ABSTRACT
Coronafacoyl phytotoxins are an important family of plant toxins that are produced by several different phytopathogenic bacteria, including the gammaproteobacterium Pseudomonas syringae and the actinobacterium Streptomyces scabiei (formerly Streptomyces scabies). The phytotoxins consist of coronafacic acid (CFA) linked via an amide bond to different amino acids or amino acid derivatives. Previous work suggested that S. scabiei and P. syringae use distinct biosynthetic pathways for producing CFA, which is subsequently linked to its amino acid partner to form the complete phytotoxin. Here, we provide further evidence that the S. scabiei CFA biosynthetic pathway is novel by characterizing the role of CYP107AK1, a predicted cytochrome P450 that has no homologue in P. syringae. Deletion of the CYP107AK1 gene abolished production of coronafacoyl-isoleucine (CFA-Ile), the primary coronafacoyl phytotoxin produced by S. scabiei. Structural elucidation of accumulated biosynthetic intermediates in the ΔCYP107AK1 mutant indicated that CYP107AK1 is required for introducing the oxygen atom that ultimately forms the carbonyl group in the CFA backbone. The CYP107AK1 gene along with two additional genes involved in CFA-Ile biosynthesis in S. scabiei were found to be associated with putative CFA biosynthetic genes in other actinobacteria but not in other organisms. Analysis of the overall genetic content and organization of known and putative CFA biosynthetic gene clusters, together with phylogenetic analysis of the core biosynthetic genes, indicates that horizontal gene transfer has played an important role in the dissemination of the gene cluster and that rearrangement, insertion, and/or deletion events have likely contributed to the divergent biosynthetic evolution of coronafacoyl phytotoxins in bacteria.
IMPORTANCE The ability of plants to defend themselves against invading pathogens relies on complex signaling pathways that are controlled by key phytohormones such as jasmonic acid (JA). Some phytopathogenic bacteria have evolved the ability to manipulate JA signaling in order to overcome host defenses by producing coronatine (COR), which functions as a potent JA mimic. COR and COR-like molecules, collectively referred to as coronafacoyl phytotoxins, are produced by several different plant-pathogenic bacteria, and this study provides supporting evidence that different biosynthetic pathways are utilized by different bacteria for production of these phytotoxins. In addition, our study provides a greater understanding of how coronafacoyl phytotoxin biosynthesis may have evolved in phylogenetically distinct bacteria, and we demonstrate that production of these compounds may be more widespread than previously recognized and that their role for the producing organism may not be limited to host-pathogen interactions.
KEYWORDS: biosynthesis, coronafacoyl phytotoxin, coronatine, cytochrome P450, gene cluster, jasmonic acid, Pseudomonas syringae, Streptomyces scabiei, plant pathogens, signaling
INTRODUCTION
The ability of plants to protect themselves from invading microbial pathogens relies on complex signaling networks that are controlled by key hormones such as salicylic acid (SA) and jasmonic acid (JA) (1, 2). SA is primarily responsible for regulating defense against biotrophic and hemibiotrophic pathogens, while JA is mainly responsible for mediating defense against necrotrophic pathogens as well as against chewing insects and other herbivores. SA and JA signaling pathways are generally antagonistic in that activation of SA-regulated pathways leads to suppression of JA-regulated pathways, and vice versa (1, 2). JA is also critical for regulating the plant response to abiotic stress, and it modulates plant growth and developmental processes and influences plant secondary metabolism (3).
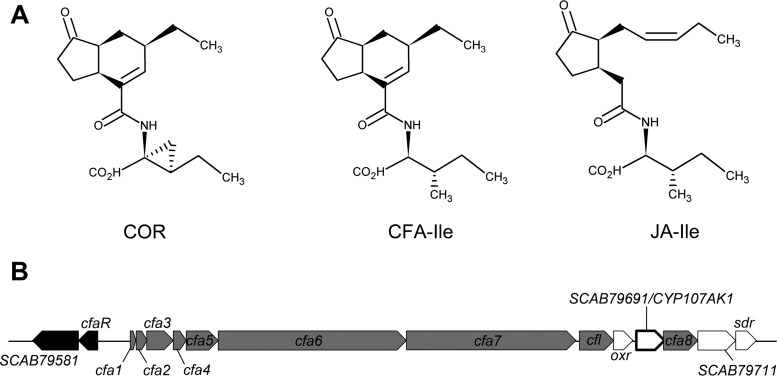
In order to overcome the plant immune response, many plant-pathogenic microorganisms have evolved strategies that allow them to manipulate the plant hormone signaling pathways for their own benefit. One of the best-studied examples of this is the production of coronatine (COR) (Fig. 1A) by several pathovars of the Gram-negative hemibiotrophic plant pathogen Pseudomonas syringae. COR functions as a molecular mimic of (+)-iso-jasmonyl-l-isoleucine (JA-Ile) (Fig. 1A), the most bioactive form of JA (4). Both JA-Ile and COR promote the binding of the F-box protein coronatine-insensitive 1 (COI1) to jasmonate zim domain (JAZ) repressor proteins, thereby leading to degradation of the JAZ proteins and activation of the JA signaling pathway (5). This, in turn, causes suppression of the SA-mediated defense response, which is important for combating P. syringae infections (6). Intriguingly, COR is ∼1,000 times more active than JA-Ile at promoting the formation of the COI1-JAZ complex and subsequent degradation of the JAZ proteins, suggesting that COR is a highly effective mimic of JA-Ile perception in plants (4, 7). Furthermore, COR suppresses callose deposition in an SA-independent manner and enables bacterial entry into the plant host by overcoming stomatal defenses (6, 8). Thus, the production of COR provides an adaptive advantage to P. syringae by serving as a multifunctional suppressor of plant defenses during infection.
FIG 1.
(A) Structure of the coronafacoyl phytotoxins coronatine (COR) and coronafacoyl-l-isoleucine (CFA-Ile) produced by P. syringae and S. scabiei, respectively, and of the bioactive plant hormone conjugate (+)-7-iso-jasmonyl-l-isoleucine (JA-Ile). (B) Organization of the CFA biosynthetic gene cluster from S. scabiei 87-22. Regulatory genes are in black, biosynthetic genes that have homologues in the P. syringae CFA biosynthetic gene cluster are in gray, and biosynthetic genes unique to the S. scabiei gene cluster are in white. The CYP107AK1 gene is highlighted in bold.
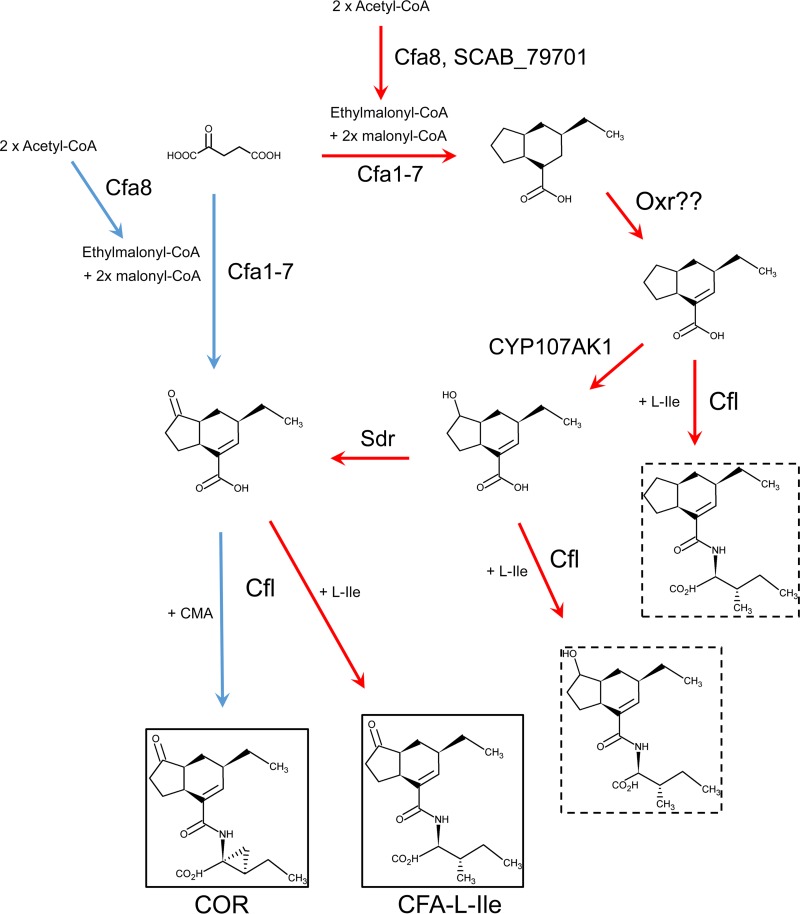
COR consists of the polyketide metabolite coronafacic acid (CFA) linked via an amide bond to coronamic acid (CMA), which is derived from l-isoleucine (9). In P. syringae, the two moieties are produced independently by the cfa and cma biosynthetic gene clusters, respectively, after which they are joined together most likely by the coronafacate ligase (Cfl) enzyme encoded within the cfa gene cluster (Fig. 2) (9). Although COR is the primary product synthesized by P. syringae and is the most toxic, other related compounds (referred to as coronatine analogs or coronatine-like compounds) in which CFA is linked to amino acids such as isoleucine or valine (9) can also be produced. Together, these compounds form a family of molecules called the coronafacoyl phytotoxins. COR and COR-like molecules are known or predicted to be produced by several plant-pathogenic bacteria other than P. syringae, including Pseudomonas savastanoi, Pectobacterium atrosepticum, Pectobacterium carotovorum subsp. carotovorum, Xanthomonas campestris pv. phormiicola, and Streptomyces scabiei (formerly Streptomyces scabies) (9–13). The conservation of this family of compounds across diverse bacterial genera suggests that the JA signaling pathway is a common target for manipulation during host-pathogen interactions.
FIG 2.
Proposed biosynthetic pathways for the production of coronafacoyl phytotoxins in P. syringae and S. scabiei. The hypothetical pathway in P. syringae is indicated by blue arrows, whereas the hypothetical pathway in S. scabiei is indicated by red arrows. Biosynthetic intermediates that have been isolated from S. scabiei biosynthetic mutants are in hatched boxes.
We have been studying the production of coronafacoyl phytotoxins by S. scabiei, which is an important causative agent of potato common scab disease worldwide. This organism produces N-coronafacoyl-l-isoleucine (CFA-Ile) (Fig. 1A) as the primary product as well as other minor coronafacoyl phytotoxins, but it does not produce COR due to the absence of the cma biosynthetic genes in the genome (11, 14). S. scabiei harbors a biosynthetic gene cluster that contains homologues of the cfa biosynthetic genes from P. syringae (Fig. 1B). Interestingly, this gene cluster also contains six additional genes that are absent from the cfa biosynthetic gene cluster in P. syringae. Two of these genes, cfaR and SCAB79581, are found at the beginning of the S. scabiei cfa biosynthetic gene cluster (Fig. 1B) and are regulatory genes that activate the production of CFA-Ile (14, 15). The four remaining unique genes are predicted to encode biosynthetic enzymes and are cotranscribed with the other biosynthetic genes in the gene cluster (14). Recently, we reported that two of the genes, SCAB79681 (also called oxr) and SCAB79721 (also called sdr), are required for normal production of CFA-Ile and that the Sdr enzyme and possibly Oxr are directly involved in the biosynthesis of the CFA moiety (Fig. 2) (16). Given that the P. syringae genome does not encode an Oxr homolog and encodes only a weakly similar Sdr homologue, our results suggested that S. scabiei utilizes a novel biosynthetic pathway for producing coronafacoyl phytotoxins (16).
In this study, we provide further evidence that the production of the coronafacoyl phytotoxins in S. scabiei involves a novel biosynthetic pathway by characterizing the function of the SCAB79691 gene (here referred to as CYP107AK1). We also utilize bioinformatics and genome sequencing data to investigate the evolutionary history of coronafacoyl phytotoxin biosynthesis in different bacteria.
RESULTS
CYP107AK1 encodes a predicted cytochrome P450 monooxygenase.
Protein BLAST and phylogenetic analyses revealed that the S. scabiei CYP107AK1 amino acid sequence is most closely related to that of a predicted cytochrome P450 (CYP) from Streptomyces sp. strain NRRL WC-3618 (WP_078960852; 87% identity and 90% similarity to CYP107AK1) and Kitasatospora azatica (WP_083975939; 88% identity and 91% similarity to CYP107AK1) and also shows similarity to other known or predicted CYPs from various species of actinomycetes. Analysis using the Pfam database showed that CYP107AK1 contains a cytochrome P450 domain (Pfam domain accession number PF00067) within the amino acid sequence. Also identified were two signature CYP motifs, the EXXR motif found in the K helix and the GXXXCXG motif found in the heme-binding loop and containing the invariant cysteine residue (17). Perusal of the Cytochrome P450 Homepage database (http://drnelson.uthsc.edu/cytochromeP450.html) revealed that CYP107AK1 was previously classified as a member of the CYP107A subfamily of CYPs. This subfamily includes P450 enzymes that are known to be associated with microbial secondary metabolism.
Gene deletion analysis confirms that CYP107AK1 plays a direct role in CFA biosynthesis in S. scabiei.
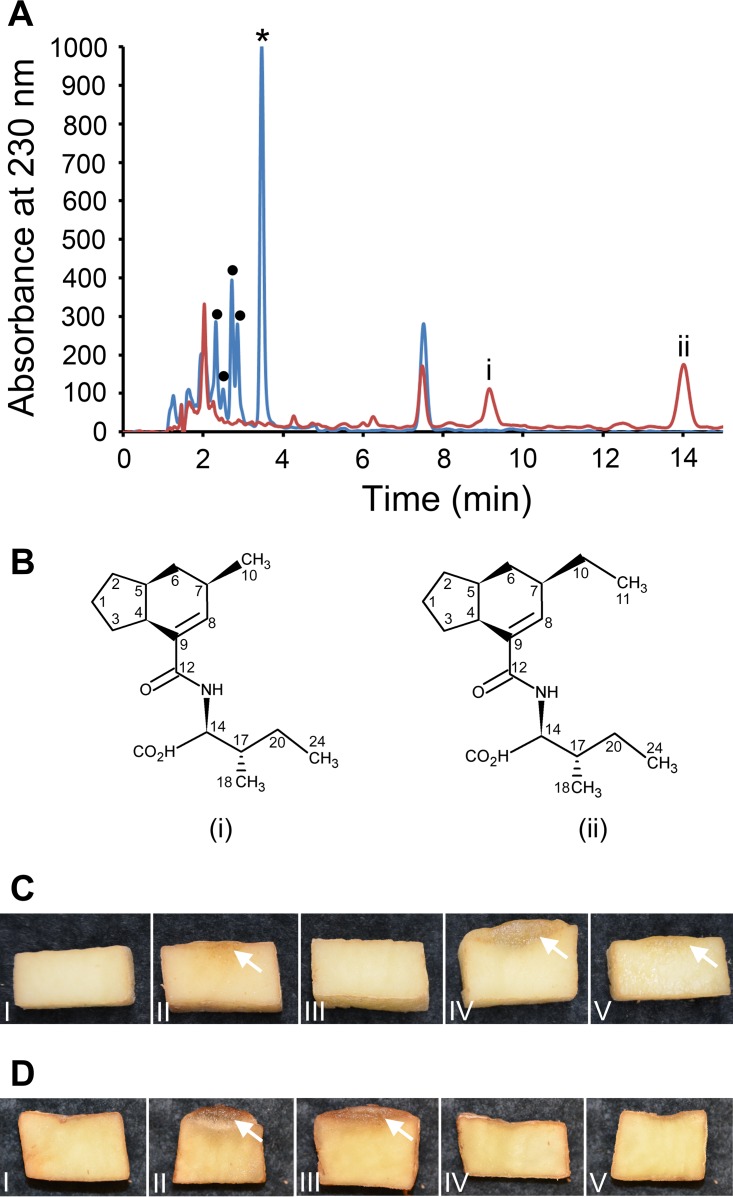
To determine the role of CYP107AK1 in the production of the coronafacoyl phytotoxins, an S. scabiei mutant in which the CYP107AK1 gene was deleted was created and the corresponding levels of phytotoxin production were examined. The ΔCYP107AK1 mutant was created in the ΔtxtA/pRLDB51-1 strain, which exhibits high production levels of coronafacoyl phytotoxins due to cfaR regulatory gene overexpression and also lacks production of the virulence-associated phytotoxin thaxtomin A due to deletion of the txtA biosynthetic gene (11, 14). The latter feature allows for the rapid detection of phytotoxic activity associated with coronafacoyl phytotoxins in culture extracts, and it also simplifies the purification of coronafacoyl phytotoxins from the extracts. Organic extraction of the culture supernatants followed by reverse-phase high-performance liquid chromatography (RP-HPLC) analysis revealed that production of CFA-Ile and of the other minor coronafacoyl phytotoxins is abolished in the ΔCYP107AK1 mutant. In addition, two new metabolites (i and ii) were observed in the mutant extract with retention times (tR) of 9.16 min and 14.02 min, respectively (Fig. 3A). Liquid chromatography/low-resolution mass spectrometry (LC-LRMS) analysis of these metabolite peaks in negative ion mode demonstrated the presence of pseudomolecular [M-H]− ions at m/z 292 and 306, which indicated a molecular mass of 293 Da for compound i and 307 Da for compound ii. The two metabolites were subsequently purified, and analysis by liquid chromatography/high-resolution mass spectrometry (LC-HRMS) in positive ion mode revealed pseudomolecular [M+H]+ and [M+Na]+ ions at m/z 294.2064 and 316.1884, respectively, for compound i, while [M+H]+ and [M+Na]+ ions at m/z 308.2219 and 330.2040, respectively, were observed for compound ii (see Fig. S1 in the supplemental material). This is consistent with molecular formulas of C17H27NO3 (calculated: m/z 294.2069 [M+H]+, Δ = −1.7 ppm) and C18H29NO3 (calculated: m/z 308.2226 [M+H]+, Δ = −2.3 ppm) for compounds i and ii, respectively.
FIG 3.
(A) RP-HPLC analysis of culture extracts from the S. scabiei ΔtxtA/pRLDB51-1 strain (blue line) and the ΔCYP107AK1 mutant (red line). The peak corresponding to the CFA-Ile coronafacoyl phytotoxin is indicated with an asterisk (*), and the peaks corresponding to minor coronafacoyl phytotoxins are indicated with a filled black circle (•). The two intermediates (i and ii) that accumulated in the ΔCYP107AK1 mutant extract are labeled. (B) Chemical structures of the purified intermediates i and ii. The carbon atoms are numbered according to the numbering scheme used for the raw data analysis (see Table S1 in the supplemental material). (C) Hypertrophy-inducing activity of organic culture extracts of S. scabiei ΔtxtA/pRLDB51-1 (IV) and ΔCYP107AK1 (V) on potato tuber tissue. Pure COR (1 nmol) was included as a positive control (II), while organic solvent (100% methanol; I) and culture extract from the CFA-Ile-deficient S. scabiei ΔtxtA Δcfa6 strain (III) were included as negative controls. The observed tissue hypertrophy is indicated by white arrows. (D) Hypertrophy-inducing activity of purified CFA-Ile (III) and of the ΔCYP107AK1 mutant intermediates i (IV) and ii (V) on potato tuber tissue. Solvent (methanol + 0.1% formic acid) served as a negative control (I), while pure COR (1 nmol) was used as a positive control (II). The observed tissue hypertrophy is indicated by white arrows.
Analysis using 1-dimensional (1H and 13C) (see Table S1 in the supplemental material) and 2-dimensional (correlation spectroscopy [COSY], heteronuclear single-quantum correlation [HSQC], heteronuclear multiple-bond correlation [HMBC], and nuclear Overhauser effect spectroscopy [NOESY]) nuclear magnetic resonance (NMR) revealed the structure of each compound as shown in Fig. 3B. Both metabolites differ from the CFA-Ile end product by the absence of the carbonyl group at position C-2 of the CFA backbone (Fig. 1A). In addition, compound i was determined to contain a methyl side chain at position C-7 instead of an ethyl side chain (Fig. 1A and 3B). This is supported by the strong HMBC correlations between the methyl H-10 and C-6, C-7, and C-8 from the CFA backbone (see Fig. S2 in the supplemental material). Also, tandem mass spectrometry (HRMS/MS) analysis of compound i revealed two prominent fragment ions in positive ion mode at m/z 163.1190 and 135.1193, which is in agreement with the proposed structure (see Fig. S3 in the supplemental material). HRMS/MS was additionally used to confirm the proposed structure of compound ii (see Fig. S4 in the supplemental material).
The attached oxygen at position C-2 is important for the bioactivity of CFA-Ile.
The bioactivity of the ΔCYP107AK1 intermediates was tested using a potato tuber disk bioassay, which detects the tissue hypertrophy-inducing activity of the coronafacoyl phytotoxins (11). Assays using total organic culture extract from the ΔCYP107AK1 mutant demonstrated that the extract exhibited very weak bioactivity compared to that of extract from the parental (ΔtxtA/pRLDB51-1) strain (Fig. 3C), and neither intermediate was able to induce any tissue hypertrophy compared to CFA-Ile when equimolar amounts (100 nmol) of the pure metabolites were used (Fig. 3D). This suggests that the enzyme activity of CYP107AK1 is critical for the hypertrophy-inducing bioactivity of the CFA-Ile end product.
The CYP107AK1 gene is conserved in a subset of bacterial CFA biosynthetic gene clusters.
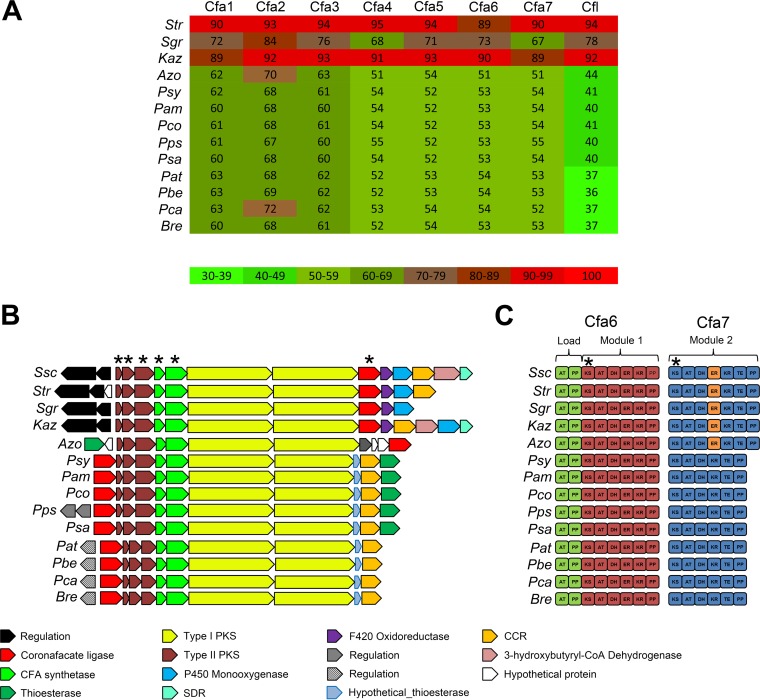
Given the requirement of CYP107AK1 for the biosynthesis of CFA-Ile in S. scabiei, we were interested to know whether the CYP107AK1 gene is associated with known or predicted CFA biosynthetic gene clusters in other organisms. Although it was already known that a CYP107AK1 homologue is absent from the CFA biosynthetic gene cluster in P. syringae pv. tomato DC3000 and in P. atrosepticum SCRI1043 (14), the availability of a large number of bacterial genome sequences in the database presented an opportunity to look for other CFA biosynthetic gene clusters that may harbor a CYP107AK1 homologue. To first identify potential CFA gene clusters, we performed a protein BLAST analysis using the core CFA biosynthetic gene products from S. scabiei (Cfa1-7, Cfl) in order to identify homologues of each protein. Using this approach, we identified the known CFA biosynthetic enzymes from different plant-pathogenic Pseudomonas species (e.g., P. syringae, P. coronafaciens, P. amygdali, P. savastanoi) as well as the predicted CFA biosynthetic enzymes from different plant-pathogenic Pectobacterium spp. (e.g., P. atrosepticum, P. carotovorum, P. betavasculorum) (Fig. 4A). Interestingly, we also found homologues of the CFA biosynthetic enzymes from a plant-pathogenic bacterium (Brenneria sp. strain EniD312) not known to produce coronafacoyl phytotoxins as well as from organisms not known to be plant pathogens, including Azospirillum sp. strain B510, Kitasatospora azatica, Pseudomonas psychrotolerans, Streptomyces griseoruber, and Streptomyces sp. strain NRRL WC-3618. A comparison of the amino acid sequences of each protein revealed that the S. scabiei proteins are most similar to the corresponding homologues from S. griseoruber, Streptomyces sp. NRRL WC-3618, and K. azatica (Fig. 4A). As expected, the corresponding cfa1 to cfa7 (cfa1–7) and cfl genes were found clustered together in all of the genome sequences analyzed (Fig. 4B). The S. griseoruber, Streptomyces sp. NRRL WC-3618, and K. azatica gene clusters were additionally found to harbor a CYP107AK1 homologue as well as a homologue of the oxr gene, which encodes a predicted F420-dependent oxidoreductase and is also involved in CFA-Ile biosynthesis in S. scabiei (Fig. 2 and 4B) (16). Although an sdr homologue was detected only in the K. azatica gene cluster, a homologue (ADL01_RS35145) was found elsewhere in the Streptomyces sp. NRRL WC-3618 genome. No homologues of oxr, sdr, and CYP107AK1 were detected in the genome sequences of the other organisms harboring CFA biosynthetic genes (Fig. 4B).
FIG 4.
(A) Heat map showing the protein BLAST identity hits to the S. scabiei 87-22 Cfa1-7 and Cfl proteins in different bacterial genomes. The actual percent amino acid identity is shown in each square. Abbreviations are as follows: Str, Streptomyces sp. NRRL WC-3618; Sgr, Streptomyces griseoruber DSM40281; Kaz, Kitasatospora azatica ATCC 9699; Azo, Azospirillum sp. B510, Psy, Pseudomonas syringae pv. tomato DC3000; Pam, Pseudomonas amygdali pv. morsprunorum, Pco, Pseudomonas coronafaciens pv. porri LMG 28495; Pps, Pseudomonas psychrotolerans NS274; Psa, Pseudomonas savastanoi pv. glycinea B076; Pat, Pectobacterium atrosepticum SCRI1043; Pbe, Pectobacterium betavasculorum NCPPB 2795; Pca, Pectobacterium carotovorum subsp. carotovorum UGC32; Bre, Brenneria sp. EniD312. (B) Organization of known and putative CFA biosynthetic gene clusters identified in sequenced bacterial genomes. Related genes are in the same color, and the known or predicted functions are indicated below. The genes that were used for construction of the concatenated phylogenetic tree (Fig. 6A) are denoted by an asterisk. Ssc, Streptomyces scabiei 87-22. (C) Domain organization of the Cfa6 and Cfa7 type I polyketide synthases encoded in the CFA biosynthetic gene clusters from different bacteria. The different modules (load, modules 1 and 2) are indicated as well as the enoyl reductase (ER) domain that is found only in a subset of Cfa7 proteins (indicated in orange). The domain nucleotide sequences that were used for construction of the concatenated phylogenetic tree (Fig. 6A) are denoted by an asterisk. Abbreviations are as follows: AT, acyltransferase; PP, phosphopantheinate; KS, ketosynthase; DH, dehydratase; KR, ketoreductase; TE, thioesterase.
Gene content and phylogenetic analyses provide insights into the evolution of coronafacoyl phytotoxin biosynthesis in bacteria.
In our previous study (16), we proposed that S. scabiei utilizes a novel biosynthetic pathway for producing coronafacoyl phytotoxins compared to P. syringae due to the involvement of the oxr and sdr genes, and work presented here on CYP107AK1 further supports this idea (Fig. 2). The conservation of oxr, sdr, and/or CYP107AK1 in known or predicted CFA biosynthetic gene clusters from some bacterial species, but not others, raises intriguing questions about the evolution of coronafacoyl phytotoxin biosynthesis in bacteria. To investigate this further, we first examined the overall gene content and architecture of the CFA biosynthetic gene clusters obtained from the database. As shown in Fig. 4B, the cfa1–3 genes encoding the type II polyketide synthase (PKS) enzymes, the cfa4 and cfa5 genes encoding the CFA synthetase enzymes, and the cfa6 and cfa7 genes encoding the type I PKS enzymes are found in identical arrangements in all of the gene clusters analyzed. In contrast, the relative position of the cfl gene encoding the coronafacate ligase enzyme was found to vary when comparing the Pseudomonas/Pectobacterium/Brenneria gene clusters and the Streptomyces/Azospirillum/Kitasatospora gene clusters (Fig. 4B). Most of the gene clusters were also determined to harbor a gene (designated cfa8 in P. syringae) encoding a predicted crotonyl coenzyme A (crotonyl-CoA) carboxylase/reductase (CCR), which most likely serves to provide a sufficient amount of the ethylmalonyl-CoA extender unit required for CFA polyketide biosynthesis (18). In addition, the S. scabiei and K. azatica gene clusters both harbor a gene encoding a predicted 3-hydroxybutyryl-CoA dehydrogenase, which presumably functions together with the CCR to synthesize the ethylmalonyl-CoA extender unit (18). A gene (designated cfa9 in P. syringae) encoding a predicted thioesterase was found within the Pseudomonas and Azospirillum gene clusters but not in the other gene clusters analyzed. Furthermore, a small gene encoding a hypothetical protein with a predicted thioesterase domain was found upstream of the CCR-encoding gene in the Pseudomonas, Pectobacterium, and Brenneria gene clusters. Many of the gene clusters additionally have regulatory genes that are divergently transcribed from the biosynthetic genes (Fig. 4B). Homologues of both cfaR and SCAB79581, which are involved in activating production of CFA-Ile in S. scabiei (14, 15), are conserved in the other Streptomyces and Kitasatospora gene clusters, suggesting that the regulation of metabolite production in these organisms is similar.
An analysis of the enzymatic domain architecture of the Cfa7 modular polyketide synthase (PKS) also revealed interesting differences among the different CFA biosynthetic gene clusters (Fig. 4C). Previously, it was reported that the S. scabiei Cfa7 protein contains an enoyl reductase (ER) domain that is absent from the corresponding Cfa7 proteins from P. syringae pv. tomato DC3000 and P. atrosepticum SCRI1043 (14), and results presented here show that this domain is also found in the Cfa7 amino acid sequences from S. griseoruber, Streptomyces sp. NRRL WC-3618, and K. azatica. All four ER domains contain an amino acid sequence that is highly similar to the conserved NADP(H) binding motif that is characteristic of these domains (HAAAGGVGMA) (19), suggesting that they are functional (Fig. 5). Intriguingly, the domain is additionally predicted to be present in the Cfa7 protein from Azospirillum sp. B510, though it is not clear whether the domain is active, due to differences in the NADP(H) binding motif compared to the consensus sequence (Fig. 5). In contrast, none of the Cfa7 homologues from the Pseudomonas spp., Pectobacterium spp., or Brenneria sp. were predicted to harbor an ER domain (Fig. 4C).
FIG 5.
NAD(P)H binding motifs from the Cfa7 ER domains of S. scabiei, Azospirillum sp. B510, K. azatica, S. griseoruber, and Streptomyces sp. WC-3618. The consensus NAD(P)H cofactor binding motif is in bold. Differences in motif sequences compared to the consensus sequence are in red.
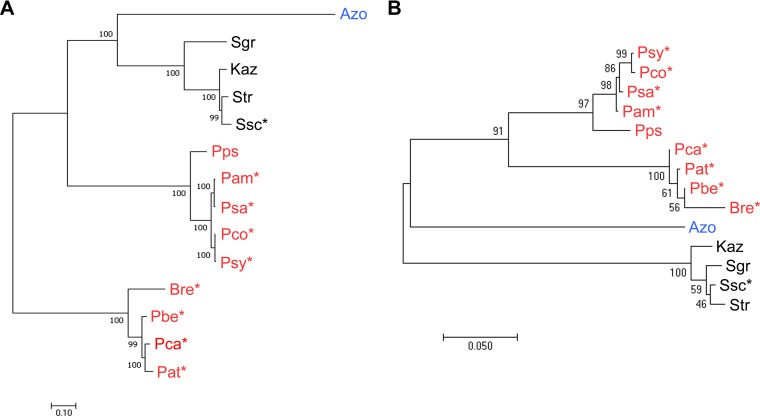
The evolutionary relationships among the different CFA biosynthetic gene clusters were further examined by constructing a concatenated phylogenetic tree using the cfa1–7 and cfl core CFA biosynthetic genes. In the case of the cfa6 and cfa7 type I PKS genes, we utilized the sequences encoding the ketosynthase (KS) domains from each gene rather than the entire gene sequence as recommended for such analyses (20). As shown in Fig. 6, the phylogeny of the core CFA biosynthetic genes is not congruent with that of the corresponding 16S rRNA genes, suggesting that horizontal gene transfer (HGT) has played a key role in the dissemination of the core biosynthetic gene cluster among the different bacteria. Notably, the CFA biosynthetic genes from the different Actinobacteria (S. scabiei, K. azatica, Streptomyces sp. WC-3618, S. griseoruber) form a distinct clade with those from the distantly related alphaproteobacterium Azospirillum sp. B510 (Fig. 6A). In turn, the genes from these organisms appear to share an ancestor with those from the Pseudomonas spp., which are members of the Gammaproteobacteria class. In contrast, the core CFA biosynthetic genes from the other Gammaproteobacteria members (Pectobacterium and Brenneria spp.) appear to form a separate clade from the Pseudomonas genes (Fig. 6).
FIG 6.
Maximum likelihood phylogeny of the core CFA biosynthetic genes (A) and the 16S rRNA gene (B) from different bacteria. The CFA biosynthetic gene cluster tree was constructed using the concatenated cfa1–5, cfl, cfa6 (KS domain), and cfa7 (KS domain) nucleotide sequences. Bootstrap values of ≥50% are shown at the respective branch points and are based on 100 repetitions for the CFA gene cluster tree and 1,000 repetitions for the 16S rRNA gene tree. The scale bars indicate the number of nucleotide substitutions per site. Gammaproteobacteria are in red, Alphaproteobacteria are in blue, and Actinobacteria are in black. Known plant-pathogenic organisms are indicated with an asterisk (*). Abbreviations are as described for Fig. 4.
DISCUSSION
This study has shown that the CYP107AK1 gene is required for coronafacoyl phytotoxin biosynthesis in the common scab pathogen S. scabiei. CYP107AK1 is predicted to encode an enzyme belonging to the CYP superfamily, members of which are heme-thiolate proteins that utilize a wide variety of substrates and are ubiquitously distributed in all domains of life (21). Most CYPs are monooxygenases that catalyze the scission of molecular oxygen and the regio- and stereospecific insertion of an oxygen atom into the substrate. Oxidation reactions attributed to CYPs include epoxidation, hydroxylation, dealkylation, N-oxidation, deamination, dehalogenation, and decarboxylation (21). In Streptomyces spp., CYPs are often associated with secondary metabolite biosynthesis gene clusters, and many have been shown to play a direct role in the biosynthesis of the resulting metabolite(s). For example, CYPs are involved in the production of the antibacterial compounds pikromycin (22) and oleandomycin (23), the antifungal/insecticidal compound nikkomycin (24), the antitumor compound pladienolide (25), and the phytotoxin thaxtomin (26, 27).
Previous work from our lab demonstrated that two additional genes, oxr and sdr, are also involved in coronafacoyl phytotoxin production in S. scabiei. A constructed Δsdr mutant strain was found to accumulate biosynthetic intermediates that contain a hydroxyl group at position C-2 of the CFA backbone, and it was proposed that CYP107AK1 may catalyze the hydroxylation of C-2 and that Sdr then reduces the hydroxyl group to form the C-2 carbonyl in the final CFA molecule (Fig. 2) (16). Work presented here supports this hypothesis, as the ΔCYP107AK1 mutant accumulated two intermediates that were both missing the oxygen attached to C-2 (Fig. 3B). Neither intermediate was able to induce potato tuber tissue hypertrophy compared to an equimolar amount of CFA-Ile (Fig. 3D), indicating that the oxygen at position C-2 is critical for the bioactivity of the phytotoxin. As previously noted with the Δsdr mutant, the CFA intermediates in the ΔCYP107AK1 mutant were isolated only as Ile conjugates, indicating that the coronafacate ligase enzyme is able to efficiently utilize the CFA intermediates as a substrate for ligation to Ile. Surprisingly, one of the ΔCYP107AK1 intermediates was found to contain a methyl group attached to the C-7 position of CFA instead of the ethyl group that is normally present (Fig. 3B). This would presumably occur as a result of the incorporation of methylmalonyl-CoA instead of ethylmalonyl-CoA during polyketide assembly of the CFA backbone by Cfa6. The selection and incorporation of extender units during polyketide biosynthesis are controlled by the acyltransferase (AT) domains of modular PKSs, and while such domains typically exhibit a high degree of specificity for their substrate (28), there is precedence for some AT domains exhibiting a broader substrate specificity. For example, it has been reported that either ethylmalonyl-CoA or methylmalonyl-CoA can be incorporated at the same elongation stage during the biosynthesis of monensin and ascomycin (FK520), and the AT5 domain involved in niddamycin biosynthesis shows a preference for ethylmalonyl-CoA but is able to accept methylmalonyl-CoA (29–31). It is noteworthy that the parental S. scabiei ΔtxtA/pRLDB51-1 strain accumulates a minor metabolite that was previously thought to be CFA conjugated to valine (11), and a predicted valine-containing biosynthetic intermediate was observed to accumulate in the Δsdr mutant (16). As these metabolites would have the exact same molecular formula as an Ile conjugate of a methyl-substituted CFA derivative (or intermediate in the case of the Δsdr mutant), it is possible that the production of different CFA derivatives occurs naturally in S. scabiei depending on the availability of the ethylmalonyl-CoA and methylmalonyl-CoA substrates during polyketide biosynthesis, an idea that warrants further investigation.
The requirement of CYP107AK1 for the production of the CFA moiety in S. scabiei provides further evidence that the CFA biosynthetic pathway in this organism is distinct from that of P. syringae since the P. syringae genome sequence does not encode any proteins showing similarity to CYP107AK1. Additionally, our study revealed that other bacteria may produce coronafacoyl phytotoxins using a biosynthetic pathway similar to that observed in S. scabiei. Protein BLAST analysis resulted in the identification of putative CFA biosynthetic gene clusters in two other Streptomyces spp. and in the actinobacterium K. azatica, and these gene clusters all contain homologues of both CYP107AK1 and oxr (Fig. 4B). An sdr homologue is additionally present in the K. azatica gene cluster, while a homologue is located elsewhere in the genome of Streptomyces sp. WC-3618 (Fig. 4B). Although known or putative CFA biosynthetic gene clusters were identified in different Pseudomonas spp., Pectobacterium spp., Brenneria sp., and Azospirillum sp. B510, none of these gene clusters appear to contain homologues of CYP107AK1, oxr, and sdr, and homologues were not identified elsewhere in the genome sequences. It appears, therefore, that the involvement of these enzymes in CFA biosynthesis is limited to members of the Actinobacteria.
This study also investigated the evolution of coronafacoyl phytotoxin biosynthesis among phylogenetically distinct bacteria. A comparison of the CFA biosynthetic gene clusters among the different members of the Gammaproteobacteria, Alphaproteobacteria, and Actinobacteria revealed that although a core set of genes (cfa1–7 and cfl) are conserved among all of the gene clusters, variations exist in the organization of such genes as well as in the presence or absence of other genes (Fig. 4B). The overall genetic architecture, together with the phylogenetic analysis of the core genes, suggests that the S. scabiei gene cluster is most closely related to the gene clusters from the other Actinobacteria (Fig. 4 and 6). As the CYP107AK1, oxr, and sdr homologues were found only in a subset of gene clusters, it is possible that these genes represent a subcluster that was recently joined to an ancestral core gene cluster. Interestingly, the core CFA gene tree also suggests that the Azospirillum sp. B510 gene cluster shares a common ancestor with the actinobacterial gene clusters, which agrees with the observation that the Cfa7 homologue from Azospirillum sp. B510 is the only homologue outside the Actinobacteria that is predicted to harbor an ER domain (Fig. 4C). How this ER domain arose in the Cfa7 homologues in these organisms is unclear, but it could be due either to an insertion event in a common ancestor or to a deletion event that resulted in loss of the domain in the other Cfa7 homologues. Overall, our analyses provide evidence that HGT has played a critical role in the propagation of the CFA biosynthetic genes among the different bacteria, with intragenic rearrangement, insertion, and/or deletion events likely contributing to the divergence of the gene clusters within these bacteria (32). It is noteworthy that the biosynthetic gene cluster for coronamic acid, which is found in the COR phytotoxin (Fig. 1A), is present only in the genome sequences of a subset of known or predicted CFA producers (Pseudomonas spp. and Azospirillum sp. B510), suggesting that the acquisition of this cluster likely occurred independently from that of the CFA biosynthetic genes.
A final revelation arising from this study is the observation that predicted CFA biosynthetic genes not only are found in plant-pathogenic bacteria but also appear to be present in bacteria that are not known to be pathogenic (Fig. 4). This may suggest that the production of JA mimics by microbes provides an evolutionary advantage that is not always directly related to host-pathogen interactions. It is known, for example, that JA plays an important role in regulating the induced systemic resistance (ISR), an enhanced state of resistance that provides plants with broad-spectrum protection against pathogens and parasites (33). ISR is elicited by exposure to certain nonpathogenic bacteria, and this primes the whole plant to mount a stronger and/or faster response upon pathogen challenge in order to better combat the invading pathogen. It is conceivable that the production of a JA mimic by a beneficial plant symbiont may help to protect the plant host by ensuring that the host is ready to mount an effective response when it encounters an invading pathogen. It is notable that the Azospirillum sp. strain B510 was originally isolated as a rice endophyte and has been shown to promote plant growth as well as resistance to the fungal and bacterial rice pathogens (34). Whether coronafacoyl phytotoxin production contributes to the beneficial features of this organism remains to be determined.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All strains, plasmids, and cosmids used in this study are listed in Table 1. Escherichia coli strains were routinely cultured at 37°C with shaking (200 rpm) in Difco LB Lennox broth (BD Biosciences), low-salt LB broth (1% [wt/vol] tryptone, 0.5% [wt/vol] yeast extract, 0.25% [wt/vol] NaCl), SOB (35), SOC (New England BioLabs), or LB (Lennox or low-salt) medium with 1.5% (wt/vol) agar (BD Biosciences). When required, the media were supplemented with the following antibiotics at the indicated final concentrations: ampicillin (100 μg ml−1), kanamycin (50 μg ml−1), hygromycin B (100 μg ml−1), chloramphenicol (25 μg ml−1). For hygromycin B, low-salt LB medium was always utilized. S. scabiei strains were routinely cultured at 28°C with shaking (200 rpm) in Trypticase soy broth (TSB; BD Biosciences) or at 25°C with shaking (125 rpm) in soy flour mannitol broth (SFMB; 2% [wt/vol] defatted soy flour, 2% [wt/vol] mannitol). Solid cultures were grown on the International Streptomyces Project Medium 4 (ISP-4; BD Biosciences), soy flour mannitol agar (SFMA) (36), Difco nutrient agar (NA; BD Biosciences), or potato mash agar (PMA) (11). When required, the media were supplemented with the following antibiotics at the indicated final concentrations: hygromycin B (50 μg ml−1), apramycin (50 μg ml−1), nalidixic acid (50 μg ml−1), kanamycin (50 μg ml−1), thiostrepton (25 μg ml−1).
TABLE 1.
Bacterial strains, cosmids, and plasmids used in this study
| Strain, plasmid, or cosmid | Description | Antibiotic resistancea | Reference(s) or source |
|---|---|---|---|
| Escherichia coli strains | |||
| BW25113/pIJ790 strain | Host for λRED-mediated PCR targeting system | Camr | 37, 46 |
| ET12567/pUZ8002 strain | Nonmethylating host (lacking genes dam, dcm, and hsdM); carries the pUZ8002 plasmid that encodes the machinery for the conjugal transfer of DNA into Streptomyces | Camr, Tetr, Kanr | 36, 47 |
| Streptomyces scabiei strains | |||
| ΔtxtA/pRLDB51-1 strain | Strain containing a deletion of the txtA gene and carrying the cfaR (scab79591) overexpression plasmid pRLDB51-1 | Thior, Aprar | 14 |
| ΔtxtA Δcfa6 strain | Strain containing a deletion of the txtA and cfa6 genes | Hygr, Aprar | 14 |
| ΔCYP107AK1 strain | ΔtxtA/pRLDB51-1 derivative containing a deletion of the CYP107AK1 (scab79691) gene | Thior, Aprar, Hygr | This study |
| Plasmid or cosmid | |||
| pIJ10700 | Template for PCR amplification of the hyg-oriT cassette used for PCR targeting | Hygr | 46 |
| Cosmid 1770 | SuperCos1 derivative containing the S. scabiei CFA biosynthetic gene cluster | Ampr, Kanr | 14 |
| 1770/ΔCYP107AK1 | Cosmid 1770 derivative in which the CYP107AK1 gene was replaced with the hyg-oriT disruption cassette | Ampr, Kanr, Hygr | This study |
Camr, Tetr, Kanr, Thior, Aprar, Hygr, and Ampr indicate chloramphenicol, tetracycline, kanamycin, thiostrepton, apramycin, hygromycin B, and ampicillin resistance, respectively.
Construction of the S. scabiei ΔCYP107AK1 gene deletion mutant.
Deletion of the CYP107AK1 gene in the S. scabiei ΔtxtA/pRLDB51-1 strain was accomplished using a λRED-mediated PCR targeting system (37). A cassette (hyg-oriT) containing the hygromycin resistance gene and origin of transfer sequence was PCR amplified using pIJ10700 (Table 1) as the template and the primers JS3 (5′-GTGGTGCTGGGCGCGGAGTTCGTGCGGAATCCGCATGAGATTCCGGGGATCCGTCGACC-3′) and YL7 (5′-CGCTGTGCCGTCCGGACGGGCAGGGACCGCAGCCCGTGGTGTAGGCTGGAGCTGCTTC-3′), which contain 39-nucleotide homology extensions for targeting of the CYP107AK1 gene. Following gel purification, the PCR product was electroporated into E. coli strain BW25113 containing the λRED expression plasmid pIJ790 and cosmid 1770, which harbors the S. scabiei CFA biosynthetic gene cluster (Table 1). Mutant cosmids in which the CYP107AK1 gene was replaced by the hyg-oriT cassette were isolated and verified by PCR using the primers YL8 (5′-GGGACAAGGAGTGCGGAGCC-3′) and YL9 (5′-CATCGGGTTTCCCCGCGCC-3′) (data not shown). A single mutant cosmid (1770/ΔCYP107AK1) was subsequently introduced into E. coli strain ET12567 containing pUZ8002 (Table 1) before being transferred to S. scabiei ΔtxtA/pRLDB51-1 via intergenic conjugation (36). Hygromycin-resistant exconjugants that arose were screened for kanamycin sensitivity, and PCR was used to confirm the resulting mutant isolates using the YL8 and YL9 primers (data not shown).
Analysis of N-coronafacoyl phytotoxin production.
Triplicate cultures of S. scabiei strains were grown in 5 ml of SFMB for 7 days in 6-well tissue culture plates, after which the culture supernatants were harvested and extracted with organic solvent as described previously (11). Detection of N-coronafacoyl phytotoxins in the extracts was by RP-HPLC as described previously (11), except that the separation was achieved using an isocratic mobile phase consisting of 50:50 acetonitrile-water with 0.1% (vol/vol) formic acid, and the method run time was 6 min with no postrun extension. LC-LRMS analysis of the S. scabiei culture extracts was performed using an Agilent 1100 series HPLC system (Agilent Technologies Inc.) interfaced to a Waters G1946A single quadrupole mass spectrometer (Waters Corporation). Separation and detection of N-coronafacoyl phytotoxins were as described previously (16), except that an isocratic mobile phase consisting of 50:50 acetonitrile-water with 0.1% (vol/vol) formic acid was used with a run time of 10 min.
Purification of N-coronafacoyl biosynthetic intermediates.
Large-scale S. scabiei cultures (1 liter) were prepared, and the supernatants were subjected to chemical extracted as described in reference 16. Purification of the metabolites was by preparative thin-layer chromatography (TLC) and semipreparative RP-HPLC as described before (11), except that an isocratic mobile phase consisting of 50:50 acetonitrile-water with 0.1% formic acid was used for the semipreparative RP-HPLC.
Structural elucidation of the N-coronafacoyl biosynthetic intermediates.
LC-HRMS analysis of the purified biosynthetic intermediate structures was performed on an Accela 1250 system (Thermo Fisher Scientific) hyphenated with an Exactive benchtop Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with an electrospray ionization (ESI) probe and the UltiMate 3000 diode array detector (DAD) (Thermo Scientific Dionex) and evaporative light scatter detector (ELSD) 3300 (Alltech). Reversed-phase separation was achieved on a Hypersil C18 column (2.1 by 50 mm, 1.9 μm; GL Sciences) with a mobile phase consisting of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B), with a linear gradient from 5% B to 100% B in 4.2 min, held for 3.3 min, with the flow rate at 500 μl/min. The mass spectra were acquired in positive ion mode. HRMS/MS of the intermediates was conducted using a Waters Q-Tof Premier mass spectrometer equipped with a lockspray ESI source with a collision energy of 20 eV. One-dimensional (1H and 13C) and 2-dimensional (COSY, HSQC, HMBC, and NOESY) NMR spectra of each metabolite were acquired at the Memorial University Centre for Chemical Analysis, Research and Training (C-CART) using a Bruker Avance II 600 spectrometer (Bruker BioSpin GmbH, Rheinstetten, Germany) operating at 600.33 MHz for proton and 150.96 MHz for carbon and equipped with a 5-mm inverse triple-resonance probe (TXI). The samples were dissolved in CD3OD with 0.1% trifluoroacetic acid-D (TFA-D). Chemical shifts were referenced to trimethylsilane for both 1H and 13C.
Potato tuber slice bioassay.
An in vitro potato tuber slice bioassay was performed using S. scabiei organic culture extracts (25 μl) or purified S. scabiei metabolites (CFA-Ile, ΔCYP107AK1 intermediates i and ii [100 nmol each]) as previously described (11). Solvent alone (HPLC grade methanol [100%] with or without formic acid [final concentration, 0.1%]) was used as a negative control, while pure COR (1 nmol; Sigma-Aldrich Canada) dissolved in 100% methanol was used as a positive control. Each bioassay was repeated at least once.
Bioinformatics analyses.
Homologues of the S. scabiei 87-22 CFA biosynthetic gene cluster were identified by using NCBI BLAST (online version, blastp, default settings) (38) with the S. scabiei Cfa1-7 and Cfl amino acid sequences. Protein domain analyses were performed using the Pfam database (39) or the Structure Based Sequence Analysis of Polyketide Synthase website (SBSPKS) (40) in the case of the modular PKS proteins. Alignment of the cfa1–5, cfa6 (KS domain), cfa7 (KS domain), and cfl nucleotide sequences from the different bacterial species was conducted using MUSCLE (alignment by codons using default parameters) within the Molecular Evolutionary Genetic Analysis (MEGA) software version 7.0.21 (41). Alignments were subsequently concatenated for phylogenetic analysis of the CFA biosynthetic genes. A model selection was performed for each partition within the concatenated alignment using jModelTest version 2.1.9 (42) to identify the best-suited model for distance estimation: cfa1, TPM3+G (42); cfa2, HKY+G (43); cfa3, TPM3uf+I+G (42); cfa4, HKY+I+G (43); cfa5, TPM3uf+I+G (42); cfa6 (KS domain), HKY+G (43); cfa7 (KS domain), HKY+I+G (43); cfl, TPM3uf+I+G (42). Phylogeny was inferred using PhyML (version 3.3.20161110) using the concatenated gene alignments. Alignment of the 16S rRNA gene sequences was conducted using MUSCLE (default parameters) within the MEGA software, and the 16S rRNA phylogenetic tree was constructed using MEGA with the maximum likelihood method and the Tamura-Nei substitution model (44). The significance of the branch order in both trees was tested using the bootstrapping method (45) with 100 replicates for the CFA gene tree and 1,000 replicates for the 16S rRNA gene tree.
Supplementary Material
ACKNOWLEDGMENTS
We thank Celine Schneider and Dave Davidson of the Memorial University C-CART facility for assistance with the NMR structural analyses and the anonymous reviewers for their helpful comments and suggestions.
This work was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant No. 386696-2010 to D.R.D.B. J.T.P.V. was supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant No. 06548-2015 to S.C.D.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01169-17.
REFERENCES
- 1.Glazebrook J. 2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 2.Thaler JS, Humphrey PT, Whiteman NK. 2012. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17:260–270. doi: 10.1016/j.tplants.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111:1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonseca S, Chini A, Hamberg M, Adie B, Porzel A, Kramell R, Miersch O, Wasternack C, Solano R. 2009. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat Chem Biol 5:344–350. doi: 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- 5.Browse J. 2009. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- 6.Xin XF, He SY. 2013. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51:473–498. doi: 10.1146/annurev-phyto-082712-102321. [DOI] [PubMed] [Google Scholar]
- 7.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. 2008. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A 105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng X, Cheng J, Gangadharan A, Mackey D. 2012. The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. Plant Cell 24:4763–4774. doi: 10.1105/tpc.112.105312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bender CL, Alarcon-Chaidez F, Gross DC. 1999. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev 63:266–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell KS, Sebaihia M, Pritchard L, Holden MT, Hyman LJ, Holeva MC, Thomson NR, Bentley SD, Churcher LJ, Mungall K, Atkin R, Bason N, Brooks K, Chillingworth T, Clark K, Doggett J, Fraser A, Hance Z, Hauser H, Jagels K, Moule S, Norbertczak H, Ormond D, Price C, Quail MA, Sanders M, Walker D, Whitehead S, Salmond GP, Birch PR, Parkhill J, Toth IK. 2004. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc Natl Acad Sci U S A 101:11105–11110. doi: 10.1073/pnas.0402424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fyans JK, Altowairish MS, Li Y, Bignell DRD. 2015. Characterization of the coronatine-like phytotoxins produced by the common scab pathogen Streptomyces scabies. Mol Plant Microbe Interact 28:443–454. doi: 10.1094/MPMI-09-14-0255-R. [DOI] [PubMed] [Google Scholar]
- 12.Qi M, Wang D, Bradley CA, Zhao Y. 2011. Genome sequence analyses of Pseudomonas savastanoi pv. glycinea and subtractive hybridization-based comparative genomics with nine pseudomonads. PLoS One 6:e16451. doi: 10.1371/journal.pone.0016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slawiak M, Lojkowska E. 2009. Genes responsible for coronatine synthesis in Pseudomonas syringae present in the genome of soft rot bacteria. Eur J Plant Pathol 124:353–361. doi: 10.1007/s10658-008-9418-7. [DOI] [Google Scholar]
- 14.Bignell DR, Seipke RF, Huguet-Tapia JC, Chambers AH, Parry R, Loria R. 2010. Streptomyces scabies 87-22 contains a coronafacic acid-like biosynthetic cluster that contributes to plant-microbe interactions. Mol Plant Microbe Interact 23:161–175. doi: 10.1094/MPMI-23-2-0161. [DOI] [PubMed] [Google Scholar]
- 15.Cheng Z, Bown L, Tahlan K, Bignell DRD. 2015. Regulation of coronafacoyl phytotoxin production by the PAS-LuxR family regulator CfaR in the common scab pathogen Streptomyces scabies. PLoS One 10:e0122450. doi: 10.1371/journal.pone.0122450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bown L, Altowairish MS, Fyans JK, Bignell DRD. 2016. Production of the Streptomyces scabies coronafacoyl phytotoxins involves a novel biosynthetic pathway with an F420-dependent oxidoreductase and a short-chain dehydrogenase/reductase. Mol Microbiol 101:122–135. doi: 10.1111/mmi.13378. [DOI] [PubMed] [Google Scholar]
- 17.Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. 1996. P450 superfamily: update on new sequences, gene mapping, accession numbers and nomenclature. Pharmacogenetics 6:1–42. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Chan YA, Podevels AM, Kevany BM, Thomas MG. 2009. Biosynthesis of polyketide synthase extender units. Nat Prod Rep 26:90–114. doi: 10.1039/B801658P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwan DH, Sun Y, Schulz F, Hong H, Popovic B, Sim-Stark JC, Haydock SF, Leadlay PF. 2008. Prediction and manipulation of the stereochemistry of enoylreduction in modular polyketide synthases. Chem Biol 15:1231–1240. doi: 10.1016/j.chembiol.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 20.Ziemert N, Jensen PR. 2012. Phylogenetic approaches to natural product structure prediction. Methods Enzymol 517:161–182. doi: 10.1016/B978-0-12-404634-4.00008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munro AW, Girvan HM, Mason AE, Dunford AJ, McLean KJ. 2013. What makes a P450 tick? Trends Biochem Sci 38:140–150. doi: 10.1016/j.tibs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Xue Y, Wilson D, Zhao L, Liu H, Sherman DH. 1998. Hydroxylation of macrolactones YC-17 and narbomycin is mediated by the pikC-encoded cytochrome P450 in Streptomyces venezuelae. Chem Biol 5:661–667. doi: 10.1016/S1074-5521(98)90293-9. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez AM, Olano C, Mendez C, Hutchinson CR, Salas JA. 1995. A cytochrome P450-like gene possibly involved in oleandomycin biosynthesis by Streptomyces antibioticus. FEMS Microbiol Lett 127:117–120. doi: 10.1111/j.1574-6968.1995.tb07459.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Hubbard BK, O'Connor SE, Walsh CT. 2002. Formation of beta-hydroxy histidine in the biosynthesis of nikkomycin antibiotics. Chem Biol 9:103–112. doi: 10.1016/S1074-5521(02)00090-X. [DOI] [PubMed] [Google Scholar]
- 25.Machida K, Arisawa A, Takeda S, Tsuchida T, Aritoku Y, Yoshida M, Ikeda H. 2008. Organization of the biosynthetic gene cluster for the polyketide antitumor macrolide, pladienolide, in Streptomyces platensis Mer-11107. Biosci Biotechnol Biochem 72:2946–2952. doi: 10.1271/bbb.80425. [DOI] [PubMed] [Google Scholar]
- 26.Barry SM, Kers JA, Johnson EG, Song L, Aston PR, Patel B, Krasnoff SB, Crane BR, Gibson DM, Loria R, Challis GL. 2012. Cytochrome P450-catalyzed L-tryptophan nitration in thaxtomin phytotoxin biosynthesis. Nat Chem Biol 8:814–816. doi: 10.1038/nchembio.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Healy FG, Krasnoff SB, Wach M, Gibson DM, Loria R. 2002. Involvement of a cytochrome P450 monooxygenase in thaxtomin A biosynthesis by Streptomyces acidiscabies. J Bacteriol 184:2019–2029. doi: 10.1128/JB.184.7.2019-2029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith S, Tsai SC. 2007. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat Prod Rep 24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatanaka H, Kino T, Miyata S, Inamura N, Kuroda A, Goto T, Tanaka H, Okuhara M. 1988. FR-900520 and FR-900523, novel immunosuppressants isolated from a Streptomyces. II. Fermentation, isolation and physico-chemical and biological characteristics. J Antibiot (Tokyo) 41:1592–1601. [DOI] [PubMed] [Google Scholar]
- 30.Liu H, Reynolds KA. 1999. Role of crotonyl coenzyme A reductase in determining the ratio of polyketides monensin A and monensin B produced by Streptomyces cinnamonensis. J Bacteriol 181:6806–6813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stassi DL, Kakavas SJ, Reynolds KA, Gunawardana G, Swanson S, Zeidner D, Jackson M, Liu H, Buko A, Katz L. 1998. Ethyl-substituted erythromycin derivatives produced by directed metabolic engineering. Proc Natl Acad Sci U S A 95:7305–7309. doi: 10.1073/pnas.95.13.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fischbach MA, Walsh CT, Clardy J. 2008. The evolution of gene collectives: how natural selection drives chemical innovation. Proc Natl Acad Sci U S A 105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pieterse CM, Zamioudis C, Berendsen RL, Weller DM, Van Wees SC, Bakker PA. 2014. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 34.Kaneko T, Minamisawa K, Isawa T, Nakatsukasa H, Mitsui H, Kawaharada Y, Nakamura Y, Watanabe A, Kawashima K, Ono A, Shimizu Y, Takahashi C, Minami C, Fujishiro T, Kohara M, Katoh M, Nakazaki N, Nakayama S, Yamada M, Tabata S, Sato S. 2010. Complete genomic structure of the cultivated rice endophyte Azospirillum sp. B510. DNA Res 17:37–50. doi: 10.1093/dnares/dsp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 36.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 37.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci U S A 100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 39.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. 2008. The Pfam protein families database. Nucleic Acids Res 36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anand S, Prasad MV, Yadav G, Kumar N, Shehara J, Ansari MZ, Mohanty D. 2010. SBSPKS: structure based sequence analysis of polyketide synthases. Nucleic Acids Res 38:W487–W496. doi: 10.1093/nar/gkq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santorum JM, Darriba D, Taboada GL, Posada D. 2014. jmodeltest.org: selection of nucleotide substitution models on the cloud. Bioinformatics 30:1310–1311. doi: 10.1093/bioinformatics/btu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 44.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. [DOI] [PubMed] [Google Scholar]
- 45.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 46.Gust B, Chandra G, Jakimowicz D, Yuqing T, Bruton CJ, Chater KF. 2004. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv Appl Microbiol 54:107–128. doi: 10.1016/S0065-2164(04)54004-2. [DOI] [PubMed] [Google Scholar]
- 47.MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61–68. doi: 10.1016/0378-1119(92)90603-M. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.