ABSTRACT
Influenza A viruses (IAVs) in swine can cause sporadic infections and pandemic outbreaks among humans, but how avian IAV emerges in swine is still unclear. Unlike domestic swine, feral swine are free ranging and have many opportunities for IAV exposure through contacts with various habitats and animals, including migratory waterfowl, a natural reservoir for IAVs. During the period from 2010 to 2013, 8,239 serum samples were collected from feral swine across 35 U.S. states and tested against 45 contemporary antigenic variants of avian, swine, and human IAVs; of these, 406 (4.9%) samples were IAV antibody positive. Among 294 serum samples selected for antigenic characterization, 271 cross-reacted with ≥1 tested virus, whereas the other 23 did not cross-react with any tested virus. Of the 271 IAV-positive samples, 236 cross-reacted with swine IAVs, 1 with avian IAVs, and 16 with avian and swine IAVs, indicating that feral swine had been exposed to both swine and avian IAVs but predominantly to swine IAVs. Our findings suggest that feral swine could potentially be infected with both avian and swine IAVs, generating novel IAVs by hosting and reassorting IAVs from wild birds and domestic swine and facilitating adaptation of avian IAVs to other hosts, including humans, before their spillover. Continued surveillance to monitor the distribution and antigenic diversities of IAVs in feral swine is necessary to increase our understanding of the natural history of IAVs.
IMPORTANCE There are more than 5 million feral swine distributed across at least 35 states in the United States. In contrast to domestic swine, feral swine are free ranging and have unique opportunities for contact with wildlife, livestock, and their habitats. Our serological results indicate that feral swine in the United States have been exposed to influenza A viruses (IAVs) consistent with those found in both domestic swine and wild birds, with the predominant infections consisting of swine-adapted IAVs. Our findings suggest that feral swine have been infected with IAVs at low levels and could serve as hosts for the generation of novel IAVs at the interface of feral swine, wild birds, domestic swine, and humans.
KEYWORDS: feral swine, influenza A virus, influenza surveillance, mixing vessel, seroprevalence, swine influenza virus, United States, avian viruses, domestic swine, wild birds
INTRODUCTION
Influenza A virus (IAV), a negative-stranded RNA virus with 8 genomic segments, can infect a wide range of hosts, including humans, wild birds and domestic poultry, swine, canines, felines, equines, mink, ferrets, sea mammals, and bats. IAVs have been recovered from at least 105 wild bird species of 26 different families (1). Migratory waterfowl, such as species of Anseriformes (e.g., ducks, geese, and swans) and Charadriiformes (e.g., gulls, terns, and waders), are considered the major natural reservoirs of IAVs (2). Sixteen IAV hemagglutinin (HA) (H1 to H16) and nine neuraminidase (NA) (N1 to N9) subtypes have been recovered from migratory waterfowl. The prevalence of IAV infection is up to 30% among wild birds (2), and virus transmission typically occurs via exposure to virus shed in the feces of infected animals (3, 4). It has been conceptually proposed that antigenic evolution in migratory waterfowl could be static (5); this theory is supported by recent studies indicating a lack of antigenic diversity among H3 and H7 IAVs in migratory waterfowl in North America (6, 7).
IAVs in domestic swine are genetically and antigenically diverse. In the past decade, the predominantly circulating domestic swine strains in the United States were IAV subtypes H1N1, H1N2, and H3N2 (8, 9). Subtypes such as H4N6, H2N3, and H3N1 were also identified in North American domestic swine (10–12), but these viruses did not become enzootic. The H1 subtypes circulating in domestic swine form four genetic clades: swH1α (classic H1N1), swH1β (reassortant H1N1-like), swH1γ (H1N2-like), and swH1δ (human-like H1). Clade swH1γ is further divided into subclades swH1γ1 and swH1γ2, and clade swH1δ is subdivided into swH1δ1 (human-like H1N2) and swH1δ2 (human-like H1N1) (13). In addition, the 2009 H1N1 pandemic virus, A(H1N1)pdm09, emerged from a classic H1N1 virus and evolved into a distinct genetic and antigenic lineage (13). There are 4 genetic clusters (I to IV) of the H3N2 subtype of IAVs present in the U.S. swine population (14–16). Cluster IV, currently the predominant IAV cluster circulating among domestic swine, can be further divided into at least 2 antigenic clusters, H3N2-α and H3N2-β (17). Antigenic characterization suggests that these genetically diverse H1 and H3 viruses are antigenically distinct, showing different extents of cross-reactivity in serological assays (17, 18). Influenza surveillance studies in domestic swine from 2009 through 2012 identified the cocirculation of H1N1, H1N2, and H3N2 IAVs, including 6 H1 genetic clades [H1α, H1β, H1γ, H1δ1, H1δ2, and A(H1N1)pdm09] and 2 H3 cluster IV antigenic clusters (H3SIVα and H3SIVβ) (19, 20).
Avian- and human-origin IAVs typically preferentially bind to receptor saccharides containing terminal α2,3-linked sialic acid-galactose (SA2,3Gal) and α2,6-linked sialic acid-galactose (SA2,6Gal), respectively (21, 22). Swine tracheal epithelium expresses both SA2,3Gal and SA2,6Gal receptors (23), and swine are therefore proposed as the intermediate host for avian IAV adaptation and as a “mixing vessel” for generating novel viruses by reassortment between avian-origin and human-origin IAVs (24–26). In addition to avian-origin H2N3 and H4N6 IAVs, which were identified in domestic swine in North America, avian-origin H1N1 (27, 28), H1N2 (29), H3N3 (30), H5N1 (31), H6N6 (32), and H9N2 (33, 34) IAVs were also identified in domestic swine in Eurasia. Among these avian-origin IAVs, only subtypes H1N1 and H3N2 have become enzootic in domestic swine; the other avian-origin IAVs caused only low seroconversion rates and have been transient in domestic swine. Nevertheless, under laboratory conditions, avian-origin IAVs of subtypes H1 to H13 can infect and replicate in swine at various susceptibilities (26).
Feral swine in the United States are domestic swine that escaped from commercial operations or were intentionally released, descendants of Eurasian wild boar introduced for hunting purposes, or hybrids of the two (35). In 2013, an estimated 5 million feral swine were found in at least 35 U.S. states, with both numbers and geographic range increasing. H1N1 and H3N2 IAVs have been recovered from feral swine, and serological surveillance conducted during 2011 and 2012 showed that 9.2% of 1,983 serum samples from feral swine in 31 states were IAV seropositive (17). Similar to domestic swine, feral swine can be infected with IAVs under laboratory conditions (36). Feral swine have opportunities to encounter wild waterfowl by frequenting the same bodies of water, feeding in the same areas, and preying or scavenging on wild waterfowl, which can provide potential for IAV transmission from wild birds to feral swine. Because feral swine are highly mobile, they can also have opportunities to come into contact with IAVs from infected domestic swine, poultry, and even humans via contaminated fomites or aerosol dispersal (37).
Our objective was to conduct a serological survey of feral swine for IAV exposure. Utilizing 8,239 serum samples collected from feral swine in 35 U.S. states between 2010 and 2013, we explored patterns of IAV seropositivity and further characterized seropositive samples' cross-reaction to 45 antigenically diverse prototype IAVs from avian, domestic swine, and human hosts.
RESULTS
IAV exposure in feral swine.
To evaluate the overall seroprevalence of IAVs among feral swine, we used 8,239 serum samples collected across 35 U.S. states from 1 October 2009 to 30 September 2013; this collection period included fiscal year (FY2010) to FY2013 (Fig. 1; Table 1). Serological testing by the IDEXX AI MultiS-Screen antibody (Ab) test suggested that 4.9% (406) of the samples were IAV positive.
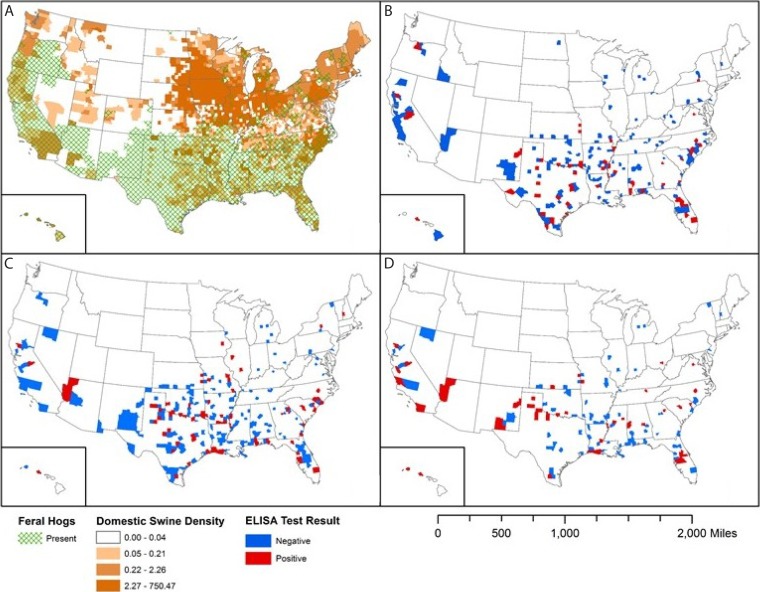
FIG 1.
Geographic distribution of swine and of influenza A virus (IAV)-positive and IAV-negative serum samples collected from feral swine across the United States during fiscal year 2010 (FY2010) to FY2013. (A) Distribution of feral and domestic swine; the domestic swine density values shown in the key are to be multiplied by 1,000,000 (e.g., 0.04 represents 40,000). (B to D) Distributions of IAV ELISA-negative and -positive feral swine serum samples collected in FY2011 (1 October 2010 to 30 September 2011) (B), FY2012 (1 October 2011 to 30 September 2012) (C), and FY2013 (1 October 2012 to 30 September 2013) (D).
TABLE 1.
Summary of feral swine serum samples used to determine the predominant source of feral swine exposure to enzootic influenza A virus in the United States
| Fiscal year samples were collecteda | No. of samples collected | No. (%) of IAV-positive samplesb | No. of positive samples selected for testing by HIc assay |
|---|---|---|---|
| 2010 | 1,818 | 112 (6.2) | 0d |
| 2011 | 2,467 | 95 (3.9) | 95 |
| 2012 | 1,846 | 111 (6.0) | 111 |
| 2013 | 2,108 | 88 (4.2) | 88 |
| Total | 8,239 | 406 (4.9) | 294 |
Fiscal years run from 1 October of one year through 30 September of the next year; the FY is named according to the second year.
Serum samples were considered IAV positive if the sample-to-negative control ratio was ≥0.681by ELISA (IDEXX AI MultiS-Screen Ab test; IDEXX, Westbrook, ME) (36).
HI, hemagglutination inhibition.
Not analyzed to make the assay more cost-effective.
An association was identified between fiscal years and IAV seroprevalence and months and IAV seroprevalence for individual feral swine (P = 0.0002 and P < 0.0001, respectively) but not for groups (P = 0.1717 and P = 0.1184, respectively). An analysis of seroprevalence by month determined that during FY2010 (1 October 2009 to 30 September 2010), the highest and lowest seroprevalence rates were detected among samples collected in April (12.3% [20 of 162 swine]) and September (1.8% [1 of 57 swine]), respectively. This temporal pattern, with a relatively higher IAV seroprevalence in spring and winter, was similar to the patterns seen in FY2011 and FY2012 and to those seen in FY2013 in our previous study (17) (Fig. 2A).
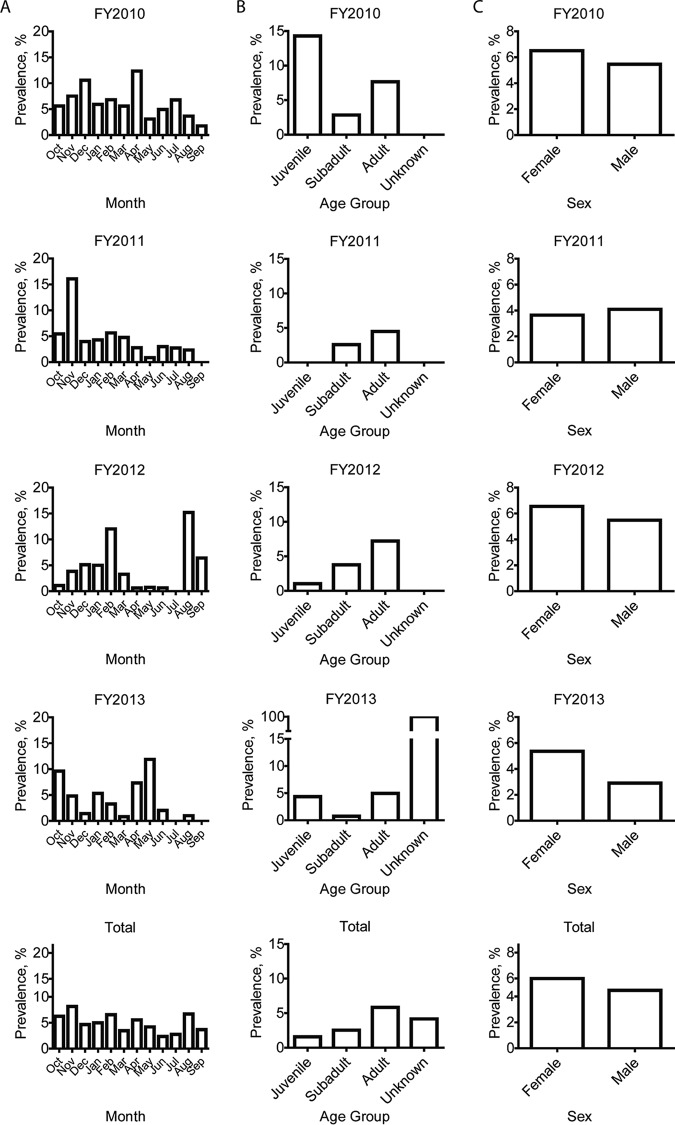
FIG 2.
Epidemiologic analyses of the percentage of influenza A virus-positive feral swine serum samples collected across the United States during FY2010 to FY2013. Samples were determined to be positive by ELISA. (A) Temporal distribution of positive serum samples. (B) Age distribution of feral swine with positive serum samples. (C) Sex distribution of feral swine with positive serum samples. FYs run from 1 October of one year through 30 September of the next year; the FY is named according to the second year.
To understand variables that are associated with IAV seropositivity among feral swine, we analyzed our results by swine age group and sex. IAV seroprevalence was highest among the adult swine (5.8% [350/5,984]) and lowest among juvenile swine (1.6% [3/190]) (Fig. 2B). This pattern was consistent for all years; 2010 had the largest percent positives only due to having the smallest sampling of juveniles (14.29% [1/7 samples]) (see Table S2 in the supplemental material). We identified an association between swine age group and IAV seroprevalence (P < 0.0001) for both individual swine and groups. In addition, IAV seroprevalence was higher among female (5.4% [233/4,333]) than male (4.5% [173/3,871]) swine; this pattern was consistent for all fiscal years except FY2011 (Table S2), in which seroprevalence was higher among male (4.1% [48/1,170]) than female (3.7% [47/1,287]) swine. However, no association between sex and IAV-positive samples for individual swine (P = 0.0583) and groups (P = 0.0665) was identified. Overall, most IAV-positive samples were from adult (86.2%) and female (57.4%) feral swine (Fig. 2C).
Although our data set is comprised of samples collected from 35 states, sample sizes were not evenly distributed because feral swine populations vary widely between states. Consequently, only 23 states had samples which tested IAV positive. The seroprevalence of IAV was highest in North Carolina (16.1% [34/211 samples]) and Texas (10.5% [164/1,561 samples]) (Table S3).
Of 438 counties, 112 (25.56%) yielded IAV-positive samples (Fig. 1). Texas had the most IAV-positive samples (40.4% [164/406 total]). Of 31 counties sampled in Texas, the highest seroprevalence rates were in Dickens County (36.2% [42/116 total]), Hall County (42.3% [22/52 total]), and Freestone County (35.7% [5/14 total]). The number of IAV-positive samples varied by year. In Hall County, for example, 0 of 1 samples were positive in 2010, 0 of 12 were positive in 2011, 6 (30.0%) of 20 were positive in 2012, and 16 (84.2%) of 19 were positive in 2013.
Distinction between swine and avian IAVs.
Hemagglutination inhibition (HI) assays were performed on 294 enzyme-linked immunosorbent assay (ELISA)-positive samples in the most recent three fiscal years (FY2011, FY2012, and FY2013) and tested against 45 IAVs, including HA subtypes H1 to H14 and diverse antigenic clusters of contemporary avian-, swine-, and human-origin IAVs (Table 2). Of note, among the tested strains, there were different extents of cross-reactivity against the reference sera against these viruses although most of these viruses are antigenically different, with ≥4-fold losses in HI activity relative to the homologous titer (Table S1).
TABLE 2.
Cross-reactivities of feral swine serum samples against influenza A viruses in hemagglutination inhibition assays
| Virus | Antigenic group | Source of virusa | No. of seropositive samples (%)b | GMT (LB–HB)c |
|---|---|---|---|---|
| A/swine/Minnesota/02093/2008 | H1N1-α | Domestic swine | 44 (14.67) | 138.85 (40–1,280) |
| A/swine/Minnesota/A01394082/2013 | H1N2-α | Domestic swine | 20 (6.67) | 80 (40–320) |
| A/swine/Nebraska/A01399642/2013 | H1N1-β | Domestic swine | 58 (19.33) | 124.49 (40–1,280) |
| A/swine/Nebraska/A01240348/2011 | H1N1-β | Domestic swine | 57 (19.00) | 146.94 (40–1,280) |
| A/swine/Indiana/13TOSU0832/2013 | H1N1-γ | Domestic swine | 54 (18.00) | 204.19 (40–1,280) |
| A/swine/Indiana/13TOSU1154/2013 | H1N1-γ | Domestic swine | 53 (17.67) | 187.19 (40–1,280) |
| A/swine/Illinois/A01076767/2010 | H1N1-γ2 | Domestic swine | 5 (1.67) | 45.95 (40–80) |
| A/swine/South Dakota/A01349306/2013 | H1N1-γ2 | Domestic swine | 63 (21.00) | 271.32 (40–1,280) |
| A/swine/Iowa/15/2013 | H1δ1 | Domestic swine | 12 (4.00) | 75.51 (40–160) |
| A/swine/Iowa/18/2013 | H1δ2 | Domestic swine | 22 (7.33) | 600.92 (40–1,280) |
| A/swine/Iowa/19/2013 | H1δ2 | Domestic swine | 12 (4.00) | 59.93 (40–320) |
| A/swine/Iowa/7/2013 | H12009p | Domestic swine | 91 (30.33) | 264.51 (40–1,280) |
| A/swine/Iowa/8/2013 | H12009p | Domestic swine | 81 (27.00) | 301.39 (40–1280) |
| A/California/04/2009 | H12009p | Human | 61 (20.33) | 212.57 (40–1,280) |
| A/mallard/Wisconsin/A00751454/2009 | H1N1 | Avian | 13 (4.33) | 75.85 (40–640) |
| A/mallard/Oregon/A0030758/2007 | H2N3 | Avian | 0 | 0 |
| A/swine/Ohio/09SW96/2009 | H3N2-α | Domestic swine | 84 (28.00) | 105.04 (40–1,280) |
| A/swine/Ohio/10SW215/2010 | H3N2-β | Domestic swine | 117 (39.00) | 140.45 (40–1,280) |
| A/swine/Ohio/11SW347/2011 | H3N2-β | Domestic swine | 93 (31.00) | 146.31 (40–1,280) |
| A/swine/Texas/A01104013/2012 | H3N2-β | Feral swine | 123 (41.00) | 64.58 (40–1,280) |
| A/Perth/16/2009 | H3N2 | Human | 177 (59.00) | 109.86 (40–1,280) |
| A/Wisconsin/112/2010 | H3N2v | Human (spillover from domestic swine) | 119 (39.67) | 153.61 (40–1,280) |
| A/Pennsylvania/14/2010 | H3N2v | Human (spillover from domestic swine) | 89 (29.67) | 89.91 (40–640) |
| A/Minnesota/10/2011 | H3N2v | Human (spillover from domestic swine) | 105 (35.00) | 106.96 (40–640) |
| A/Iowa/07/2011 | H3N2v | Human (spillover from domestic swine) | 118 (39.33) | 148.24 (40–1,280) |
| A/Victoria/361/2011 | H3N2 | Human | 195 (65.00) | 154.18 (40–1,280) |
| A/mallard/Wisconsin/A00661712/2009 | H3N2 | Avian | 1 (0.33) | 80 (±0.00) |
| A/blue-winged teal/Colorado/A00170379/2006 | H3N8 | Avian | 0 | 0 |
| A/mallard/Washington/A00714770/2009 | H4N6 | Avian | 0 | 0 |
| A/mallard/Wisconsin/10os3845/2010 | H5N2 | Avian | 0 | 0 |
| A/mallard/Oregon/A00571208/2007 | H6N1 | Avian | 0 | 0 |
| A/mallard/Ohio/648/2002 | H6N2 | Avian | 1 (0.33) | 40 (±0.00) |
| A/bufflehead/Virginia/A00120022/2008 | H7N2 | Avian | 1 (0.33) | 40 (±0.00) |
| A/American black duck/Delaware/A00870108/2010 | H7N3 | Avian | 0 | 0 |
| A/northern shoveler/Illinois/10os3632/2010 | H8N4 | Avian | 0 | 0 |
| A/mallard/Minnesota/10os4670/2010 | H9N2 | Avian | 0 | 0 |
| A/northern shoveler/Arkansas/11os386/2011 | H9N2 | Avian | 0 | 0 |
| A/mallard/South Dakota/A00536114/2007 | H10N7 | Avian | 0 | 0 |
| A/mallard/Illinois/10OS3249/2010 | H11N2 | Avian | 0 | 0 |
| A/mallard/Wisconsin/10OS2889/2010 | H11N9 | Avian | 0 | 0 |
| A/American green-winged teal/Missouri/10OS4622/2010 | H12N4 | Avian | 0 | 0 |
| A/bufflehead/Wisconsin/10OS3204/2010 | H12N5 | Avian | 0 | 0 |
| A/hooded merganser/New Brunswick/03750/2009 | H13N6 | Avian | 0 | 0 |
| A/white-winged scooter/Wisconsin/10OS3922/2010 | H14 | Avian | 0 | 0 |
| A/long-tailed duck/Wisconsin/10OS3912/2010 | H14N6 | Avian | 0 | 0 |
The host from which the virus was isolated.
Serum samples were determined to be positive against a testing virus if the associated HI titer was ≥1:40.
The geometric mean titer (GMT) was calculated for each group of positive samples. HB, high boundary of HI titer; LB, low boundary of HI titer.
Of the 294 feral swine serum samples tested, 271 from 71 counties within 21 states tested positive by HI assay for at least one virus in the reference panel, and 23 samples from 17 counties within 13 states tested negative to all viruses in the reference panel. Of the feral swine samples tested, 38.4% were positive against H1 swine IAVs (113 out of 294), and 53.7% were positive against H3 swine IAVs (158 out of 294). In total, 52 (17.7%) were positive against both H1 and H3 swine IAVs. Of the total 294 ELISA-positive samples, 106 (36.1%) and 233 (79.3%) were also positive against H1 and H3 human IAVs, respectively (Fig. 3). The serological characterization suggests that the swine-origin IAVs in H3α and H3β clusters cross-reacted with the ferret reference sera against H3 seasonal and A(H3N2)v human IAVs, and vice versa (Table S4).
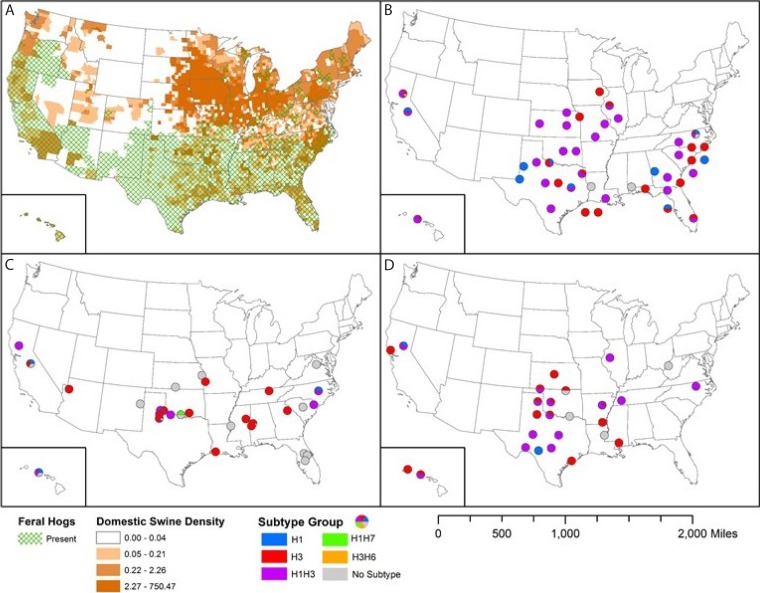
FIG 3.
Geographic distribution of domestic and feral swine across the United States and distribution of influenza A virus (IAV)-positive serum samples (by antigenic characterization) collected from feral swine during FY2011 to FY2013. (A) Distribution of feral and domestic swine; the domestic swine density values shown in the key are to be multiplied by 1,000,000 (e.g., 0.04 represents 40,000). (B to D) Antigenic characterization was determined by hemagglutination inhibition assay for FY2011 (B), FY2012 (C), and FY2013 (D). Dots (pie charts) indicate U.S. counties where samples positive for different IAV subtypes were collected.
Although 271 of the 294 feral swine serum samples tested HI positive against swine and human IAV, only 16 (5.4%) cross-reacted to one of the four avian IAVs included in the reference panel: 13 (4.4%) samples cross-reacted to subtype H1 virus, 1 (0.3%) cross-reacted to H3 virus, 1 (0.3%) cross-reacted to H6 virus, and 1 (0.3%) cross-reacted to H7 virus (Table 2). HI with reference sera showed that H1 avian IAVs cross-reacted with ferret reference sera against H1N1 human IAVs and that avian subtypes H3N2, H6N2, and H7N3 did not cross-react with the reference sera against the tested human and swine IAVs. To confirm the HI results, we performed neutralization assays, which also showed that the 16 feral swine serum samples were indeed cross-reactive against avian IAVs (Table S4).
Distribution of avian IAV-positive feral swine sera.
Sixteen feral swine serum samples from 10 counties in six U.S. states were positive for avian IAV (Fig. 3). The states with the highest number of positive samples were Texas (5 samples), California (4 samples), and Hawaii (4 samples); the remaining samples were from Iowa, Kansas, and Ohio. The surveillance year with the highest number of avian IAV-positive samples was 2013 (7 samples), followed by 2011 (6 samples) and 2012 (3 samples). Thirteen samples were positive for avian subtype H1. Linn County, IA, was the only location with an avian H3 subtype. Colusa County, CA, was the only location with an avian H6 subtype, and Jefferson County, OK, was the only place where an avian H7 subtype was identified.
Factors associated with IAV seroprevalance in feral swine.
Using manual forward variable selection to determine factors that affect the likelihood for IAV exposure in feral swine, we first tested four variables using a multivariable model: the numbers of domestic swine farms, small domestic swine farms, and poultry farms and the human population per state. We considered farms with 1,000 or fewer domestic swine to be small farms. Small farms were of interest due to lower biosecurity and increased chance for contact with feral swine. Individually, the numbers of domestic swine farms, small domestic swine farms, and poultry farms were significant (P < 0.05), but the number of small domestic swine farms had a better fit based on quasilikelihood under the independence model criterion (QIC; 2.29). The number of domestic swine farms was highly correlated with the other variables and were confounded, making each variable nonsignificant. There were more IAV-seropositive samples from states with more small domestic swine farms.
Feral swine are susceptible to H3 and H6 avian IAVs.
For the feral swine that were inoculated with A/mallard/Wisconsin/A00661712/2009 (H3N2), viral shedding was detected from 1 to 6 days postinfection (dpi) from 5 of 8 pigs with viral titers of 0.625 to 2.5 log10 50% egg infective doses (EID50) (Table S5). The results from HI assays demonstrated that one feral swine seroconverted, with HI titers of 1:80 at 21 dpi (Table S6). The two treatment swine that were necropsied at 5 dpi both had viral titers of 1.333 to 3.5 log10 EID50/g/ml in all tissues collected, except turbinate of one feral swine (Table S7). Neither of the control swine had any viral titers in the tissues collected.
For the four feral swine inoculated with H6N2 avian IAV, all pigs had seroconverted at 21 dpi, with titers ranging from 1:20 to 1:80 (Table S8). All control pigs remained seronegative against IAVs during this experiment. Clinical signs were not observed in any of the experimental feral swine.
DISCUSSION
Feral swine are a potential reservoir for infectious pathogens of domestic swine, including IAVs, because bidirectional transmission of pathogens occurs through direct and indirect contact between feral and domestic swine, primarily through backyard farming operations with poor biosecurity (38). A previous study suggested that IAVs circulating among feral swine are antigenically and genetically similar to those circulating among domestic swine (17). Laboratory experiments have demonstrated that swine IAVs can infect feral swine and transmit efficiently among them (36). In addition, feral swine may be exposed to avian IAVs through direct and indirect contact with wild birds via scavenging or preying and by using common sources of water and forage. Our study findings confirm that although feral swine may be exposed to swine or avian IAVs, exposure to swine IAVs is much more common, especially with subtypes H1 and H3. Exposure to avian IAV was rare from our findings reported here, yet there is concern that feral swine could have a mixed IAV infection and generate reassortants between swine and avian IAVs that could ultimately be transmitted to domestic swine or humans.
Our analyses focused only on those serum samples in the most recent 3 years studied (FY2011, FY2012, and FY2013), in which H3N2v emerged in domestic swine and caused outbreaks in humans (17, 39, 40). Our prior studies have demonstrated that H3N2-β-like viruses were predominant in feral swine in FY2012. This study suggested that even though there is no apparent temporal pattern, a relatively higher IAV seroprevalence in the summer could be due to an increased change of contact between domestic and feral swine, e.g., through an increase of pasture time for noncommercial swine. Additionally, the highest seroprevalence in adults can be explained by the larger number of opportunities for IAV exposure than for that of younger swine. Because there was no significant difference in IAV seroprevalence for sex, we can assume that there was equal opportunity for IAV exposure.
Based on serological evidence, our findings suggest that IAV-positive feral swine in the United States were predominantly exposed to subtypes H1 and H3. H3N2 and H1N1 IAVs have been isolated from feral swine and are genetically close to enzootic domestic swine IAVs (17, 41). It is unclear if there is an epidemiological link; however, this finding is consistent with a scenario in which domestic swine IAVs occasionally spill over into feral swine populations. Feral swine may be more likely to have contact with domestic swine in backyards or small farming operations that have less biosecurity than large swine operations, and it is possible that direct or indirect (i.e., through fomites) transmission occurs between feral swine and domestic swine. Another possible source of domestic-like IAVs in feral swine could be escaped infected domestic swine; however, the rate of recruitment of domestic swine into the feral swine population is not clear. Additionally, some feral swine serum samples cross-reacted with both H1 and H3 subtypes of IAVs, which suggests that these swine could have been exposed to more than one IAV and is consistent with previously reported findings (17). These possibilities need to be investigated by isolating IAVs currently circulating in feral swine and comparing their genetics to those of nearby IAVs circulating in the domestic swine populations.
A significant portion of the tested feral swine serum samples (78.57% [231/294]) cross-reacted with human-origin IAVs, including H1N1 and H3N2 viruses. The source of feral swine exposure to human H1N1 and H3N2 viruses is unknown. In 1934, Elkeles demonstrated the susceptibility of swine to human influenza virus strains (42). In the past 5 decades, genomic analyses have suggested at least 20 introductions of IAVs from humans to swine, the majority being human seasonal subtype H3N2 viruses (43). Human-to-swine transmission of influenza A(H1N1)pdm09 virus was detected in domestic swine approximately 1 month after the virus was detected in humans (44). After this “reverse zoonosis” event, A(H1N1)pdm09 virus cocirculated with endemic swine influenza virus, including triple-reassortant H3N2, human-origin H1N2 (H1δ1), and classical H1N1 (H1γ) swine influenza viruses (19), resulting in reassortment events (45–48). Additionally, in our panel of reference sera, H3N2 seasonal viruses did not cross-react with any reference sera against any contemporary H3 swine IAVs; this is consistent with the H3 human-like viruses found in domestic swine (49). As early as 2010, within the domestic swine population, novel HAs of H3 viruses emerged and were most genetically similar to human H3N2 strains from the 2010-2011 season; this spillover of human H3N2 into swine is currently being sustained within the domestic swine population (49).
Recently, through evaluating dynamics of serological responses in feral swine inoculated with influenza A viruses, we optimized the sample-to-negative-control ratio (S/N) cutoff for using IDEXX AI MultiS-Screen Ab test to determine the seropositivity for serum samples from feral swine, and a cutoff of S/N ≥0.681 was determined (36). In this study, we adapted this cutoff of S/N for all feral swine serum samples collected in this study. Because this cutoff was more stringent than the one used in another study (17), a lower seroprevalence was obtained. For example, for those samples collected in FY2012, a seroprevalence of 6.0% was obtained in this study, compared with a seroprevalence of 9.2% reported in the prior study (17).
The serological data present in this study could be affected by limitations of serological assays. We could have false-negative results because the avian IAVs usually induce a low serological titer that may decrease with time, resulting in serum samples below the threshold of detection. In addition, we could not test all possible antigenic variants in contemporary avian IAVs. Furthermore, for those samples that cross-react with the IAVs of both avian and swine hosts, it will be difficult to conclude that the IAV exposures were from avian or swine hosts.
In summary, feral swine were predominantly exposed to H1 and H3 swine IAVs, but 5.4% of IAV seropositive samples cross-reacted with avian IAVs. Thus, there is potential for feral swine to generate novel IAVs by hosting and reassorting IAVs from wild birds with those from domestic swine, facilitating adaptation of avian IAVs before their spillover to other hosts, including humans. Continued surveillance is warranted to monitor the distribution and genomic/antigenic diversity of IAVs in feral swine to assess their risk to human health and commercial livestock producers.
MATERIALS AND METHODS
Sample collection and serology testing.
From 1 October 2009 to 30 September 2013, the U.S. Department of Agriculture (USDA) Animal and Plant Health Inspection Service's Wildlife Services collected postmortem serum samples from 8,239 individual feral swine across 35 U.S. states. This collection period included fiscal years 2010 (1 October 2009 to 30 September 2010; 1,818 samples), 2011 (1 October 2010 to 30 September 2011; 2,467 samples), 2012 (1 October 2011 to 30 September 2012; 1,846 samples), and 2013 (1 October 2012 to 30 September 2013; 2,108 samples) (Table 1). Serum samples from 1 October 2011 to 30 September 2012 were previously reported at a total of 1,989, yet 143 serum samples had duplicate information and were ignored for this study. The date of collection, geographic location, age (juvenile, subadult, adult, and unknown), and sex were recorded. Antibody status was determined with the IDEXX AI MultiS-Screen Ab test (IDEXX, Westbrook, ME). Serum samples with a sample-to-negative-control ratio of ≤0.681 were determined to be IAV positive (36). The ELISA results for a subset of feral swine samples (76 of 111) from FY2012 are reported elsewhere (17) in an assessment of the seroprevalence of subtype H3 IAV in feral swine. To ensure complete results, we included all feral swine serum samples collected for the study. Of the samples tested, 406 were identified as IAV positive; from these 406 samples, all of 294 IAV positive samples collected in the most recent 3 years (FY2011, FY2012, and FY2013) were selected for subtyping by HI assay.
Viruses and reference sera.
A total of 45 IAVs were selected to represent the following antigenic groups of contemporary IAVs: enzootic swine IAVs H1 (α, β, γ1, γ2, δ1, and δ2) and H3 cluster IV (α and β), human influenza viruses [A(H1N1)pdm09, swine-origin influenza viruses A(H3N2)v, and seasonal H3N2], and avian influenza viruses H1 to H14 (Table 2). Swine viruses were propagated in Madin-Darby canine kidney (MDCK) epithelial cells, and avian viruses were propagated in specific-pathogen-free (SPF) embryonic eggs. These viruses were used in the serological characterization. The reference swine, ferret, and chicken sera (Table S1) used to assess cross-reactivity among tested viruses were generated as described elsewhere (7, 20, 50).
Hemagglutination and HI assays.
Hemagglutination and hemagglutination inhibition (HI) assays were performed according to the World Health Organization Global Influenza Surveillance Network Manual for the laboratory diagnosis and virologic surveillance of influenza (51). In brief, we treated 1 volume of feral swine serum with 3 volumes of receptor-destroying enzyme (RDE; Denka Seiken Co., Japan) overnight at 37°C and then heat inactivated the serum at 56°C for 30 min. After returning to room temperature, treated antisera were diluted with 6 volumes of 1× phosphate-buffered saline (PBS; pH 7.4). To minimize nonspecific agglutination, we treated RDE-treated serum samples with 0.5% turkey red blood cells (RBCs) (52–54) and then incubated them at 4°C for 1 h, followed by centrifugation at 1,200 rpm for 10 min; we then collected the serum samples without disturbing the packed RBCs. RBC absorption was repeated until no nonspecific agglutination was associated with any serum sample. In the HI assay, 0.5% turkey RBCs were used for absorption; serum samples were determined to be positive against a specific virus if the HI titer was ≥1:40, as described previously (17, 55).
Virus neutralization assays.
RDE-treated feral swine serum was serially diluted 1:2 in a microtiter plate, 100 μl of virus at 100 50% tissue culture infectious doses (TCID50) was added to each well, and then the plate was incubated at 37°C for 1 h. The serum-virus mixture was then incubated with MDCK cells for 1 h in a 96-well tissue culture plate (USA Scientific, Ocala, FL), washed twice with 200 μl 1× PBS (pH 7.4), washed with 200 μl of Opti-MEM (Thermo Fisher, Waltham, MA), and then incubated for 96 h at 37°C in 5% CO2. Detection of nonneutralized virus was conducted using HA assays with 0.5% turkey RBCs. Serum samples were determined to be positive against a specific virus if the neutralization titer was ≥1:40 (56).
Feral swine experiments.
All work was registered and conducted under the supervision of the USDA, NWRC Institutional Animal Care and Use Committee, using approved protocols to ensure humane handling and use. A total of 16 juvenile feral swine (body weight, 16 to 22 kg) were trapped in a rural area of Oktibbeha County, MS, transported to the research facilities, and housed as described elsewhere (36). The captured swine were quarantined for 1 week; before the experiments, all animals tested seronegative to brucellosis, pseudorabies, and IAV by ELISA.
Eight animals were used to test susceptibility of feral swine to A/mallard/Wisconsin/A00661712/2009 (H3N2), four animals were used to test susceptibility of feral swine to A/mallard/Ohio/648/2002 (H6N2), and four additional feral swine were used as negative controls. For H3N2 virus, eight feral swine, housed in four individual pens (two per pen), were intranasally inoculated with 106 TCID50 of A/mallard/Wisconsin/A00661712/2009 (H3N2). Nasal wash samples were collected from all eight feral swine daily from 1 to 10 days postinfection (dpi) and titrated in SPF embryonic eggs, and serum was collected at 0, 7, 14, and 21 dpi to determine seroconversion. To detect pathogenesis of virus in swine, two infected and one control swine were necropsied at 5 and 7 dpi, respectively; turbinate, trachea, and lung of each feral swine were collected and virus titers in these tissues were detected in SPF eggs. For H6N2 virus, four feral swine were intranasally inoculated with 106 TCID50 of influenza A/mallard/Ohio/648/2002 (H6N2) virus in a 1-ml volume. Serum was collected at 0, 5, 7, and 21 dpi to determine seroconversion. The animals used as the negative control were inoculated with sterile PBS. Swine were monitored daily for subjective signs of influenza virus infection (e.g., lethargy, nasal discharge, coughing, and dyspnea) and objective signs (e.g., body temperature) until 14 dpi.
Data analyses.
To understand the risk factors (i.e., seasonality and host factors) affecting IAV seroprevalence in feral swine, chi-square tests were used to assess the differences of IAV seroprevalence between sex, age group, and month, year, and state of sample collection. We developed a multilevel multivariable logistic regression model to test potential risk factors associated with IAV seroprevalence in feral swine, focusing on the population sizes of domestic swine and poultry. The logistic regression model was developed using generalized estimation equations with binomial distribution and logit link function and accounted for clustering of pig samples on the same date and location. Variables were manually selected if they contributed significance to the likelihood ratio statistic for type 3 analysis at an alpha level of <0.05. IAV seroprevalence for sex and age group of feral swine and month, fiscal year, and state of sample collection were analyzed as individuals and feral swine groups, and best fit was assigned based on quasilikelihood under the independence model criterion (QIC). Feral swine groups were defined to eliminate confounding variables. We grouped samples if collected on the same date at the same location; in theory, swine from the same group would be exposed to the same virus. In addition, an odds ratio (OR) was estimated using the GLIMMIX procedure for generalized linear mixed models with binomial distribution and logit link function. Observations from states with fewer than 100 samples were excluded from the analysis. We obtained population data for domestic swine and poultry per state from the 2012 USDA Census of Agriculture (https://www.agcensus.usda.gov/Publications/2012/). Small domestic swine farms were considered those which had less than or equal to a total of 1,000 swine. All statistical analyses were conducted in SAS 9.5 (SAS Institute Inc., Cary, NC).
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the National Institutes of Health (grant P20GM103646) and by the U.S. Department of Agriculture (cooperative agreements 15-729-1146CA and 14-7428-1041-CA).
Additional thanks go to the USDA Swine Influenza Surveillance Program for providing swine isolates.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01346-17.
REFERENCES
- 1.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. 2006. Global patterns of influenza A virus in wild birds. Science 312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 2.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stallknecht DE, Kearney MT, Shane SM, Zwank PJ. 1990. Effects of pH, temperature, and salinity on persistence of avian influenza viruses in water. Avian Dis 34:412–418. doi: 10.2307/1591429. [DOI] [PubMed] [Google Scholar]
- 4.Ito T, Okazaki K, Kawaoka Y, Takada A, Webster RG, Kida H. 1995. Perpetuation of influenza A viruses in Alaskan waterfowl reservoirs. Arch Virol 140:1163–1172. doi: 10.1007/BF01322743. [DOI] [PubMed] [Google Scholar]
- 5.Sturm-Ramirez KM, Hulse-Post DJ, Govorkova EA, Humberd J, Seiler P, Puthavathana P, Buranathai C, Nguyen TD, Chaisingh A, Long HT, Naipospos TS, Chen H, Ellis TM, Guan Y, Peiris JS, Webster RG. 2005. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol 79:11269–11279. doi: 10.1128/JVI.79.17.11269-11279.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey E, Long LP, Zhao N, Hall JS, Baroch JA, Nolting J, Senter L, Cunningham FL, Pharr GT, Hanson L, Slemons R, DeLiberto TJ, Wan XF. 2016. Antigenic characterization of H3 subtypes of avian influenza A viruses from North America. Avian Dis 60(1 Suppl):346–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Bailey E, Spackman E, Li T, Wang H, Long LP, Baroch JA, Cunningham FL, Lin X, Jarman RG, DeLiberto TJ, Wan XF. 2016. Limited antigenic diversity in contemporary H7 avian-origin influenza A viruses from North America. Sci Rep 9:20688. doi: 10.1038/srep20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vincent AL, Ma W, Lager KM, Janke BH, Richt JA. 2008. Swine influenza viruses: a North American perspective. Adv Virus Res 72:127–154. doi: 10.1016/S0065-3527(08)00403-X. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan BS, DeBeauchamp J, Stigger-Rosser E, Franks J, Crumpton JC, Turner J, Darnell D, Jeevan T, Kayali G, Harding A, Webby RJ, Lowe JF. 2015. Influenza virus surveillance in coordinated swine production systems, United States. Emerg Infect Dis 21:1834–1836. doi: 10.3201/eid2110.140633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karasin AI, Brown IH, Carman S, Olsen CW. 2000. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J Virol 74:9322–9327. doi: 10.1128/JVI.74.19.9322-9327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webby RJ, Rossow K, Erickson G, Sims Y, Webster R. 2004. Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res 103:67–73. doi: 10.1016/j.virusres.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Ma W, Gramer M, Rossow K, Yoon KJ. 2006. Isolation and genetic characterization of new reassortant H3N1 swine influenza virus from pigs in the midwestern United States. J Virol 80:5092–5096. doi: 10.1128/JVI.80.10.5092-5096.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorusso A, Vincent AL, Harland ML, Alt D, Bayles DO, Swenson SL, Gramer MR, Russell CA, Smith DJ, Lager KM, Lewis NS. 2011. Genetic and antigenic characterization of H1 influenza viruses from United States swine from 2008. J Gen Virol 92:919–930. doi: 10.1099/vir.0.027557-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hause BM, Oleson TA, Bey RF, Stine DL, Simonson RR. 2010. Antigenic categorization of contemporary H3N2 swine influenza virus isolates using a high-throughput serum neutralization assay. J Vet Diagn Invest 22:352–359. doi: 10.1177/104063871002200302. [DOI] [PubMed] [Google Scholar]
- 15.Olsen CW, Karasin AI, Carman S, Li Y, Bastien N, Ojkic D, Alves D, Charbonneau G, Henning BM, Low DE, Burton L, Broukhanski G. 2006. Triple reassortant H3N2 influenza A viruses, Canada, 2005. Emerg Infect Dis 12:1132–1135. doi: 10.3201/eid1207.060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Richt JA, Lager KM, Janke BH, Woods RD, Webster RG, Webby RJ. 2003. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J Clin Microbiol 41:3198–3205. doi: 10.1128/JCM.41.7.3198-3205.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng Z, Baroch JA, Long LP, Xu Y, Cunningham FL, Pedersen K, Lutman MW, Schmit BS, Bowman AS, Deliberto TJ, Wan XF. 2014. Influenza A subtype H3 viruses in feral swine, United States, 2011-2012. Emerg Infect Dis 20:843–846. doi: 10.3201/eid2005.131578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis NS, Russell CA, Langat P, Anderson TK, Berger K, Bielejec F, Burke DF, Dudas G, Fonville JM, Fouchier RA, Kellam P, Koel BF, Lemey P, Nguyen T, Nuansrichy B, Peiris JM, Saito T, Simon G, Skepner E, Takemae N, ESNIP3 Consortium, Webby ERJ, Van Reeth K, Brookes SM, Larsen L, Watson SJ, Brown IH, Vincent AL. 2016. The global antigenic diversity of swine influenza A viruses. eLife 5:e12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL. 2013. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir Viruses 7(Suppl 4):S42–S51. doi: 10.1111/irv.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng Z, Gomez J, Bowman AS, Ye J, Long LP, Nelson SW, Yang J, Martin B, Jia K, Nolting J, Cunningham F, Cardona C, Zhang J, Yoon KJ, Slemons R, Wan X-F. 2013. Antigenic characterization of H3N2 influenza A viruses from Ohio agricultural fairs. J Virol 87:7655–7667. doi: 10.1128/JVI.00804-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers GN, Paulson JC. 1983. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127:361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 22.Rogers GN, D'Souza BL. 1989. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 23.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y. 1998. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol 72:7367–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma W, Kahn RE, Richt JA. 2008. The pig as a mixing vessel for influenza viruses: human and veterinary implications. J Mol Genet Med 3:158–166. [PMC free article] [PubMed] [Google Scholar]
- 25.Scholtissek C. 1990. Pigs as ‘mixing vessels’ for the creation of new pandemic influenza A viruses. Med Princ Pract 2:64–71. doi: 10.1159/000157337. [DOI] [Google Scholar]
- 26.Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge KF, Kawaoka Y, Webster RG. 1994. Potential for transmission of avian influenza viruses to pigs. J Gen Virol 75(Part 9):2183–2188. [DOI] [PubMed] [Google Scholar]
- 27.Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. 1981. Evidence for the natural transmission of influenza A virus from wild ducks to swine and its potential importance for man. Bull World Health Organ 59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 28.Guan Y, Shortridge KF, Krauss S, Li PH, Kawaoka Y, Webster RG. 1996. Emergence of avian H1N1 influenza viruses in pigs in China. J Virol 70:8041–8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karasin AI, Olsen CW, Anderson GA. 2000. Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana. J Clin Microbiol 38:2453–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karasin AI, West K, Carman S, Olsen CW. 2004. Characterization of avian H3N3 and H1N1 influenza A viruses isolated from pigs in Canada. J Clin Microbiol 42:4349–4354. doi: 10.1128/JCM.42.9.4349-4354.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi YK, Nguyen TD, Ozaki H, Webby RJ, Puthavathana P, Buranathal C, Chaisingh A, Auewarakul P, Hanh NT, Ma SK, Hui PY, Guan Y, Peiris JS, Webster RG. 2005. Studies of H5N1 influenza virus infection of pigs by using viruses isolated in Vietnam and Thailand in 2004. J Virol 79:10821–10825. doi: 10.1128/JVI.79.16.10821-10825.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang G, Kong W, Qi W, Long LP, Cao Z, Huang L, Qi H, Cao N, Wang W, Zhao F, Ning Z, Liao M, Wan XF. 2011. Identification of an H6N6 swine influenza virus in southern China. Infect Genet Evol 11:1174–1177. doi: 10.1016/j.meegid.2011.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peiris JS, Guan Y, Markwell D, Ghose P, Webster RG, Shortridge KF. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J Virol 75:9679–9686. doi: 10.1128/JVI.75.20.9679-9686.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu C, Fan W, Wei R, Zhao H. 2004. Isolation and identification of swine influenza recombinant A/Swine/Shandong/1/2003(H9N2) virus. Microbes Infect 6:919–925. doi: 10.1016/j.micinf.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 35.Gipson PS, Hlavachick B, Berger T, Lee CD. 1997. Explanations for recent range expansions by wild hogs into midwestern states, p 148– Proceedings of the Great Plains Wildlife Damage Control Workshop. [Google Scholar]
- 36.Sun H, Cunningham FL, Harris J, Xu Y, Long LP, Hanson-Dorr K, Baroch JA, Fioranelli P, Lutman MW, Li T, Pedersen K, Schmit BS, Cooley J, Lin X, Jarman RG, DeLiberto TJ, Wan XF. 2015. Dynamics of virus shedding and antibody responses in influenza A virus-infected feral swine. J Gen Virol 96:2569–2578. doi: 10.1099/jgv.0.000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kristensen CS, Botner A, Takai H, Nielsen JP, Jorsal SE. 2004. Experimental airborne transmission of PRRS virus. Vet Microbiol 99:197–202. doi: 10.1016/j.vetmic.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Wyckoff AC, Henke SE, Campbell TA, Hewitt DG, VerCauteren KC. 2009. Feral swine contact with domestic swine: a serologic survey and assessment of potential for disease transmission. J Wildl Dis 45:422–429. doi: 10.7589/0090-3558-45.2.422. [DOI] [PubMed] [Google Scholar]
- 39.Centers for Disease Control and Prevention. 2011. Swine-origin influenza A (H3N2) virus infection in two children—Indiana and Pennsylvania, July–August 2011. MMWR Morb Mortal Wkly Rep 60:1213–1215. [PubMed] [Google Scholar]
- 40.Biggerstaff M, Reed C, Epperson S, Jhung MA, Gambhir M, Bresee JS, Jernigan DB, Swerdlow DL, Finelli L. 2013. Estimates of the number of human infections with influenza A(H3N2) variant virus, United States, August 2011–April 2012. Clin Infect Dis 57(Suppl 1):S12–S15. doi: 10.1093/cid/cit273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clavijo A, Nikooienejad A, Esfahani MS, Metz RP, Schwartz S, Atashpaz-Gargari E, Deliberto TJ, Lutman MW, Pedersen K, Bazan LR, Koster LG, Jenkins-Moore M, Swenson SL, Zhang M, Beckham T, Johnson CD, Bounpheng M. 2013. Identification and analysis of the first 2009 pandemic H1N1 influenza virus from U.S. feral swine. Zoonoses Public Health 60:327–335. doi: 10.1111/zph.12006. [DOI] [PubMed] [Google Scholar]
- 42.Elkeles G. 1934. Experimentelle Untersuchungen zur Aetiologie der Influenza (in German). Mededeelingen Instit Praeventieve Geneeskunde 1934:60–79. [Google Scholar]
- 43.Nelson MI, Wentworth DE, Culhane MR, Vincent AL, Viboud C, LaPointe MP, Lin X, Holmes EC, Detmer SE. 2014. Introductions and evolution of human-origin seasonal influenza A viruses in multinational swine populations. J Virol 88:10110–10119. doi: 10.1128/JVI.01080-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howden KJ, Brockhoff EJ, Caya FD, McLeod LJ, Lavoie M, Ing JD, Bystrom JM, Alexandersen S, Pasick JM, Berhane Y, Morrison ME, Keenliside JM, Laurendeau S, Rohonczy EB. 2009. An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can Vet J 50:1153–1161. [PMC free article] [PubMed] [Google Scholar]
- 45.Ducatez MF, Hause B, Stigger-Rosser E, Darnell D, Corzo C, Juleen K, Simonson R, Brockwell-Staats C, Rubrum A, Wang D, Webb A, Crumpton JC, Lowe J, Gramer M, Webby RJ. 2011. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg Infect Dis 17:1624–1629. doi: 10.3201/1709.110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lam TT, Zhu H, Wang J, Smith DK, Holmes EC, Webster RG, Webby R, Peiris JM, Guan Y. 2011. Reassortment events among swine influenza A viruses in China: implications for the origin of the 2009 influenza pandemic. J Virol 85:10279–10285. doi: 10.1128/JVI.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL, Smith GJ, Peiris JS, Guan Y. 2010. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328:1529. doi: 10.1126/science.1189132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Starick E, Lange E, Fereidouni S, Bunzenthal C, Hoveler R, Kuczka A, grosse Beilage E, Hamann HP, Klingelhofer I, Steinhauer D, Vahlenkamp T, Beer M, Harder T. 2011. Reassorted pandemic (H1N1) 2009 influenza A virus discovered from pigs in Germany. J Gen Virol 92:1184–1188. doi: 10.1099/vir.0.028662-0. [DOI] [PubMed] [Google Scholar]
- 49.Rajão DS, Gauger PC, Anderson TK, Lewis NS, Abente EJ, Killian ML, Perez DR, Sutton TC, Zhang J, Vincent AL. 2015. Novel reassortant human-like H3N2 and H3N1 influenza A viruses detected in pigs are virulent and antigenically distinct from swine viruses endemic to the United States. J Virol 89:11213–11222. doi: 10.1128/JVI.01675-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balzli C, Lager K, Vincent A, Gauger P, Brockmeier S, Miller L, Richt JA, Ma W, Suarez D, Swayne DE. 2016. Susceptibility of swine to H5 and H7 low pathogenic avian influenza viruses. Influenza Other Respir Viruses 10:346–352. doi: 10.1111/irv.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.WHO. 2011. Manual for the laboratory diagnosis and virological surveillance of influenza. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 52.WHO. 2013. The serological detection of avian influenza A(H7N9) virus infections by turkey haemagglutination-inhibition assay. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 53.Pawar SD, Parkhi SS, Koratkar SS, Mishra AC. 2012. Receptor specificity and erythrocyte binding preferences of avian influenza viruses isolated from India. Virology J 9:251. doi: 10.1186/1743-422X-9-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Killian ML. 2008. Hemagglutination assay for the avian influenza virus, p 47–52. In Spackman E. (ed), Avian influenza virus. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 55.Hobson D, Curry RL, Beare AS, Ward-Gardner A. 1972. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg (Lond) 70:767–777. doi: 10.1017/S0022172400022610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Potter CW, Oxford JS. 1979. Determinants of immunity to influenza infection in man. Br Med Bull 35:69–75. doi: 10.1093/oxfordjournals.bmb.a071545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.