Ninety-six-well plates (MicroWell plates; Maxisorp Nunc, Roskilde, Denmark) were coated with a solution of 500 μg/ml of fibronectin (Sigma) in 100 μl PBS at pH 7.4 (i.e., 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4). Unbound protein was removed by washing the plates twice with PBS, followed by blocking the wells by rinsing with 1% BSA in PBS for 30 min at 37°C. The blocking buffer was removed, and the wells were washed twice prior to the addition of bacterial cells in a 100-μl final volume. Incubation with bacteria was performed for 1 h at 37°C. The wells were washed with PBS, and the bacterial cells that adhered to the wells were collected by scraping them into PBS with 0.5% (vol/vol) Triton X-100; serial dilutions were plated onto MRS or GM17 agar plates. The number of adherent bacteria (CFU per milliliter) was determined by counting the resulting colonies in duplicate. Adherence data are also expressed as the percentage of adherent bacteria recovered from triplicate wells, and the means were gathered from two independent experiments. Statistical differences are expressed as the P value determined by a paired Student's t test.
Identification of bifidobacterial SD fimbriome components in gut metagenomic and metatranscriptomic data sets.
All identified bifidobacterial PEL were aligned with microbiota sequenced reads belonging to three projects previously deposited in the NCBI Sequence Read Archive (SRA). These metagenomic data were obtained from shotgun sequencing of fecal samples of healthy and preterm infants from the Metagenome from Infant Gut Samples Project (BioProject identification [ID] no. PRJNA63661), children administered with antibiotics from the Child Gut Microbiome under Antibiotics Project (BioProject ID no. PRJEB11685), and healthy adults from the Human Microbiome Project (BioProject ID no. PRJNA48479). Metagenomic data sets were filtered using the fastq-mcf script (https://expressionanalysis.github.io/ea-utils/) (minimum mean quality score, 20; window size, 5; quality threshold, 25; and minimum length, 80) to exclusively retrieve high-quality reads. The resulting reads were aligned against the human genome using the Burrows-Wheeler Aligner program (65) (BWAMEM algorithm with trigger reseeding, 1.5; minimum seed length, 19; matching score, 1; mismatch penalty, 4; gap open penalty, 6; and gap extension penalty, 1) and further processed with the SAMtools software package (66) to remove human reads. The mapping against the bifidobacterial PEL and COPGs was performed using Bowtie 2 (67) through multiple-hit mapping and “very-sensitive” policy. The mapping was performed using a minimum score threshold function (-score-min C,-13,0) in order to limit reads with at least 98% full-length identity. The software employed to calculate read counts corresponding to bifidobacterial PEL and COPGs was HTSeq (68) (running in union mode). The percentages of bifidobacterial PEL and COPG for each sample were based on the total amount of filtered reads.
Accession number(s).
From 18 novel bifidobacterial strains selected for PEL inspection, sequences for 81 genes have been deposited in GenBank under accession no. MF043305 to MF043385.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the EU Joint Programming Initiative—a Healthy Diet for a Healthy Life (JPI HDHL, http://www.healthydietforhealthylife.eu/) and the MIUR to M.V. We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. D.v.S. is a member of The APC Microbiome Institute funded by Science Foundation Ireland (SFI), through the Irish Government's National Development Plan (grant no. SFI/12/RC/2273). Part of this research was conducted using the High Performance Computing (HPC) facility of the University of Parma.
The authors declare that they have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01295-17.
REFERENCES
- 1.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turroni F, van Sinderen D, Ventura M. 2009. Bifidobacteria: from ecology to genomics. Front Biosci 14:4673–4684. doi: 10.2741/3559. [DOI] [PubMed] [Google Scholar]
- 3.O'Callaghan A, van Sinderen D. 2016. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol 7:925. doi: 10.3389/fmicb.2016.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. 2012. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol 20:467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Turroni F, Ventura M, Butto LF, Duranti S, O'Toole PW, Motherway MO, van Sinderen D. 2014. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell Mol Life Sci 71:183–203. doi: 10.1007/s00018-013-1318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, Lugli GA, Ferrario C, Gioiosa L, Ferrarini A, Li J, Palanza P, Delledonne M, van Sinderen D, Ventura M. 2016. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J 10:1656–1668. doi: 10.1038/ismej.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arboleya S, Watkins C, Stanton C, Ross RP. 2016. Gut bifidobacteria populations in human health and aging. Front Microbiol 7:1204. doi: 10.3389/fmicb.2016.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. 2009. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Kline KA, Dodson KW, Caparon MG, Hultgren SJ. 2010. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol 18:224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proft T, Baker EN. 2009. Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell Mol Life Sci 66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishnan V. 2015. Pilins in Gram-positive bacteria: a structural perspective. IUBMB Life 67:533–543. doi: 10.1002/iub.1400. [DOI] [PubMed] [Google Scholar]
- 12.Danne C, Dramsi S. 2012. Pili of Gram-positive bacteria: roles in host colonization. Res Microbiol 163:645–658. doi: 10.1016/j.resmic.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Spirig T, Weiner EM, Clubb RT. 2011. Sortase enzymes in Gram-positive bacteria. Mol Microbiol 82:1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foroni E, Serafini F, Amidani D, Turroni F, He F, Bottacini F, Motherway MO, Viappiani A, Zhang Z, Rivetti C, van Sinderen D, Ventura M. 2011. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb Cell Fact 10(Suppl 1):S16. doi: 10.1186/1475-2859-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott JR, Zahner D. 2006. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol 62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- 16.Danelishvili L, Yamazaki Y, Selker J, Bermudez LE. 2010. Secreted Mycobacterium tuberculosis Rv3654c and Rv3655c proteins participate in the suppression of macrophage apoptosis. PLoS One 5:e10474. doi: 10.1371/journal.pone.0010474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schreiner HC, Sinatra K, Kaplan JB, Furgang D, Kachlany SC, Planet PJ, Perez BA, Figurski DH, Fine DH. 2003. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc Natl Acad Sci U S A 100:7295–7300. doi: 10.1073/pnas.1237223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomich M, Planet PJ, Figurski DH. 2007. The tad locus: postcards from the widespread colonization island. Nat Rev Microbiol 5:363–375. doi: 10.1038/nrmicro1636. [DOI] [PubMed] [Google Scholar]
- 19.Motherway MO, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kachlany SC, Planet PJ, DeSalle R, Fine DH, Figurski DH. 2001. Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol 9:429–437. doi: 10.1016/S0966-842X(01)02161-8. [DOI] [PubMed] [Google Scholar]
- 21.Turroni F, Serafini F, Foroni E, Duranti S, Motherway MO, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sanchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A 110:11151–11156. doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turroni F, Serafini F, Mangifesta M, Arioli S, Mora D, van Sinderen D, Ventura M. 2014. Expression of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions. FEMS Microbiol Lett 357:23–33. doi: 10.1111/1574-6968.12509. [DOI] [PubMed] [Google Scholar]
- 23.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, Delcenserie V, Barrangou R, Margolles A, van Sinderen D, Ventura M. 2014. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, Sanchez B, Ferrario C, Viappiani A, Mangifesta M, Mancino W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2015. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ Microbiol 17:2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 25.Duranti S, Milani C, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Sanchez B, Margolles A, van Sinderen D, Ventura M. 2016. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci Rep 6:23971. doi: 10.1038/srep23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnett TC, Scott JR. 2002. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J Bacteriol 184:2181–2191. doi: 10.1128/JB.184.8.2181-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klemm P, Schembri MA. 2000. Bacterial adhesins: function and structure. Int J Med Microbiol 290:27–35. doi: 10.1016/S1438-4221(00)80102-2. [DOI] [PubMed] [Google Scholar]
- 28.Ton-That H, Schneewind O. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol 50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 29.Mishra A, Das A, Cisar JO, Ton-That H. 2007. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J Bacteriol 189:3156–3165. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillaspy AF, Lee CY, Sau S, Cheung AL, Smeltzer MS. 1998. Factors affecting the collagen binding capacity of Staphylococcus aureus. Infect Immun 66:3170–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snodgrass JL, Mohamed N, Ross JM, Sau S, Lee CY, Smeltzer MS. 1999. Functional analysis of the Staphylococcus aureus collagen adhesin B domain. Infect Immun 67:3952–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liddington R. 2010. Catching pneumonia. Structure 18:6–8. doi: 10.1016/j.str.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Deivanayagam CC, Rich RL, Carson M, Owens RT, Danthuluri S, Bice T, Hook M, Narayana SV. 2000. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure 8:67–78. doi: 10.1016/S0969-2126(00)00081-2. [DOI] [PubMed] [Google Scholar]
- 34.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Podolsky DK. 1985. Oligosaccharide structures of human colonic mucin. J Biol Chem 260:8262–8271. [PubMed] [Google Scholar]
- 37.Shaik MM, Maccagni A, Tourcier G, Di Guilmi AM, Dessen A. 2014. Structural basis of pilus anchoring by the ancillary pilin RrgC of Streptococcus pneumoniae. J Biol Chem 289:16988–16997. doi: 10.1074/jbc.M114.555854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pavkov T, Egelseer EM, Tesarz M, Svergun DI, Sleytr UB, Keller W. 2008. The structure and binding behavior of the bacterial cell surface layer protein SbsC. Structure 16:1226–1237. doi: 10.1016/j.str.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Boraston AB, Healey M, Klassen J, Ficko-Blean E, Lammerts van Bueren A, Law V. 2006. A structural and functional analysis of alpha-glucan recognition by family 25 and 26 carbohydrate-binding modules reveals a conserved mode of starch recognition. J Biol Chem 281:587–598. doi: 10.1074/jbc.M509958200. [DOI] [PubMed] [Google Scholar]
- 40.Filloux A. 2010. A variety of bacterial pili involved in horizontal gene transfer. J Bacteriol 192:3243–3245. doi: 10.1128/JB.00424-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bottacini F, Milani C, Turroni F, Sanchez B, Foroni E, Duranti S, Serafini F, Viappiani A, Strati F, Ferrarini A, Delledonne M, Henrissat B, Coutinho P, Fitzgerald GF, Margolles A, van Sinderen D, Ventura M. 2012. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 7:e44229. doi: 10.1371/journal.pone.0044229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, Ferrario C, Mangifesta M, Hevia A, Viappiani A, Scholz M, Arioli S, Sanchez B, Lane J, Ward DV, Hickey R, Mora D, Segata N, Margolles A, van Sinderen D, Ventura M. 2015. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep 5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, Ferrario C, van Sinderen D, Ventura M. 2015. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol 82:980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorsey CW, Laarakker MC, Humphries AD, Weening EH, Baumler AJ. 2005. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol Microbiol 57:196–211. doi: 10.1111/j.1365-2958.2005.04666.x. [DOI] [PubMed] [Google Scholar]
- 45.Duranti S, Lugli GA, Mancabelli L, Turroni F, Milani C, Mangifesta M, Ferrario C, Anzalone R, Viappiani A, van Sinderen D, Ventura M. 2017. Prevalence of antibiotic resistance genes among human gut-derived bifidobacteria. Appl Environ Microbiol 83:e02894-16. doi: 10.1128/AEM.02894-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morrow AL, Lagomarcino AJ, Schibler KR, Taft DH, Yu Z, Wang B, Altaye M, Wagner M, Gevers D, Ward DV, Kennedy MA, Huttenhower C, Newburg DS. 2013. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome 1:13. doi: 10.1186/2049-2618-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, de Vos WM. 2016. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 7:10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lugli GA, Milani C, Turroni F, Tremblay D, Ferrario C, Mancabelli L, Duranti S, Ward DV, Ossiprandi MC, Moineau S, van Sinderen D, Ventura M. 2016. Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ Microbiol 18:2196–2213. doi: 10.1111/1462-2920.13154. [DOI] [PubMed] [Google Scholar]
- 49.Lugli GA, Milani C, Turroni F, Duranti S, Ferrario C, Viappiani A, Mancabelli L, Mangifesta M, Taminiau B, Delcenserie V, van Sinderen D, Ventura M. 2014. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl Environ Microbiol 80:6383–6394. doi: 10.1128/AEM.02004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nuriel-Ohayon M, Neuman H, Koren O. 2016. Microbial changes during pregnancy, birth, and infancy. Front Microbiol 7:1031. doi: 10.3389/fmicb.2016.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, Kubota H, Swinkels S, Sakai T, Oishi K, Kushiro A, Knol J. 2016. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 11:e0158498. doi: 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yassour M, Vatanen T, Siljander H, Hamalainen AM, Harkonen T, Ryhanen SJ, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, Lander ES, Knip M, DIABIMMUNE Study Group, Xavier RJ. 2016. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med 8:343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avershina E, Lundgard K, Sekelja M, Dotterud C, Storro O, Oien T, Johnsen R, Rudi K. 2016. Transition from infant- to adult-like gut microbiota. Environ Microbiol 18:2226–2236. doi: 10.1111/1462-2920.13248. [DOI] [PubMed] [Google Scholar]
- 54.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. 2010. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156:3329–3341. doi: 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- 55.Sakaguchi K, He JL, Tani S, Kano Y, Suzuki T. 2012. A targeted gene knockout method using a newly constructed temperature-sensitive plasmid mediated homologous recombination in Bifidobacterium longum. Appl Microbiol Biotechnol 95:499–509. doi: 10.1007/s00253-012-4090-4. [DOI] [PubMed] [Google Scholar]
- 56.O'Connell KJ, Motherway MO, Liedtke A, Fitzgerald GF, Ross RP, Stanton C, Zomer A, van Sinderen D. 2014. Transcription of two adjacent carbohydrate utilization gene clusters in Bifidobacterium breve UCC2003 is controlled by LacI- and repressor open reading frame kinase (ROK)-type regulators. Appl Environ Microbiol 80:3604–3614. doi: 10.1128/AEM.00130-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jacomy M, Venturini T, Heymann S, Bastian M. 2014. ForceAtlas2, a continuous graph layout algorithm for handy network visualization designed for the Gephi software. PLoS One 9:e98679. doi: 10.1371/journal.pone.0098679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 61.Turroni F, Foroni E, Montanini B, Viappiani A, Strati F, Duranti S, Ferrarini A, Delledonne M, van Sinderen D, Ventura M. 2011. Global genome transcription profiling of Bifidobacterium bifidum PRL2010 under in vitro conditions and identification of reference genes for quantitative real-time PCR. Appl Environ Microbiol 77:8578–8587. doi: 10.1128/AEM.06352-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sambrook J, Fritsch EF, Maniatis T (ed). 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 63.Turroni F, Strati F, Foroni E, Serafini F, Duranti S, van Sinderen D, Ventura M. 2012. Analysis of predicted carbohydrate transport systems encoded by Bifidobacterium bifidum PRL2010. Appl Environ Microbiol 78:5002–5012. doi: 10.1128/AEM.00629-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turroni F, Ozcan E, Milani C, Mancabelli L, Viappiani A, van Sinderen D, Sela DA, Ventura M. 2015. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front Microbiol 6:1030. doi: 10.3389/fmicb.2015.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.