ABSTRACT
The oral biofilm is a multispecies community in which antagonism and mutualism coexist among friends and foes to keep an ecological balance of community members. The pioneer colonizers, such as Streptococcus gordonii, produce H2O2 to inhibit the growth of competitors, like the mutans streptococci, as well as strict anaerobic middle and later colonizers of the dental biofilm. Interestingly, Veillonella species, as early colonizers, physically interact (coaggregate) with S. gordonii. A putative catalase gene (catA) is found in most sequenced Veillonella species; however, the function of this gene is unknown. In this study, we characterized the ecological function of catA from Veillonella parvula PK1910 by integrating it into the only transformable strain, Veillonella atypica OK5, which is catA negative. The strain (OK5-catA) became more resistant to H2O2. Further studies demonstrated that the catA gene expression is induced by the addition of H2O2 or coculture with S. gordonii. Mixed-culture experiments further revealed that the transgenic OK5-catA strain not only enhanced the growth of Fusobacterium nucleatum, a strict anaerobic periodontopathogen, under microaerophilic conditions, but it also rescued F. nucleatum from killing by S. gordonii. A potential role of catalase in veillonellae in biofilm ecology and pathogenesis is discussed here.
IMPORTANCE Veillonella species, as early colonizers, can coaggregate with many bacteria, including the initial colonizer Streptococcus gordonii and periodontal pathogen Fusobacterium nucleatum, during various stages of oral biofilm formation. In addition to providing binding sites for many microbes, our previous study also showed that Veillonella produces nutrients for the survival and growth of periodontal pathogens. These findings indicate that Veillonella plays an important “bridging” role in the development of oral biofilms and the ecology of the human oral cavity. In this study, we demonstrated that the reducing activity of Veillonella can rescue the growth of Fusobacterium nucleatum not only under microaerophilic conditions, but also in an environment in which Streptococcus gordonii is present. Thus, this study will provide a new insight for future studies on the mechanisms of human oral biofilm formation and the control of periodontal diseases.
KEYWORDS: Fusobacterium nucleatum, oxidative stress, Streptococcus gordonii, veillonellae, anaerobes, transformable strain
INTRODUCTION
The human oral biofilm is a multispecies community colonized by more than 700 bacterial species, and 200 species may present in each human mouth (1, 2). Formation of the dental biofilm involves a sequential process (3–5): on a newly emerged or professionally cleaned tooth surface, pioneer colonizers consisting mostly of the mitis streptococci (i.e., Streptococcus gordonii and Streptococcus sanguinis) attach to the tooth surface via specific interactions with the acquired pellicle on the surface. Growth and metabolic activity of the pioneer colonizers create an environment conducive to colonization by the bridging species, some of which are species of the Veillonella genus. Growth of the bridging species then modifies the local environment or generates nutrients, such as heme/hemin, facilitating the growth of later colonizers, many of which are periodontopathogens (6). Eventually, through cell growth and coadhesion, a mature biofilm is formed (3, 5). Under normal conditions, this biofilm community keeps an ecological homeostasis; however, environmental perturbation can disrupt this balance, leading to dysbiosis, and then oral diseases, such as dental caries and periodontitis, ensue (7–9). As bridging species play such an important role in oral biofilm development, understanding their interaction with both pioneer and later colonizers would generate a knowledge base leading to developments in disease prevention.
The mitis streptococci, such as S. gordonii, produce abundant hydrogen peroxide via multiple pathways (4, 10–12). Kreth et al. reported that the concentrations of H2O2 generated by S. sanguinis and S. gordonii are sufficient to inhibit the growth of many oral bacteria, such as the cariogenic Streptococcus mutans (13). Interestingly, our previous study demonstrated that adding Veillonella parvula strain PK1910 (formerly Veillonella atypica PK1910 [14]) to the S. gordonii-S. mutans mixed culture could rescue S. mutans from the inhibition of S. gordonii (15), implying that V. parvula PK1910 may employ some strategies to counteract the killing effect of H2O2.
Veillonellae, as some of the most predominant bacteria in oral microbiota (16–18), possess two characteristics that make them some of the most important bridging species in the oral biofilm community. One is the use of lactate, generated mainly by streptococci, as their primary carbon and energy source (19). Thus, it is not surprising that veillonellae might also encounter a high level of streptococcus-produced H2O2. Interestingly, our studies revealed that veillonellae, although anaerobic, have an extremely high capacity to tolerate oxygen stress (P. Zhou, unpublished data). The other characteristic is the ability to coaggregate with numerous initial, early, middle, and later colonizers (20–23). In vitro, Veillonella species have been shown to physically coaggregate with streptococci and other periodontal pathogens, such as Fusobacterium nucleatum (20, 23, 24).
F. nucleatum (25), a strict anaerobe and middle colonizer, is often present in the early biofilm community (17). How does this pathogen deal with the high concentration of H2O2 produced by mitis streptococci? It has been shown that fusobacteria play a role in protecting against atmospheric oxygen and hydrogen peroxide in the oral biofilm and even support the growth of Porphyromonas gingivalis under aerated conditions (26, 27). However, compared to fusobacteria, Veillonella species are more tolerant to oxygen stress (P. Zhou, unpublished data). Thus, we hypothesized that veillonellae might be able to protect downstream strict anaerobes, such as F. nucleatum, against oxygen stress. In this study, we identified a functional catalase in V. parvula PK1910 and hypothesized that the catalase activity of this strain may play a crucial role in protecting F. nucleatum from oxygen stress and in turn facilitate its persistence in early biofilm community. Recently, we have successfully developed a genetic transformation system in a clinical isolate of Veillonella atypica OK5 (28, 29), which made it possible to test the effect of Veillonella catalase in biofilm ecology. Due to the absence of catalase in the V. atypica OK5 strain, in this study, we transferred the catalase gene from V. parvula PK1910 to OK5 and tested its effect on Veillonella resistance to H2O2, as well as on the growth of F. nucleatum in the presence of S. gordonii under microaerophilic conditions.
RESULTS
V. parvula PK1910 produces active catalase.
In our previous study, we demonstrated that the presence of V. parvula PK1910 in a S. mutans-S. gordonii mixed culture could rescue S. mutans from inhibition by S. gordonii (15). To determine the cause of this effect, we first analyzed catalase activity of PK1910, because in nature, many bacteria have evolved to produce catalase as a protective strategy against H2O2 produced by hosts or neighboring microbial species (30, 31). To detect catalase activity, V. parvula PK1910 cells in mid-log phase were pelleted by centrifugation, and H2O2 was dropped on top of the cell pellet. Bubbling was observed immediately after H2O2 addition, indicating possible catalase activity (data not shown).
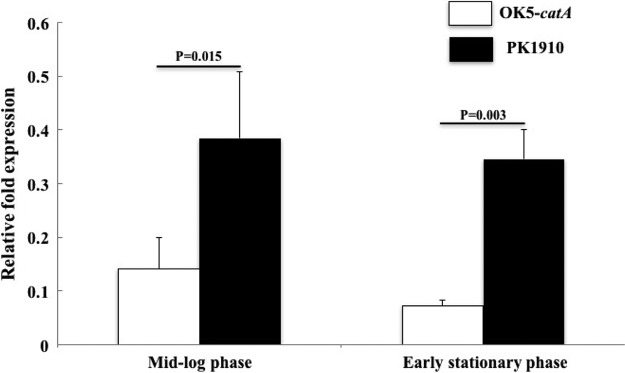
Additional proof of the existence of catalase production came from analyzing the draft genome sequence of PK1910 (P. Zhou and F. Qi, unpublished data). The predicted functions of available genes in all contigs revealed a single copy of a putative catalase gene (catA). A quantitative PCR (qPCR) assay using PCR primers designed from the predicted sequence indicated that gene-specific mRNA was present in V. parvula PK1910 (Fig. 1). Having confirmed the presence and transcription of a catA gene, we measured enzyme activity produced by V. parvula PK1910 in liquid culture. Using mid-log cells grown in brain heart infusion supplemented with 0.6% sodium lactate (BHIL) broth, approximately 30 U/ml catalase enzyme activity was detected (data not shown).
FIG 1.
Detection of catA expression in OK5-catA and PK1910 using qPCR assay. Expression of the catA gene was normalized to the expression of the reference gene gyrA, and relative fold expression was calculated using the formula 2−ΔCT (catA-gyrA). All experiments were repeated at least 3 times, and the results are shown as mean ± standard deviation (SD).
V. parvula catA is functional in V. atypica OK5.
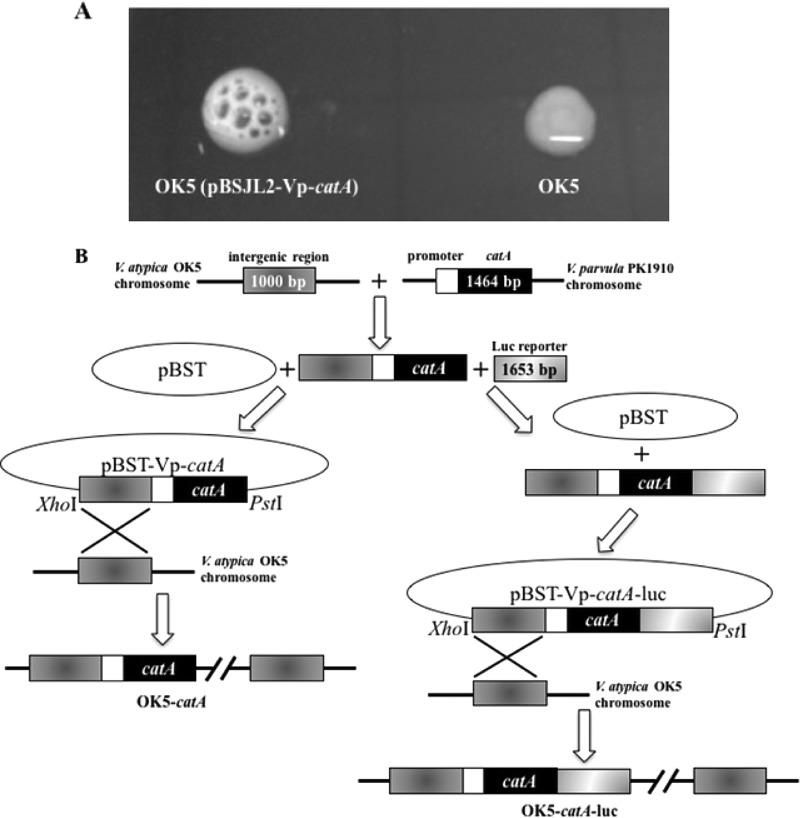
To study the function of catA in Veillonella species, we chose the only transformable strain, V. atypica OK5, as a model (28, 32), because a catalase gene does not exist in the V. atypica OK5 genome (33), and transformation is not feasible with V. parvula PK1910. Using the shuttle vector pBSJL2 for V. atypica (29), we constructed plasmid pBSJL2-Vp-catA, carrying the catA gene under the control of its native promoter, and transformed it into OK5. Transformants showed strong catalase activity when tested with externally added H2O2 (Fig. 2A), suggesting that the V. parvula catA is functional in V. atypica.
FIG 2.
(A) Catalase test for OK5-pBSJL2-catA (left) and OK5 wild type. (B) Construction of OK5-pBSJL2-catA, catA-integrated, and catA-luc-integrated strains in V. atypica.
Having confirmed the function of catA in V. atypica by the shuttle plasmid pBSJL2-Vp-catA, we then wanted to integrate this gene into the chromosomal DNA of OK5, because putting catA into a multicopy shuttle vector is not suitable when studying gene expression and regulation. We chose an intergenic region to integrate the catA gene into the chromosome of V. atypica OK5 (see Materials and Methods and Fig. 2B). Using the previously mentioned H2O2 drop assay, obvious bubble formation was detected for this strain, but the effect was not as obvious as when the same assay was performed using the strain containing the shuttle plasmid pBSJL2-Vp-catA (data not shown). This is likely due to different copy numbers of catA between the two strains. To test if this strain, named OK5-catA, can be a surrogate for V. parvula PK1910 to study the function of catalase in the Veillonella genus, a qPCR assay was utilized to compare catA expression in OK5-catA and V. parvula PK1910. The data showed that the expressive pattern was similar in the two strains at mid-log- and early stationary phases, although the expression of catA in OK5-catA was lower than that in V. parvula PK1910 (Fig. 1). Thus, OK5-catA was used in subsequent studies.
OK5-catA was more tolerant to H2O2 than its parental strain.
To determine the sensitivities of OK5, OK5-catA, and PK1910 to H2O2, the MIC was measured using hydrogen peroxide challenge assays. All three Veillonella strains were treated as described in Materials and Methods. As expected, the strain most susceptible to H2O2 inhibition was the OK5 wild type (MIC, 15.63 μM). Similar to PK1910 (MIC, 31.25 μM), OK5-catA (MIC, 31.25 μM) showed more tolerance to hydrogen peroxide, thus suggesting that catalase might be an important factor contributing to the difference in H2O2 sensitivities between V. atypica and V. parvula.
Expression of catA was induced by chemical and S. gordonii-produced H2O2.
As the function of catalase is to eliminate H2O2 in cells, we speculated that the expression of the catA gene in Veillonella species might be inducible by H2O2. To test this hypothesis, a luciferase reporter was transcriptionally fused with catA and then integrated into the same intergenic region as in OK5-catA (see Materials and Methods and Fig. 2B). The resulting strain, designated OK5-catA-luc, displayed the same level of catalase activity as in OK5-catA (data not shown), suggesting that luciferase gene fusion did not affect catA gene expression.
As expected, the expression of the catA-luc gene was low when grown anaerobically (Fig. 3) and obviously less than that with exposure to oxygen. To test if H2O2 could induce the catA gene, strain OK5-catA-luc was allowed to preincubate with various concentrations of H2O2 in the range of 0 to 2,400 μM for 30 min at room temperature. The highest increase in catA-luc expression was observed at 300 μM H2O2, indicating that the expression of the catA gene in OK5 could be induced by a low concentration of H2O2. In contrast, catA-luc expression was unable to be detected at a high concentration of H2O2 (more than 1,200 μM), perhaps due to Veillonella dying under this detrimental condition (Fig. 4).
FIG 3.
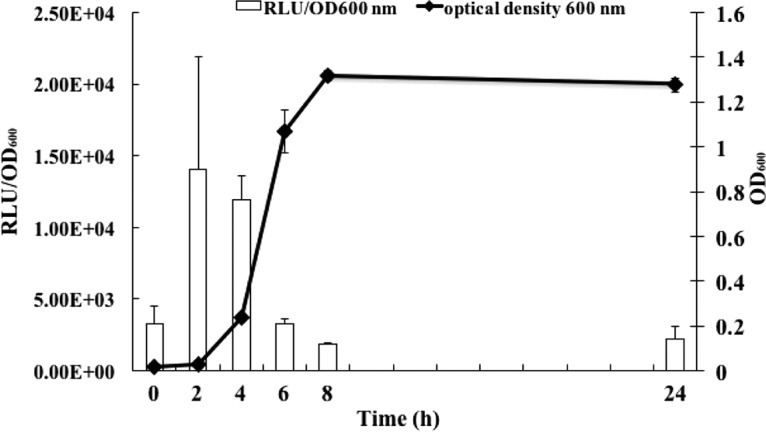
Expression of the catA gene in OK5-catA-luc. Cells were grown in BHIL anaerobically at 37°C, and samples were taken at 2-h intervals. Luciferase activity was measured and normalized to cell density (OD600), shown as RLU/OD600. The results are shown as the means ± SD of the results from three independent experiments.
FIG 4.
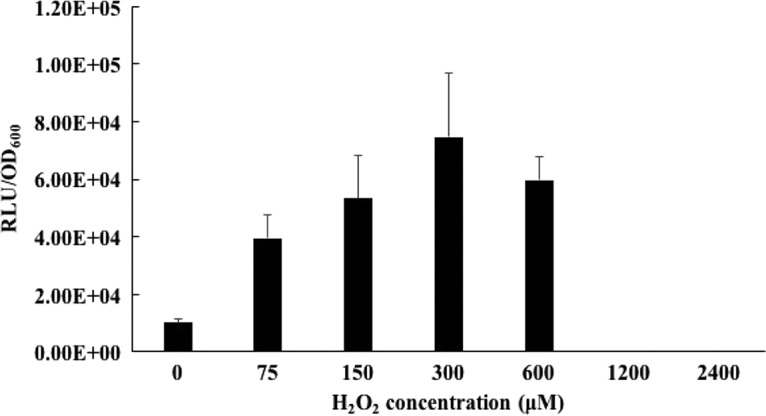
Expression of the catA gene by H2O2 treatment. OK5-catA-luc was precultured overnight in BHIL medium, centrifuged to remove supernatant, and resuspended in fresh BHIL broth to an OD600 of ∼1.0. The suspension was diluted 1:20 into fresh BHIL and the culture grown to mid-log phase (OD600, 0.7 to ∼0.8). The cells were treated with different concentrations of H2O2 for 30 min at room temperature, the cells were centrifuged to remove the supernatants, and the cell pellets were resuspended into fresh BHIL broth. Luciferase activity was measured and normalized to the OD600, expressed as RLU/OD600. The culture without H2O2 treatment was used as a negative control. The results are shown as the means ± SD of the results from three independent experiments.
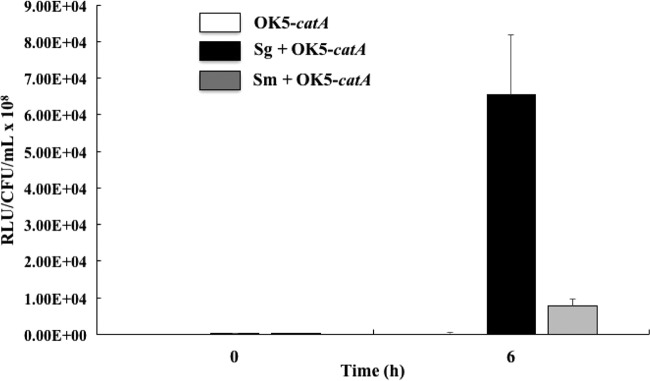
Having demonstrated that catA-luc gene expression was induced by chemical H2O2, we reasoned that coculture with S. gordonii, which generates plentiful H2O2 via different pathways, should also upregulate the expression of catA-luc gene. To test this, OK5-catA-luc was cocultured with S. gordonii under microaerophilic conditions, samples were taken at 0 h and 6 h after coincubation, and luciferase activity was measured. A mixed culture with S. mutans was used as a control because S. mutans was known not to produce H2O2. At 6 h postincubation, luciferase activity in the mixed culture with S. gordonii increased 180-fold compared with OK5-catA-luc monoculture and 8.5-fold compared with mixed culture with S. mutans (Fig. 5), suggesting that the expression of catA was induced by coculturing with S. gordonii.
FIG 5.
Expression of the catA gene in the presence of different streptococcal species. Luciferase activity was measured at 0 h and 6 h. The CFU per milliliter of OK5-catA was obtained by plate counting on the BHIL plate supplied with tetracycline (2.5 μg/ml). Luciferase activity was expressed as RLU/CFU/ml × 108. The results are shown as the means ± SD of the results from three independent experiments. Sg, S. gordonii; Sm, S. mutans.
OK5-catA rescued the growth of F. nucleatum under microaerophilic conditions.
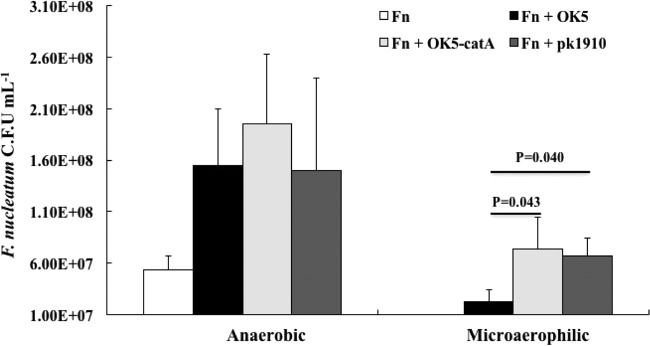
F. nucleatum, known as a strict anaerobe, can also be found in early biofilm plaque, which is primarily an aerobic or microaerophilic niche (17). How does this strict anaerobe deal with oxygen stress? Based on our previous observation that Veillonella could rescue S. mutans from the inhibitory effect of S. gordonii (15), we speculate that Veillonella might exert a crucial impact that facilitates the growth of F. nucleatum under hyperoxic conditions. To test this assumption, we first measured catA expression in OK5-catA-luc under different conditions, and a noticeable increased expression was observed when the strain was grown in a candle jar compared to growth in the anaerobic chamber (data not shown). Next, we carried out V. atypica-F. nucleatum coculture assays, as described in Materials and Methods. As shown in Fig. 6, monoculture of F. nucleatum grew to 5.30 × 107 CFU/ml, and a 3-fold increase in F. nucleatum CFU/ml was observed when grown in the presence of each of the three tested Veillonella strains/species under anaerobic conditions. In contrast, F. nucleatum single culture was unable to grow under microaerophilic conditions, but as expected, the presence of OK5-catA or PK1910 rescued F. nucleatum growth. Interestingly, the OK5 wild-type strain also enhanced the survival rate of F. nucleatum, although not as well as the catalase-positive strains (Fig. 6), suggesting the catalase in Veillonella plays a protective role for F. nucleatum against oxidative stress.
FIG 6.
Growth of F. nucleatum in monoculture and cocultures with OK5 wild type, OK5-catA, and PK1910 under anaerobic (left) and microaerophilic (right) conditions. The CFU per milliliter of F. nucleatum were obtained by plate counting on blood agar plates. The results are shown as the means ± SD of the results from at least three independent experiments. Fn, F. nucleatum.
OK5-catA protected F. nucleatum from inhibition by S. gordonii.
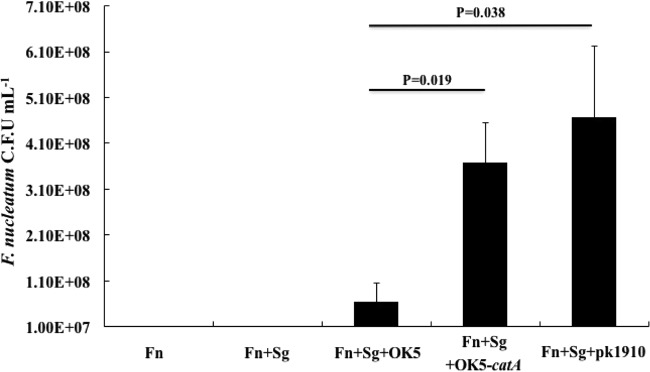
It has been well documented that Veillonella, as an early colonizer, coaggregates with initial, early/middle, and later colonizers, such as S. gordonii and F. nucleatum (14, 23, 24), so this microorganism was believed not only to play a “bridging” role in the formation of oral biofilm community, but it can also optimize the local microniche, for instance, by reducing an oxidative environment, to facilitate the growth of later colonizers. To further confirm that catalase in Veillonella species can protect F. nucleatum from the inhibitory effect of H2O2 generated by initial colonizers, a 3-species mixed culture system, including S. gordonii-V. atypica-F. nucleatum, was used in this study. Under microaerophilic conditions, F. nucleatum failed to grow in either monoculture or as a S. gordonii-F. nucleatum coculture. As expected, the addition of OK5-catA or PK1910 in 3-species cultures supported the growth of F. nucleatum (Fig. 7). This demonstrated that veillonellae utilize catalase to eliminate S. gordonii-produced H2O2 and generate a reducing environment required for the growth of F. nucleatum. Interestingly, the presence of OK5 protected F. nucleatum from killing by S. gordonii (Fig. 7), implying that another strategy might be employed by V. atypica to antagonize oxidative stress.
FIG 7.
Growth of F. nucleatum in monoculture, coculture with S. gordonii, and 3-species culture with different Veillonella strains under microaerophilic conditions. The CFU per milliliter of F. nucleatum were obtained by plate counting on blood agar plates. The results are shown as the means ± SD of the results from at least three independent experiments. Fn, F. nucleatum; Sg, S. gordonii.
DISCUSSION
All organisms living in an aerobic environment have to confront oxidative stress. Oxygen molecules, which are relatively inert, can be easily converted into reactive oxygen species (ROS), such as hydrogen peroxide (H2O2), superoxide (O2˙−), and hydroxyl radicals (OH˙−). The damaging effects of ROS on various biological macromolecules are known to be associated with aging, mutagenesis, and tumorigenesis (34, 35). Aerobes generally use two enzymes, superoxide dismutase (SOD) and catalase, to deal with oxidative stress. Superoxide anions are dismuted by SOD to H2O2, which is then decomposed to water and oxygen by catalase (30, 36). The antioxidative mechanisms employed by anaerobes involve the reduction of superoxide or H2O2 using reduced rubredoxin as an electron donor. Superoxide reductase (SOR) reduces superoxide to H2O2, and afterward, rubrerythrin can catalyze the reduction of hydrogen peroxide to water (37–40). In fact, intact H2O2 is relatively harmless to cells, but Fenton reactions, which can be triggered by ferrous or copper, can result in the conversion of H2O2 into hydroxyl radicals, which are extremely deleterious due to its ability to indiscriminately destroy all biological macromolecules (34, 41, 42). Therefore, catalase and rubrerythrin play vital roles in defense against ROS through the elimination of H2O2.
The development of human oral biofilm is a sequential process (3–5). As initial colonizers, streptococci play a vital role in the colonization of early/middle and later colonizers (3, 43); however, the mitis streptococci, such as S. gordonii, produce plentiful H2O2 to inhibit the growth of downstream bacteria, most of which are extremely sensitive to hyperoxic stress. Therefore, how do these anaerobes deal with the high concentration of H2O2 produced by streptococci? By searching the genome database of veillonellae on the Human Oral Microbiome Database (HOMD) (44), we found most Veillonella species, except for all V. atypica strains, to possess a putative catalase. Our previous study showed that V. parvula PK1910 could reduce the growth-inhibitory effect of S. gordonii on S. mutans (15), implying that catalase, encoded by the catA gene in V. parvula PK1910, might play a crucial role not only in the competition between the two streptococcal species but also in supporting the growth of downstream obligate anaerobes. To prove our hypothesis, we used the only transformable strain of Veillonella, V. atypica OK5 (29), to study the function of catalase in the Veillonella genus.
In the present study, we integrated the V. parvula PK1910 catA gene into V. atypica OK5 chromosomal DNA and confirmed that the catA gene expresses functional catalase in V. atypica. Although catA expression in V. atypica is lower than that in original species, expressive patterns in both species are similar, indicating OK5-catA can be a surrogate for studying the function of Veillonella catalase in human oral biofilm.
The oral biofilm plays a crucial role in human health and disease. Numerous epidemiological studies have suggested that homeostasis of the biofilm community contributes to oral health, while dysbiosis of the community gives rise to the development of diseases. Thus, studying the biological and environmental elements that affect the ecological balance of the biofilm has significant implications in disease prevention. As some of the bridging species, veillonellae are biologically significant for the development of the oral biofilm community, because they not only provide “site and food” to oral bacteria, such as streptococci, F. nucleatum, and P. gingivalis via cell-cell coaggregation (6, 22–24), but they can also optimize the microenvironment to ensure the survival of downstream pathogens. For example, we have shown in this study that OK5-catA and V. parvula PK1910 can improve the growth of F. nucleatum under low-oxygen conditions; however, F. nucleatum alone failed to survive under the same condition. Interestingly and unexpectedly, the OK5 wild type supported F. nucleatum growth as well. This is likely due to the antioxidant role of rubrerythrin in V. atypica OK5. Rubrerythrins, encoded by the rbr gene, are nonheme iron proteins that play vital roles in antioxidant defense in anaerobic bacteria and archaea and catalyze the reduction of H2O2 to H2O (45). The genome database shows that the rbr gene exists in all Veillonella species, implying it has an important role for antioxidant defense in these microorganisms. The expression of rbr in OK5 and OK5-catA was measured by qPCR and showed at least 1,000-fold higher in transcript abundance than in PK1910, indicating that rubrerythrin rather than catalase plays a vital role in defense against oxygen stress, especially hydrogen peroxide, in V. atypica species (P. Zhou, unpublished data). Furthermore, the extremely high expression of rbr (P. Zhou, unpublished data) in OK5 can be an explanation for its support with the growth of F. nucleatum in a low-oxygen environment. The synergy of catalase and rubrerythrin should render OK5-catA a higher capability of eliminating and then tolerating to H2O2; thus, this could explain why OK5-catA was better than PK1910 in supporting F. nucleatum growth under microaerophilic conditions (Fig. 6).
When grown anaerobically, F. nucleatum alone can grow and reach to 5.30 × 107 CFU/ml, while both V. atypica and V. parvula can boost the growth of F. nucleatum growth by 3-fold (Fig. 6). This increase in growth output is likely ascribed to the nutrients produced by Veillonella supporting the growth of F. nucleatum. A similar phenomenon was also reported by Periasamy and Kolenbrander (46): Veillonella sp. strain PK1910 increased F. nucleatum biomass in a two-species biofilm using a flow cell system.
Compared to a Veillonella-F. nucleatum coculture assay, the growth of F. nucleatum was better in a 3-species (S. gordonii-Veillonella-F. nucleatum) mixed culture in the presence of Veillonella species/strains (Fig. 6 and 7). There could be two explanations for this phenomenon: (i) S. gordonii can produce lactate, a major nutrient for Veillonella growth, and this increase in growth of Veillonella species might in turn contribute to F. nucleatum blooming; (ii) Veillonella catA and rbr genes can be induced by S. gordonii under low-oxygen conditions (Fig. 5, and P. Zhou, unpublished data), thus establishing a better anaerobic niche for luxuriant growth of F. nucleatum.
In sum, as crucial bridging species, Veillonella spp. not only coaggregate with various bacteria in the oral cavity but can also optimize the microniche to support the growth of late periodontopathogens, such as Fusobacterium species. Thus, this study provided new insights for future studies on the mechanisms of human oral biofilm formation and the control of periodontal diseases.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used or constructed in this study are listed in Table 1. V. atypica OK5 and V. parvula PK1910 were grown in brain heart infusion broth (BHI; autoclave 20 min at 121°C; Difco) supplemented with 0.6% sodium lactate (BHIL), a semichemically defined medium (47) without glucose but supplemented with 0.6% sodium lactate and 0.1% peptone (SCDM; sterilized through a 0.22-μm filter), or on BHIL agar plates. F. nucleatum ATCC 23726 was grown in Columbia broth (CB; autoclave 20 min at 121°C; Difco) supplemented with vitamin K (1.2 μM) and hemin (7.7 μM). S. gordonii DL1 and S. mutans UA140 were grown in BHI broth. All bacterial strains were grown anaerobically (85% N2, 10% CO2, 5% H2) at 37°C. The plasmids were transformed into V. atypica OK5 by using electroporation, as described previously (32). For transformant selection, cells were grown in Todd-Hewitt (TH) broth (autoclave 20 min at 121°C; Difco) with 0.6% sodium lactate (THL) plus tetracycline (2.5 μg · ml−1; Sigma-Aldrich). Escherichia coli DH5α cells were grown in Luria-Bertani (LB; autoclave 20 min at 121°C; Difco) medium with aeration at 37°C. E. coli strains carrying plasmids were grown in LB medium containing tetracycline (10 μg · ml−1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | Cloning strain | |
| V. atypica OK5 | Wild type | 29 |
| OK5-pBSJL2-catA | OK5 transformed by shuttle plasmid pBSJL2-catA; Tcr | This work |
| OK5-catA | OK5 catA-integrated strain; Tcr | This work |
| OK5-catA-luc | OK5 catA-integrated strain with luc reporter; Tcr | This work |
| V. parvula PK1910 | Wild type | 43 |
| S. gordonii DL1 | Wild type | 48 |
| S. mutans UA140 | Wild type | 49 |
| F. nucleatum ATCC 23726 | Wild type | This work |
| Plasmids | ||
| pBST | Suicide vector of V. atypica, the beta-lactamase gene in pBluescript II KS(+) was replaced by tetM cassette; Tcr | 23 |
| pBSJL2 | Shuttle vector of V. atypica, for complementation; Tcr | 29 |
| pBSJL2-Vp-catA | pBSJL2+ catA; Tcr | This work |
| pBST-Vp-catA | See Materials and Methods for catA integration | This work |
| pBST-Vp-catA-luc | See Materials and Methods for catA reporter; Tcr | This work |
Tcr, tetracycline resistance.
General DNA manipulation.
Phusion DNA polymerase, Taq DNA polymerase, restriction enzymes, and T4 DNA ligase were all purchased from New England BioLabs. Phusion DNA polymerase was used for overlapping PCR. Taq DNA polymerase was used for screening clones.
RNA isolation and real-time RT-PCR analysis.
Overnight cultures of Veillonella spp. were centrifuged, adjusted to an optical density at 600 nm (OD600) of ∼1.0, and then diluted 1:100 into fresh BHIL broth. Cultures were incubated anaerobically at 37°C, and 10-ml samples were taken at an OD600 of ∼0.5 and 1.0. Cells were harvested by centrifugation. Total RNA isolation and real-time RT-PCR were performed as previously described (47). The primers used are shown in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Purpose |
|---|---|---|
| gyrA-q-F | CAGAAGCAGGTTCCCGTAACTC | Reference gene for qPCR |
| gyrA-q-R | GCCTACCGCAAGTGGCAATA | Reference gene for qPCR |
| catA-q-F | AAACTAACAACAGCCTTTGGTG | qPCR for catA |
| catA-q-R | AGAATTTTACGGCAAAGCCA | qPCR for catA |
| pBSJL2-catA-F | CCGCTCGAGATTGAATGAAAACACCTTG | V. parvula catA amplification |
| pBSJL2-catA-R | AAACTGCAGTTATTTTACATCGCTAAAATC | V. parvula catA amplification |
| catA-in-up-F | CCGCTCGAGCTGTACATTGTACATCGTCA | catA integration |
| catA-in-up-R | CTTGTAGATCACAAGGTGTTTTCATTCAATATTAGGTAGTGTTAATAATATTGTG | catA integration |
| catA-in-dn-R | AAACTGCAGTTATTTTACATCGCTAAAATC | catA integration |
| catA-luc-in-up-R | TCTTCCATATTTACCTCCTCGATTATTTTACATCGCTAAAATCGA | catA-luc integration |
| catA-luc-in-dn-F | TCGATTTTAGCGATGTAAAATAATCGAGGAGGTAAATATGGAAGA | catA-luc integration |
| catA-luc-in-dn-R | TTACTGCAGTTACAATTTGGACTTTCCGCCCTTCTTGGCCTTTA | catA-luc integration |
Construction of the shuttle plasmid with catA gene.
The PCR primers used in this study are listed in Table 2. To construct the shuttle plasmid with the catA gene, the catA coding region with its native promoter was amplified using the chromosome DNA of V. parvula PK1910 as the template and the primer pair pBSJL2-catA-F/pBSJL2-catA-R. The PCR product was double digested with XhoI and PstI and then ligated with plasmid pBSJL2 (29), which was digested with the same enzymes. The recombinant plasmid pBSJL2-Vp-catA was confirmed by sequencing. The confirmed plasmid was then transformed into V. atypica OK5 using the established protocol (29). Positive transformants were confirmed by extracting the plasmids and restriction enzyme digestions.
Construction of integrated suicide plasmid for catA gene.
To construct the catA integrated plasmid, a 1,000-bp intergenic region of V. atypica OK5 chromosome DNA was amplified by PCR using the primer pair catA-in-up-F/catA-in-up-R. This PCR amplicon and plasmid pBSJL2-Vp-catA were used as the templates, and overlapping PCR was implemented to create a catA integrated cassette using the primer pair catA-in-up-F/catA-in-dn-R. The PCR product was double digested with XhoI and PstI and then ligated with plasmid pBST (23), which was digested with the same enzymes. The recombinant plasmid pBST-Vp-catA was confirmed by sequencing.
Construction of integrated suicide plasmid for luciferase reporter of catA gene.
To construct the integrated plasmid of catA-luc, the catA integrated cassette and luciferase region were amplified by PCR using plasmids pBST-Vp-catA and pFW5-luc as the templates and primer pairs catA-in-up-F/catA-luc-in-up-R and catA-luc-in-dn-F/catA-luc-in-dn-R, respectively. The two amplicons were then ligated by overlapping PCR to create a catA-luc integrated cassette using the primer pair catA-in-up-F/catA-luc-in-dn-R. The PCR product was double digested with XhoI and PstI and then ligated with plasmid pBST (23), which was digested with the same enzymes. The recombinant plasmid pBST-Vp-catA-luc was confirmed by sequencing.
Generation of the catA and catA-luc integrated strains.
The confirmed plasmids pBST-Vp-catA and pBST-Vp-catA-luc were transformed into V. atypica OK5 using the established protocol (29). The resulting transformants were selected on tetracycline plates and confirmed by PCR and sequencing.
H2O2 tolerance assays of OK5-catA.
A 4,000 μM H2O2 solution in BHIL was prepared, and 100 μl of BHIL medium was added into each well of a 96-well plate (Falcon). Next, 100 μl of H2O2 solution (4,000 μM) was added into the first well, and then different concentrations of H2O2 were prepared by 2-fold serial dilution. As a control, the last well had no H2O2. The overnight monocultures of OK5, OK5-catA, and PK1910 were centrifuged and resuspended with fresh BHIL medium to an OD600 of ∼1.0. The optical density at 600 nm was measured with a spectrophotometer (SmartSpec 3000; Bio-Rad). Resuspended bacterial cultures were diluted 1:500 into fresh BHIL medium, and then 100-μl cultures were added into the H2O2 solution in 96-well plate (final dilution ratio is 1:1,000). The plates were incubated in the anaerobic chamber at 37°C for 24 h. The MIC to inhibit the growth of Veillonella strains was determined. The experiments were performed three times.
Luciferase assays in OK5-catA-luc and streptococcus mixed cultures.
Overnight cultures of the OK5-catA-luc reporter strain, S. gordonii, and S. mutans were centrifuged and resuspended with fresh BHI broth to an OD600 of ∼1.0. As a control, resuspended OK5-catA-luc culture was diluted 1:100 into fresh BHIL. For coculture tests, resuspended OK5-catA-luc culture was diluted 1:100 into fresh BHI medium, and then S. gordonii or S. mutans was diluted into the same culture with streptococci–OK5-catA-luc at a ratio of 5:1. All cultures were grown in a candle jar, and the luciferase activity of OK5-catA-luc was measured at 6 h. Luciferase assays were performed by adding 25 μl of 1 mM d-luciferin (Sigma-Aldrich) solution (suspended in 0.1 M citrate buffer [pH 6.0]) into 100-μl samples, and luciferase activities were measured using a TD 20/20 luminometer (Turner Biosystems, Sunnyvale, CA). For mixed-species cultures, the CFU per milliliter of OK5-catA-luc were obtained by plate counting on the BHIL plate supplied with tetracycline (2.5 μg/ml). Luciferase activity was expressed in relative light units (RLU)/CFU/ml × 108.
Assay of OK5-catA supporting the growth of F. nucleatum.
Overnight, the cultures of the OK5 wild-type strain, OK5-catA, V. parvula PK1910, and F. nucleatum ATCC 23726 were centrifuged to remove the supernatants, and the cell pellets were washed by SCDM twice and then resuspended in fresh SCDM supplemented with vitamin K (1.2 μM) and hemin (7.7 μM). The cell suspensions were adjusted to an OD600 of ∼1.0, F. nucleatum was 1:50 diluted into fresh SCDM supplemented with vitamin K (1.2 μM), hemin (7.7 μM) and peptone (0.1%), and then Veillonella species were mixed with F. nucleatum at a ratio of 2:1. The monocultures of species were included as a control, and 0.6% lactate was added to the cocultures to support Veillonella growth. All cultures were incubated in an anaerobic chamber or candle jar at 37°C for 24 h. Ten-microliter aliquots of all cultures were spotted on blood agar plates (Hardy Diagnostics). The plates were incubated in the anaerobic chamber at 37°C for 72 h. F. nucleatum colonies were obviously distinguished from Veillonella spp. by colony morphology and color. The number of CFU was determined from at least three independent experiments.
Assay of OK5-catA supporting F. nucleatum growth in three-species culture.
The overnight cultures of the OK5 wild-type strain, OK5-catA, V. parvula PK1910, F. nucleatum ATCC 23726, and S. gordonii were centrifuged to remove the supernatants, and the cell pellets were washed by SCDM twice and then resuspended in fresh SCDM supplemented with vitamin K (1.2 μM) and hemin (7.7 μM). The cell suspensions were adjusted to an OD600 of ∼1.0, F. nucleatum was diluted 1:50 into fresh SCDM supplemented with vitamin K (1.2 μM), hemin (7.7 μM), and peptone (0.1%), and then a monoculture of F. nucleatum was included as a control. S. gordonii was mixed with F. nucleatum at a ratio of 5:1, and 0.5% glucose was added to the coculture medium to support the growth of S. gordonii. Next, Veillonella species were mixed with F. nucleatum at a ratio of 2:1, resulting in a final ratio of a 3-species culture (S. gordonii/Veillonella spp./F. nucleatum) of 5:2:1. After 24 h of incubation in a candle jar at 37°C, each 10-μl aliquot of each cultures was spotted on blood agar plates. The plates were incubated in the anaerobic chamber at 37°C for 72 h. F. nucleatum colonies were distinguished from Veillonella and S. gordonii by colony morphology and color. The number of CFU was determined from at least three independent experiments.
Statistical analysis.
The Student's test was used for statistical analyses, and significance was determined at a P value of ≤0.05.
ACKNOWLEDGMENTS
This work was supported by NIH/NIDCR grants R21DE024235 and R15DE019940 to F.Q.
REFERENCES
- 1.Huang R, Li M, Gregory RL. 2011. Bacterial interactions in dental biofilm. Virulence 2:435–444. doi: 10.4161/viru.2.5.16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hajishengallis G, Lamont RJ. 2012. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 27:409–419. doi: 10.1111/j.2041-1014.2012.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolenbrander PE, Palmer RJ Jr, Periasamy S, Jakubovics NS. 2010. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol 8:471–480. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 4.Kolenbrander PE, Palmer RJ Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. 2006. Bacterial interactions and successions during plaque development. Periodontol 2000 42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 5.Diaz PI, Chalmers NI, Rickard AH, Kong C, Milburn CL, Palmer RJ Jr, Kolenbrander PE. 2006. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol 72:2837–2848. doi: 10.1128/AEM.72.4.2837-2848.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou P, Li X, Qi F. 2016. Identification and characterization of a heme biosynthesis locus in Veillonella. Microbiology 162:1735–1743. doi: 10.1099/mic.0.000366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsh PD. 2006. Dental plaque as a biofilm and a microbial community—implications for health and disease. BMC Oral Health 6(Suppl 1):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsh PD. 1999. Microbiologic aspects of dental plaque and dental caries. Dent Clin North Am 43:599–614, v–vi. [PubMed] [Google Scholar]
- 9.Marsh PD, Bradshaw DJ. 1995. Dental plaque as a biofilm. J Ind Microbiol 15:169–175. doi: 10.1007/BF01569822. [DOI] [PubMed] [Google Scholar]
- 10.Okahashi N, Nakata M, Sumitomo T, Terao Y, Kawabata S. 2013. Hydrogen peroxide produced by oral streptococci induces macrophage cell death. PLoS One 8:e62563. doi: 10.1371/journal.pone.0062563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Ge X, Dou Y, Wang X, Patel JR, Xu P. 2011. Identification of hydrogen peroxide production-related genes in Streptococcus sanguinis and their functional relationship with pyruvate oxidase. Microbiology 157:13–20. doi: 10.1099/mic.0.039669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coykendall AL. 1989. Classification and identification of the viridans streptococci. Clin Microbiol Rev 2:315–328. doi: 10.1128/CMR.2.3.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreth J, Zhang Y, Herzberg MC. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol 190:4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes CV, Roseberry CA, Kolenbrander PE. 1990. Isolation and characterization of coaggregation-defective mutants of Veillonella atypica. Arch Oral Biol 35(Suppl):123S–125S. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Wu C, Huang IH, Merritt J, Qi F. 2011. Differential response of Streptococcus mutans towards friend and foe in mixed-species cultures. Microbiology 157:2433–2444. doi: 10.1099/mic.0.048314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, Leys EJ, Paster BJ. 2008. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol 46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. 2005. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol 43:5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker MR, Paster BJ, Leys EJ, Moeschberger ML, Kenyon SG, Galvin JL, Boches SK, Dewhirst FE, Griffen AL. 2002. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol 40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rogosa M. 1964. The genus Veillonella. I. General cultural, ecological, and biochemical considerations. J Bacteriol 87:162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes CV, Kolenbrander PE, Andersen RN, Moore LV. 1988. Coaggregation properties of human oral Veillonella spp.: relationship to colonization site and oral ecology. Appl Environ Microbiol 54:1957–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chalmers NI, Palmer RJ Jr, Cisar JO, Kolenbrander PE. 2008. Characterization of a Streptococcus sp.-Veillonella sp. community micromanipulated from dental plaque. J Bacteriol 190:8145–8154. doi: 10.1128/JB.00983-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Periasamy S, Kolenbrander PE. 2010. Central role of the early colonizer Veillonella sp. in establishing multispecies biofilm communities with initial, middle, and late colonizers of enamel. J Bacteriol 192:2965–2972. doi: 10.1128/JB.01631-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P, Liu J, Merritt J, Qi F. 2015. A YadA-like autotransporter, Hag1 in Veillonella atypica is a multivalent hemagglutinin involved in adherence to oral streptococci, Porphyromonas gingivalis, and human oral buccal cells. Mol Oral Microbiol 30:269–279. doi: 10.1111/omi.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolenbrander PE, Andersen RN, Moore LV. 1989. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect Immun 57:3194–3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Signat B, Roques C, Poulet P, Duffaut D. 2011. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol 13:25–36. [PubMed] [Google Scholar]
- 26.Diaz PI, Zilm PS, Rogers AH. 2002. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology 148:467–472. doi: 10.1099/00221287-148-2-467. [DOI] [PubMed] [Google Scholar]
- 27.Kraus FW, Nickerson JF, Perry WI, Walker AP. 1957. Peroxide and peroxidogenic bacteria in human saliva. J Bacteriol 73:727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou P, Li X, Qi F. 2015. Establishment of a counter-selectable markerless mutagenesis system in Veillonella atypica. J Microbiol Methods 112:70–72. doi: 10.1016/j.mimet.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Xie Z, Merritt J, Qi F. 2012. Establishment of a tractable genetic transformation system in Veillonella spp. Appl Environ Microbiol 78:3488–3491. doi: 10.1128/AEM.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spolarics Z, Wu JX. 1997. Role of glutathione and catalase in H2O2 detoxification in LPS-activated hepatic endothelial and Kupffer cells. Am J Physiol 273:G1304–G1311. [DOI] [PubMed] [Google Scholar]
- 31.Das D, Bishayi B. 2009. Staphylococcal catalase protects intracellularly survived bacteria by destroying H2O2 produced by the murine peritoneal macrophages. Microb Pathog 47:57–67. doi: 10.1016/j.micpath.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Merritt J, Qi F. 2011. Genetic transformation of Veillonella parvula. FEMS Microbiol Lett 322:138–144. doi: 10.1111/j.1574-6968.2011.02344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou P, Xie G, Li X, Liu J, Qi F. 2017. Complete genome sequence of Veillonella atypica OK5, the first transformable strain in the species. Genome Announc 5(22):e00391-17. doi: 10.1128/genomeA.00391-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou P, Liu L, Tong H, Dong X. 2012. Role of operon aaoSo-mutT in antioxidant defense in Streptococcus oligofermentans. PLoS One 7:e38133. doi: 10.1371/journal.pone.0038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farr SB, Kogoma T. 1991. Oxidative stress responses in Escherichia coli and Salmonella Typhimurium. Microbiol Rev 55:561–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yesilkaya H, Kadioglu A, Gingles N, Alexander JE, Mitchell TJ, Andrew PW. 2000. Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect Immun 68:2819–2826. doi: 10.1128/IAI.68.5.2819-2826.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lumppio HL, Shenvi NV, Summers AO, Voordouw G, Kurtz DM Jr. 2001. Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J Bacteriol 183:101–108. doi: 10.1128/JB.183.1.101-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch MC, Kuramitsu HK. 1999. Role of superoxide dismutase activity in the physiology of Porphyromonas gingivalis. Infect Immun 67:3367–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jenney FE Jr, Verhagen MF, Cui X, Adams MW. 1999. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science 286:306–309. doi: 10.1126/science.286.5438.306. [DOI] [PubMed] [Google Scholar]
- 40.Sztukowska M, Bugno M, Potempa J, Travis J, Kurtz DM Jr. 2002. Role of rubrerythrin in the oxidative stress response of Porphyromonas gingivalis. Mol Microbiol 44:479–488. doi: 10.1046/j.1365-2958.2002.02892.x. [DOI] [PubMed] [Google Scholar]
- 41.Barré O, Mourlane F, Solioz M. 2007. Copper induction of lactate oxidase of Lactococcus lactis: a novel metal stress response. J Bacteriol 189:5947–5954. doi: 10.1128/JB.00576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez R, Redman R. 2005. Balancing the generation and elimination of reactive oxygen species. Proc Natl Acad Sci U S A 102:3175–3176. doi: 10.1073/pnas.0500367102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou P, Liu J, Li X, Takahashi Y, Qi F. 2015. The sialic acid binding protein, Hsa, in Streptococcus gordonii DL1 also mediates intergeneric coaggregation with Veillonella species. PLoS One 10:e0143898. doi: 10.1371/journal.pone.0143898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. 2010. The Human Oral Microbiome Database: a Web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coulter ED, Kurtz DM Jr. 2001. A role for rubredoxin in oxidative stress protection in Desulfovibrio vulgaris: catalytic electron transfer to rubrerythrin and two-iron superoxide reductase. Arch Biochem Biophys 394:76–86. doi: 10.1006/abbi.2001.2531. [DOI] [PubMed] [Google Scholar]
- 46.Periasamy S, Kolenbrander PE. 2009. Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleatum and Veillonella species in saliva. Infect Immun 77:3542–3551. doi: 10.1128/IAI.00345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He X, Wu C, Yarbrough D, Sim L, Niu G, Merritt J, Shi W, Qi F. 2008. The cia operon of Streptococcus mutans encodes a unique component required for calcium-mediated autoregulation. Mol Microbiol 70:112–126. doi: 10.1111/j.1365-2958.2008.06390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pakula R, Walczak W. 1963. On the nature of competence of transformable streptococci. J Gen Microbiol 31:125–133. doi: 10.1099/00221287-31-1-125. [DOI] [PubMed] [Google Scholar]
- 49.Loesche WJ. 1986. Role of Streptococcus mutans in human dental decay. Microbiol Rev 50:353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]