ABSTRACT
We recently demonstrated that cow's milk fermented with the probiotic Lactobacillus paracasei CBA L74 (FM-CBAL74) reduces the incidence of respiratory and gastrointestinal tract infections in young children attending school. This effect apparently derives from a complex regulation of non-immune and immune protective mechanisms. We investigated whether FM-CBAL74 could regulate gut microbiota composition and butyrate production. We randomly selected 20 healthy children (12 to 48 months) from the previous randomized controlled trial, before (t0) and after 3 months (t3) of dietary treatment with FM-CBAL74 (FM) or placebo (PL). Fecal microbiota was profiled using 16S rRNA gene amplicon sequencing, and the fecal butyrate concentration was also measured. Microbial alpha and beta diversities were not significantly different between groups prior to treatment. FM-CBAL74 but not PL treatment increased the relative abundance of Lactobacillus. Individual Blautia, Roseburia, and Faecalibacterium oligotypes were associated with FM-CBAL74 treatment and demonstrated correlative associations with immune biomarkers. Accordingly, PICRUSt analysis predicted an increase in the proportion of genes involved in butyrate production pathways, consistent with an increase in fecal butyrate observed only in the FM group. Dietary supplementation with FM-CBAL74 induces specific signatures in gut microbiota composition and stimulates butyrate production. These effects are associated with changes in innate and acquired immunity.
IMPORTANCE The use of a fermented milk product containing the heat-killed probiotic strain Lactobacillus paracasei CBAL74 induces changes in the gut microbiota, promoting the development of butyrate producers. These changes in the gut microbiota composition correlate with increased levels of innate and acquired immunity biomarkers.
KEYWORDS: gut microbiota, immune system, fecal butyrate
INTRODUCTION
Common infectious diseases, affecting the respiratory and gastrointestinal tracts, are an important problem for young children attending preschool or day care centers (1). Young children are especially prone to infection, and this susceptibility is thought to be driven by immaturity in organ function, immune response, and also potentially in the gut microbiota (2). Functional foods, based on the fermentation of cow's milk with probiotics, have been proposed as an effective strategy to reduce the incidence of infectious diseases in children, but the results are still conflicting (3–8). These discrepancies could derive mainly from different study designs and population studies and from different functional properties of the investigated fermented foods. The efficacy of fermented foods is believed to be strain specific and dose dependent. Therefore, additional research is required to understand the mode of action and the impact of each product, and clinical trials are needed to determine efficacy of claims in human populations.
Bacteria associated with fermented foods may influence gut-associated microbial composition and function by direct competition, by metabolic interaction, through direct immune activation, or via the production of bioactive molecules, such as the short-chain fatty acids (SCFAs), that are able to influence the host health regulating a number of immune and non-immune protective mechanisms (9–13).
Many fermented foods are processed such that viable bacteria are inactivated at the time of consumption (9). Postbiotics containing dead bacterial cells have been shown to exert biological effects on the host immune system and to stimulate the production of anti-inflammatory cytokines (14–16). We previously demonstrated that a fermented cow's milk product with heat-killed Lactobacillus paracasei CBA L74 (FM-CBAL74) efficiently protects schooled children against respiratory and gastrointestinal tract infections and that this protective effect is associated with a significant stimulation of innate and acquired immunity (17).
In order to assess the possible association of these effects with the structure of the gut microbiota, we designed this study to determine the effects of FM-CBAL74 treatment on gut microbiota composition and butyrate production.
RESULTS
Study subjects.
The main features of the study populations are reported in Table 1. All children were from families of middle socioeconomic status from the same urban area. The dietary habits were very similar between the two study groups—energy (kcal ± the standard deviations [SD]), 1,420 ± 51 versus 1,388 ± 59; carbohydrate (in grams ± the SD), 225.9 ± 7.3 versus 215.1 ± 8; protein, 31.8 ± 2.7 versus 30.6 ± 2.9; fat, 51.28 ± 4.3 versus 51.46 ± 4; and fiber, 9.3 ± 3.6 versus 11.9 ± 2.7—in MILK (dietary product deriving from cow's milk fermented with L. paracasei CBA L74)- and placebo (PL)-treated groups, respectively. All children were nonfebrile at inclusion, and none was suffering from respiratory tract or gastrointestinal symptoms. The vaccination status was identical among the two groups. The interventions were well accepted by the children, and the compliance was good in all study subjects. No differences were detected in the daily intake of the active and placebo products between the study groups. No modifications in blood sugar and insulin levels were observed upon treatment. Significant increases in α-defensin (HNP1-3), β-defensin 2 (HBD-2), and cathelicidin (LL-37) were observed only in children treated with FM-CBAL74, as well as an increasing trend in secretory IgA (sIgA) (see Fig. S1 in the supplemental material). No differences in the average weighted Unifrac distances were detected between the MILK and PL groups at the beginning of the treatment (P > 0.05).
TABLE 1.
Main anamnestic, demographic, and immunological features of the study populationa
| Parameter | Mean ± SDa |
|
|---|---|---|
| MILK | PL | |
| Demographic data | ||
| No. of subjects | 10 | 10 |
| No. (%) of male subjects | 8 (80) | 5 (50) |
| Age (mo) | 34.3 ± 8.9 | 37.2 ± 8.7 |
| No. (%) breastfeeding | 10 (100) | 7 (70) |
| Duration (mo) of breastfeeding | 6.2 ± 5.9 | 10 ± 6.6 |
| Wt (kg) | ||
| t0 | 15.7 ± 3.3 | 14.9 ± 2.2 |
| t3 | 16.1 ± 2.8 | 15.6 ± 2.4 |
| Ht (cm) | ||
| t0 | 95.9 ± 8.5 | 95.7 ± 6.2 |
| t3 | 97.6 ± 7.6 | 98 ± 6.1 |
| Level | ||
| Alpha-defensin at t0 (ng/ml) | 1.5 ± 1.4 | 1.2 ± 1.2 |
| Alpha-defensin at t3 (ng/ml) | 4.2 ± 1.9 | 1.6 ± 1.3 |
| Beta-defensin 2 at t0 (ng/ml) | 28.7 ± 25.1 | 32.3 ± 13.2 |
| Beta-defensin 2 at t3 (ng/ml) | 46.8 ± 21.1 | 38.6 ± 15 |
| LL-37 at t0 (ng/ml) | 13.3 ± 6.9 | 16 ± 9.3 |
| LL-37 at t3 (ng/ml) | 32 ± 16.3 | 19.4 ± 14.5 |
| sIgA at t0 (μg/ml) | 24.1 ± 9.9 | 30.8 ± 18.2 |
| sIgA at t3 (μg/ml) | 42.8 ± 14 | 32.5 ± 16.4 |
Values are reported as means ± the standard deviations, except as noted otherwise in column 1. Treatment groups: MILK, cow's milk fermented with L. paracasei CBA L74; PL, placebo.
Effects of FM-CBAL74 on gut microbiota composition.
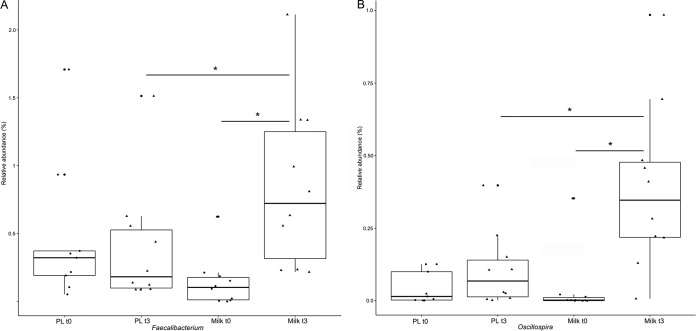
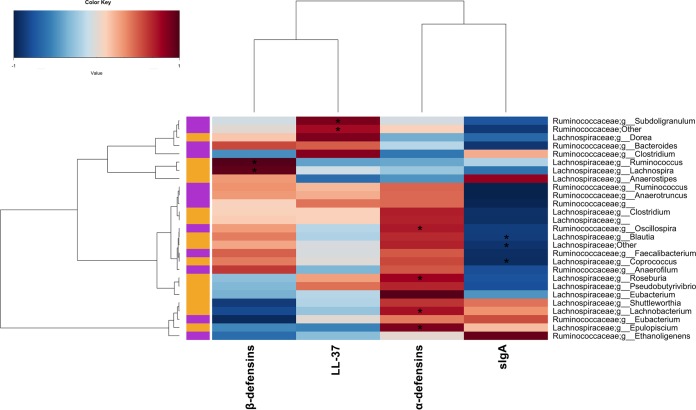
We did not observe significant differences in alpha and beta diversities between children belonging to the two groups at the baseline (see Fig. S2 and S3 in the supplemental material). Treatment with FM-CBAL74 affected gut microbiota composition. Although a great variability was observed, multivariate analysis of variance (MANOVA) showed significant differences in the gut microbiota composition between the two study groups following intervention (P < 0.05). Nevertheless, principal-coordinate analysis did not show any clustering of the subjects according to the treatment received (see Fig. S3 in the supplemental material). The relative proportion of Lactobacillus and Ruminococcaceae significantly increased following FM-CBAL74 treatment (P < 0.05), with specific significant increases in Oscillospira and Faecalibacterium (see Table S1 in the supplemental material and Fig. 1, P < 0.05). In addition, we found positive correlations between the relative abundance of several genera belonging to Ruminococcaceae and fecal LL-37 level, whereas Lachnospira and Ruminococcus (Lachnospiraceae) correlated with HBD-2 levels (Fig. 2). The gut microbiotas of PL subjects were significantly different from those of FM-CBAL74-treated subjects after 3 months of treatment, with significantly higher levels of Bacteroides (see Table S1 in the supplemental material, P < 0.05).
FIG 1.
Oscillospira and Faecalibacterium levels. Box plots show the abundance of Oscillospira and Faecalibacterium in the studied population at baseline (t0) and after 3 months of treatment (t3) with fermented milk (MILK) and placebo (PL). Boxes represent the interquartile range (IQR) between the first and third quartiles, and the line inside represents the median (second quartile). Whiskers denote the lowest and highest values within 1.5 × IQR from the first and third quartiles, respectively. Asterisks indicate a significant difference as obtained by a pairwise Wilcoxon test (P < 0.05).
FIG 2.
Lachnospiraceae and Ruminococcaceae abundance correlates with innate and acquired immunity. A heat plot shows the Spearman correlations between genera belonging to Lachnospiraceae and Ruminococcaceae and the levels of immunity biomarkers. Rows and columns are clustered by Euclidean distance and Ward linkage hierarchical clustering. The intensity of the colors represents the degree of association, as measured by Spearman correlations. Only genera occurring in at least 20% of the samples were included. Asterisks indicate a significant correlation after Benjamini-Hochberg correction.
Effect of fermented milk on gut microbiota at subgenus level.
In order to explore the possible effect of FM-CBAL74 at a subgenus level, we carried out oligotyping on sequences of Bacteroides and genera belonging to Ruminococcaceae and Lachnospiraceae, since these bacterial groups are well-known butyrate producers. Only Bacteroides, Blautia, and Roseburia oligotype patterns showed significant changes after dietary intervention, as shown by MANOVA (P < 0.05). Specific Roseburia oligotypes were promoted by FM-CBAL74 treatment (Roseburia oligotype 1 [see Fig. S4A in the supplemental material]) and showed positive correlations with sIgA (ρ = 0.63, P = 0.038) and β-defensin (ρ = 0.87, P = 0.023). Blautia oligotypes 5 and 13 also increased with FM-CBAL74 treatment and were positively correlated with α-defensin (ρ = 0.84, P = 0.007; ρ = 0.58, P = 0.040) (see Fig. S4B in the supplemental material). Finally, Bacteroides oligotypes 12 and 19 increased after FM-CBAL74 treatment (see Fig. S4C in the supplemental material), but only Bac12 (ρ = 0.67, P = 0.042) was positively correlated with α-defensin.
FM-CBAL74 treatment promotes butyrate production in the gut.
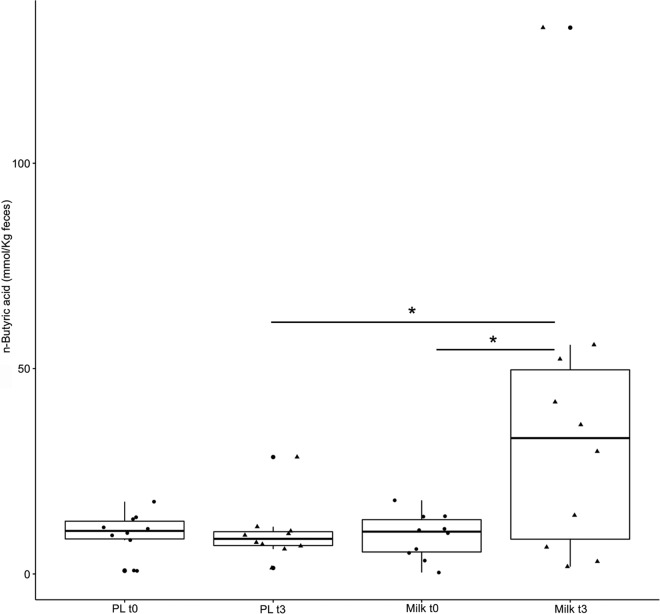
FM-CBAL74 treatment resulted in an increase in the relative abundance of predicted genes involved in butyrate synthesis (PICRUSt-predicted metagenomes), especially genes encoding butyryl coenzyme A (butyryl-CoA) transferase (EC 2.8.3.8) and butyrate kinase (EC 2.7.2.7) (see Fig. S5 in the supplemental material, P < 0.05). Consistently, a significant increase in fecal butyrate levels in children consuming FM-CBAL74 was observed (Fig. 3, P < 0.05).
FIG 3.
Fecal butyrate concentration. Box plots show the abundance of fecal butyrate in the study population at baseline (t0) and after 3 months of treatment (t3) with fermented milk (MILK) and placebo (PL). Asterisks indicate a significant difference. as obtained by pairwise Wilcoxon test (P < 0.05). See the legend to Fig. 1 for a description of the box plots.
DISCUSSION
It is increasingly understood that the outcomes of microbial fermentation provide additional properties to fermented foods beyond basic nutrition (9). During fermentation, the metabolic activity of microorganisms can change the nutritional and bioactive properties of food matrices with possible beneficial consequences for human health (9). The gut microbiota plays a key role in the development and function of the immune system (18) and some of the health benefits of fermented foods could derive also from their impact on gut microbial composition and function (9).
In a randomized controlled trial, we demonstrated that a cow's milk fermented product containing the heat-killed probiotic strain L. paracasei CBA L74 is effective in reducing the incidence respiratory and gastrointestinal tract infections in young children (17). Interestingly, these results have been recently confirmed by a multicenter trial with a similar study design (19). Here, we tested the effect of this specific fermented product on gut microbiota composition and butyrate production. Distinctive traits of the gut microbiota, with an increase in genera known as butyrate producers such as Oscillospira and Faecalibacterium (20, 21), were observed in children receiving the fermented product. It has been previously shown that Faecalibacterium strains exert a stimulating effect on the immune system and on T cell differentiation (22). In addition, an increase in Lactobacillus abundance was observed upon treatment with the fermented product, which is unlikely due to the presence of heat-killed lactobacilli in the fermented product and more probably derives from a stimulatory effect of this product on such populations of lactic acid bacteria.
We also demonstrated an effect of the fermented milk product beyond the genus level. In fact, specific oligotypes of Roseburia and Blautia were boosted by the treatment, suggesting an effect at the subgenus level and highlighting a possible different effect of the fermented product on species and strains belonging to these genera, as previously pointed out for other common members of the gut microbiota (23, 24). The dietary treatment resulted in an increase in the relative abundance of predicted genes involved in butyrate synthesis, especially genes encoding butyryl-CoA transferase (EC 2.8.3.8) and butyrate kinase (EC 2.7.2.7), and an associated significant increase in fecal butyrate levels likely deriving from lactate catabolism, one of the primary pathways for butyrate production by gut bacteria (25). Altogether, these results suggest that a shift in the relative proportion of certain bacterial genera and oligotypes may be associated with an enhanced butyrate synthesis. Butyrate regulates several non-immune and immune defense mechanisms against infections, including the regulation of luminal pH in the gut, mucus production, cell growth and differentiation, the modulation of gut permeability and of transepithelial ion transport, the modulation of the inflammatory response, and the stimulation of innate and acquired immunity (21, 26–28). Although we investigated only butyrate, it cannot be excluded that also the production of other SCFAs could be regulated by this particular fermented product.
We found positive correlations between specific gut microbiota signatures and the fecal levels of innate and acquired immunity biomarkers. These data are in line with the results obtained using a fermented milk formula containing two heat-inactivated probiotic strains in preterm infants (4).
The results of this study support the concept of a mutualistic interaction occurring between gut microbiota and immune system, where gut microbiota influences immune system development and function, and the immune system shapes gut microbiota composition (29, 30). It is possible to speculate that the effect of this particular fermented food on gut microbiota could derive at least in part by a modulation of innate immunity peptides. Indeed, evidence on the positive correlations between specific members of the gut microbiota and immunity peptides has been obtained in this study. The final result is the establishment and consolidation of a microbiota composition that can be responsible for a protective action against infectious diseases. In line with this view, we have recently demonstrated that, through a direct interaction with human enterocytes, FM-CBAL74 stimulates the synthesis of β-defensin 2 and LL-37 (31). Future studies are necessary to define the components of this particular fermented product that are involved in this effects. In this light, comparison with different types of placebo, such as milk without the addition of bacteria or fermented by other lactobacilli, would provide useful information.
It is important to recognize that this study investigated a dietary product fermented with a specific probiotic strain, a well-defined dose, and age group and that our findings cannot be extrapolated for other products containing different probiotic strains. Indeed, it cannot be excluded that similar results could be obtained with milk products fermented by phylogenetically close lactic acid bacteria. In this study, the similar dietary habits of treated and placebo groups strongly suggest that the effects observed on gut microbiota composition and butyrate production derive from the administration of the fermented milk used. In this study, children enrolled in the placebo group, received maltodextrins. Maltodextrins, even at higher doses, are commonly used as placebos in clinical trials (32, 33). Contrasting data suggest that maltodextrins could influence gut microbiota composition and immune system (34–36). Here, we did not observe significant changes in the gut microbiota composition, fecal butyrate levels, and innate and acquired immunity biomarkers in children enrolled in the placebo group, which supports the use of maltodextrins in placebo treatments.
In conclusion, although additional research should be focused on the specific molecular mechanisms involved, we have shown that FM-CBA L74 induces positive regulation of the mutual interaction between the immune system and gut microbiota.
MATERIALS AND METHODS
Study subjects.
Detailed description of screening and recruitment of study population has been provided elsewhere (17). Briefly, consecutive healthy children (12 to 48 months of age) attending day care or preschool at least 5 days a week, were invited to participate to the study. The exclusion criteria were as follows: age ≤12 months or ≥48 months, concomitant chronic systemic diseases, congenital cardiac defects, gastrointestinal or urinary or respiratory tract surgery, active tuberculosis, autoimmune diseases, immunodeficiency, chronic inflammatory bowel diseases, cystic fibrosis, metabolic diseases, history of suspected or challenge-proved food allergy, lactose intolerance, malignancy, chronic pulmonary diseases, malformations of gastrointestinal or urinary or respiratory tract, severe malnutrition (z score for weight-for-height < 3 SD scores), and the use of pre/pro/synbiotics, antibiotics, or immune stimulating products in the 2 weeks before the enrollment. From the original study population enrolled (17), we randomly selected 10 subjects per group through a random number generator (Randomness and Integrity Services, Ltd., Dublin, Ireland [https://www.random.org]). Anamnestic, demographic, and clinical features, including innate and acquired immunity biomarkers data, as well as information regarding dietary habits, assessed by a 3-day food diary collected every week for the entire study duration, were available for the entire cohort (17). The sample size was calculated, taking into account the size effect estimated from our previous data on butyrate levels (37). We calculated that 10 children per group were needed to detect an increase of at least 50% above baseline mean fecal butyrate level with a power of 0.80 at an alpha level of 0.05 (t test for two independent samples with common variance two-tailed test). This study was approved by the Ethics Committee of the University of Naples Federico II and was registered in the Clinical Trials Protocol Registration System (ClinicalTrials.gov) with the identifier NCT01909128.
Intervention.
The investigators were blinded to the treatment at all times, i.e., allocation, intervention, laboratory analysis, and statistical analysis (17). The study subjects were distributed into two groups according to a computer-generated randomization list. The investigators assigned each child the next available number on entry into the trial. Investigators, parents, and children were not aware of the dietary treatment assigned. Subjects were supplemented daily for 3 months with either a dietary product deriving from cow's milk fermented with L. paracasei CBA L74 (MILK) or a placebo (PL).
The composition of the dietary products used is reported in Table 2. They were provided in powder by Heinz Italia SpA, Latina, Italy, an affiliate of H. J. Heinz Company, Pittsburgh, PA. The fermented milk was prepared from skimmed milk fermented by L. paracasei CBA L74. The fermentation was started in the presence of 106 bacteria, reaching 5.9 × 109 CFU/g after a 15-h incubation at 37°C. After heating at 85°C for 20 s in order to inactivate the live bacteria, the formula was spray-dried. Thus, the final fermented milk powder contained only bacterial bodies and fermentation products and no living microorganisms. The placebo consisted of maltodextrins with similar energy content of the fermented milk. Study products were provided in tins containing 400 g of powder, and the packaging was similar. Study products were stored at room temperature and in a dry environment.
TABLE 2.
Composition of the study dietary products
| Component | Composition (per 100 g of product) for each treatment group |
|
|---|---|---|
| MILK | PL | |
| Energy (kcal) | 367 | 388 |
| Protein (g) | 24.0 | 0 |
| Carbohydrate (g) | 66.4 | 97 |
| Fat (g) | 0.6 | 0 |
| L. paracasei CBA L74 (CFU)a | 5.9 × 1011 | |
That is, the CFU of killed bacteria.
The investigators instructed parents about the daily amount of the assigned study product and the method of preparation. All subjects received 7 g/day of study products diluted in a maximum of 150 ml of cow's milk or water. Parents were encouraged to contact the investigators if necessary and to maintain the habitual diet of the child, but to exclude prebiotics, probiotics, postbiotics, synbiotics, and immune stimulating products during the 3-month study period.
Compliance was defined as the consumption of at least 80% of the assigned treatment during the study and was evaluated by counting and weighing the returned tins and by the notes on the diary compiled by parents. At the enrollment and at the end of the trial, a stool sample (∼3 g) was obtained from all study subjects and stored at −80°C prior to further analysis.
DNA extraction and 16S rRNA gene sequencing.
Fecal samples (∼1 g) were fully homogenized in STE buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]) and centrifuged (1,000 rpm × 1 min) in order to pellet debris. The supernatant was centrifuged again (12,000 × g, 2 min), and the pellet was used for DNA extraction by using a PowerFecal DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA). The V3-V4 region of the 16S rRNA gene was amplified by using the primers S-D-Bact-0341F5′-CCTACGGGNGGCWGCAG and S-D-Bact-0785R5′-GACTACHVGGGTATCTAATCC (38). Library multiplexing, pooling, and sequencing were carried out according to the Illumina 16S metagenomic sequencing library preparation protocol on a MiSeq platform and using the MiSeq Reagent kit v2, yielding 2×250-bp, paired-end reads.
Bioinformatics and statistical analysis.
Demultiplexed, forward, and reverse reads were joined by using FLASH (39). Joined reads were quality trimmed (Phred score < 20) and short reads (<250 bp) were discarded by using Prinseq (40). High-quality reads were then imported into QIIME (41). Operational taxonomic units (OTU) were picked using a de novo approach and the uclust method, and taxonomic assignments were obtained by using the RDP classifier and the Greengenes (42) database, following a pipeline previously reported (43). In order to avoid biases due to the different sequencing depth, OTU tables were rarefied to the lowest number of sequences per sample. Bray-Curtis distance matrix and alpha diversity indices were computed by QIIME on rarefied OTU tables. PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States [http://picrust.github.io/picrust]) (44) was used to predict the functional profiles of the samples, as recently reported (45).
Statistical analyses and plotting were carried out in an R environment (https://www.r-project.org).
Permutational multivariate analysis of variance (nonparametric MANOVA) based on Jaccard and Bray-Curtis distance matrices was carried out using 999 permutations to detect significant differences in the overall microbial community or oligotype patterns, by using the “adonis” function in the vegan package. The Bioconductor statistical package DeSeq2 (46) was used to find taxa differentially abundant between the groups. Spearman's pairwise correlations were computed between OTU and other quantitative variables (the “corr.test” function in the psych package) and plotted by using the “heatplot” function in the made4 package. P values were corrected for multiple testing using the Benjamini-Hochberg procedure (47).
Oligotyping analysis.
Reads assigned to Bacteroides and to genera within Ruminococcaceae and Lachnospiraceae with an abundance of >5% in at least 10% of the samples were extracted, and entropy analysis and oligotyping were carried out (48). Only Bacteroides, Roseburia, and Blautia oligotype patterns were significantly affected by treatment. After the initial round of oligotyping, high-entropy positions were chosen (-C option): 2, 27, 30, 31, 32, 94, 114, 120, and 291 (Bacteroides); 1, 2, 12, 27, 28, 30, 56, 57, 58, 61, 82, 101, 103, 157, 160, 170, 172, 174, 176, 184, 191, 213, 215, 220, 235, 237, 273, 274, 293, 343, 347, 371, and 409 (Blautia); and 2, 28, 57, 215, 272, 369, 370, 409, and 410 (Roseburia). To minimize the impact of sequencing errors, we required an oligotype to be represented by at least 100 reads (-M option). Moreover, rare oligotypes present in fewer than 10 samples were discarded (-s option). These parameters led to 56, 59, and 52 samples and 90,195, 298,288, and 24,503 sequences left in the Bacteroides, Blautia, and Roseburia data sets, respectively. BLASTn was used to query the representative sequences against the NCBI nr database, and the top hit was considered for taxonomic assignment. Statistical analyses and plotting were carried out in R.
Fecal butyrate analysis.
One gram of frozen feces was diluted with saline buffer, vortexed, and centrifuged (12,000 × g) for 10 min in 2-ml tubes. The supernatant was filtered (0.45-μm pore size) and stored at −20°C until analysis. Frozen fecal extracts were acidified with 20 μl of 85% (wt/vol) phosphoric acid and 0.5 ml of ethyl acetate, mixed and centrifuged (12,000 × g) for 1 h, and extracted in duplicate. About 0.5 ml of the pooled extract containing the acidified butyrate was transferred into a 2-ml glass vial and loaded onto an Agilent Technologies (Santa Clara, CA) 7890 gas chromatograph (GC) system with an automatic loader/injector. The GC column was a J&W DB-FFAP (Agilent Technologies) of 30 m, with an internal diameter of 0.25 mm and a film thickness of 0.25 μm. The GC was programmed to achieve the following run parameters: an initial temperature of 90°C, a hold for 0.5 min, and ramp of 20°C min−1 up to a final temperature of 190°C; a total run time of 8.0 min; a gas flow of 7.7 ml min−1 split less to maintain 3.26 lb/in2 column head pressure; and a septum purge of 2.0 ml min−1. Detection was achieved using a flame ionization detector. Peaks were identified using a mixed external standard and quantified by using a peak height/internal standard ratio.
Assessment of innate and acquired immunity biomarkers.
For all study subjects, data related to the fecal levels of α-defensin (HNP1-3), β-defensin 2 (HBD-2), cathelicidin (LL-37), and sIgA were available. The determinations were performed as previously described (17). The results are expressed as ng/ml for α-defensin, β-defensin, and LL-37 and as μg/ml of supernatants for sIgA.
Accession number(s).
The 16S rRNA gene sequences produced in this study are available at the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI) under accession number SRP100769.
Supplementary Material
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01206-17.
REFERENCES
- 1.Maldonado J, Cañabate F, Sempere L, Vela F, Sánchez AR, Narbona E, López-Huertas E, Geerlings A, Valero AD, Olivares M, Lara-Villoslada F. 2012. Human milk probiotic Lactobacillus fermentum CECT5716 reduces the incidence of gastrointestinal and upper respiratory tract infections in infants. J Pediatr Gastroenterol Nutr 54:55–61. doi: 10.1097/MPG.0b013e3182333f18. [DOI] [PubMed] [Google Scholar]
- 2.Prodeus A, Niborski V, Schrezenmeir J, Gorelov A, Shcherbina A, Rumyantsev A. 2016. Fermented milk consumption and common infections in children attending day-care centers: a randomized trial. J Pediatr Gastroenterol Nutr 63:534–543. doi: 10.1097/MPG.0000000000001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merenstein D, Murphy M, Fokar A, Hernandez RK, Park H, Nsouli H, Sanders ME, Davis BA, Niborski V, Tondu F, Shara NM. 2010. Use of a fermented dairy probiotic drink containing Lactobacillus casei (DN-114 001) to decrease the rate of illness in kids: the DRINK study. A patient-oriented, double-blind, cluster-randomized, placebo-controlled, clinical trial. Eur J Clin Nutr 64:669–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campeotto F, Suau A, Kapel N, Magne F, Viallon V, Ferraris L, Waligora-Dupriet AJ, Soulaines P, Leroux B, Kalach N, Dupont C, Butel MJ. 2011. A fermented formula in pre-term infants: clinical tolerance, gut microbiota, downregulation of faecal calprotectin and up-regulation of faecal secretory IgA. Br J Nutr 105:1843–1851. doi: 10.1017/S0007114510005702. [DOI] [PubMed] [Google Scholar]
- 5.Nagata S, Asahara T, Ohta T, Yamada T, Kondo S, Bian L, Wang C, Yamashiro Y, Nomoto K. 2011. Effect of the continuous intake of probiotic-fermented milk containing Lactobacillus casei strain Shirota on fever in a mass outbreak of norovirus gastroenteritis and the faecal microflora in a health service facility for the aged. Br J Nutr 106:549–556. doi: 10.1017/S000711451100064X. [DOI] [PubMed] [Google Scholar]
- 6.Thibault H, Aubert-Jacquin C, Goulet O. 2004. Effects of long-term consumption of a fermented infant formula (with Bifidobacterium breve c50 and Streptococcus thermophilus 065) on acute diarrhea in healthy infants. J Pediatr Gastroenterol Nutr 39:147–152. doi: 10.1097/00005176-200408000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Mullié C, Yazourh A, Thibault H, Odou MF, Singer E, Kalach N, Kremp O, Romond MB. 2004. Increased poliovirus-specific intestinal antibody response coincides with promotion of Bifidobacterium longum-infantis and Bifidobacterium breve in infants: a randomized, double-blind, placebo-controlled trial. Pediatr Res 56:791–795. doi: 10.1203/01.PDR.0000141955.47550.A0. [DOI] [PubMed] [Google Scholar]
- 8.Agostoni C, Goulet O, Kolacek S, Koletzko B, Moreno L, Puntis J, Rigo J, Shamir R, Szajewska H, Turck D, ESPGHAN Committee on Nutrition . 2007. Fermented infant formulae without live bacteria. J Pediatr Gastroenterol Nutr 44:392–397. doi: 10.1097/01.mpg.0000258887.93866.69. [DOI] [PubMed] [Google Scholar]
- 9.Marco ML, Heeney D, Binda S, Cifelli CJ, Cotter PD, Foligné B, Gänzle M, Kort R, Pasin G, Pihlanto A, Smid EJ, Hutkins R. 2017. Health benefits of fermented foods: microbiota and beyond. Curr Opin Biotechnol 44:94–102. doi: 10.1016/j.copbio.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Tamang JP, Watanabe K, Holzapfel WH. 2016. Review: Diversity of microorganisms in global fermented foods and beverages. Front Microbiol 7:377. doi: 10.3389/fmicb.2016.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unno T, Hisada T, Takahashi S. 2015. Hesperetin modifies the composition of fecal microbiota and increases cecal levels of short-chain fatty acids in rats. J Agric Food Chem 63:7952–7957. doi: 10.1021/acs.jafc.5b02649. [DOI] [PubMed] [Google Scholar]
- 12.Derrien M, van HylckamaVlieg JE. 2015. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol 23:354–366. doi: 10.1016/j.tim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Corrêa-Oliveira R, Fachi JL, Vieira A, Takeo Sato F, Vinolo MAR. 2016. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunol 5:e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vieira AT, Rocha VM, Tavares L, Garcia CC, Teixeira MM, Oliveira SC, Cassali GD, Gamba C, Martins FS, Nicoli JR. 2015. Control of Klebsiella pneumoniae pulmonary infection and immunomodulation by oral treatment with the commensal probiotic Bifidobacterium longum 5(1A). Microbes Infect 18:180–189. doi: 10.1016/j.micinf.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Vieira AT, Fukumori C, Ferreira CM. 2016. New insights into therapeutic strategies for gut microbiota modulation in inflammatory diseases. Clin Transl Immunol 5:e87. doi: 10.1038/cti.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asama T, Kimura Y, Kono T, Tatefuji T, Hashimoto K, Benno Y. 2016. Effects of heat-killed Lactobacillus kunkeei YB38 on human intestinal environment and bowel movement: a pilot study. Benef Microbes 7:337–344. doi: 10.3920/BM2015.0132. [DOI] [PubMed] [Google Scholar]
- 17.Nocerino R, Paparo L, Terrin G, Pezzella V, Amoroso A, Cosenza L, Cecere G, De Marco G, Micillo M, Albano F, Nugnes R, Ferri P, Ciccarelli G, Giaccio G, Spadaro R, Maddalena Y, Berni Canani F, Berni Canani R. 2015. Cow's milk and rice fermented with Lactobacillus paracasei CBA L74 prevent infectious diseases in children: a randomized controlled trial. Clin Nutr 36:118–125. doi: 10.1016/j.clnu.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. 2016. How colonization by microbiota in early life shapes the immune system. Science 352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corsello G, Carta M, Marinello R, Picca M, De Marco G, Micillo M, Ferrara D, Vigneri P, Cecere G, Ferri P, Roggero P, Bedogni G, Mosca F, Paparo L, Nocerino R, Berni Canani R. 2017. Preventive effect of cow's milk fermented with L. paracasei CBA L74 on common infectious diseases in children: a multicenter randomized controlled trial. Nutrients 9:669. doi: 10.3390/nu9070669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konikoff T, Gophna U. 2016. Oscillospira: a central, enigmatic component of the human gut microbiota. Trends Microbiol 24:523–524. doi: 10.1016/j.tim.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 21.Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de los Reyes-Gavilán CG, Salazar N. 2016. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossi O, van Berkel LA, Chain F, Tanweer Khan M, Taverne N, Sokol H, Duncan SH, Flint HJ, Harmsen HJ, Langella P, Samsom JN, Wells JM. 2016. Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci Rep 6:18507. doi: 10.1038/srep18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Filippis F, Pellegrini N, Laghi L, Gobbetti M, Ercolini D. 2016. Unusual subgenus associations of faecal Prevotella and Bacteroides with specific dietary patterns. Microbiome 4:57. doi: 10.1186/s40168-016-0202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ley RE. 2016. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat Rev Gastroenterol Hepatol 13:69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 25.Duncan SH, Louis P, Flint HJ. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 70:5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berni Canani R, Di Costanzo M, Leone L, Pedata M, Meli R, Calignano A. 2011. Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J Gastroenterol 17:1519–1528. doi: 10.3748/wjg.v17.i12.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keku TO, Dulal S, Deveaux A, Jovov B, Han X. 2015. The gastrointestinal microbiota and colorectal cancer. Am J Physiol Gastrointest Liver Physiol 308:G351–G363. doi: 10.1152/ajpgi.00360.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macfarlane GT, Macfarlane S. 2012. Bacteria, colonic fermentation, and gastrointestinal health. J AOAC Int 95:50–60. doi: 10.5740/jaoacint.SGE_Macfarlane. [DOI] [PubMed] [Google Scholar]
- 29.Honda K. 2015. TFH-IgA responses keep microbiota in check. Cell Host Microbe 17:144–146. doi: 10.1016/j.chom.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Jenke AC, Postberg J, Mariel B, Hensel K, Foell D, Däbritz J, Wirth S. 2013. S100A12 and hBD2 correlate with the composition of the fecal microflora in ELBW infants and expansion of Escherichia coli is associated with NEC. Biomed Res Int 2013:150372. doi: 10.1155/2013/150372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paparo L, Aitoro R, Nocerino R, Maddalena Y, Pezzella V, Amoroso A, Di Scala C, Siciliano E, Buono B, Berni Canani R. 2015. Direct effects of fermented cow's milk with Lactobacillus paracasei CBA L74 on human enterocytes. 48th Annu Meet Eur Soc Paediatr Gastroenterol Hepatol Nutr, Amsterdam, Netherlands, 6 to 9 May 2015. [Google Scholar]
- 32.Kolida S, Meyer D, Gibson GR. 2007. A double-blind placebo-controlled study to establish the bifidogenic dose of inulin in healthy humans. Eur J Clin Nutr 61:1189–1195. doi: 10.1038/sj.ejcn.1602636. [DOI] [PubMed] [Google Scholar]
- 33.Abrams SA, Griffin IJ, Hawthorne KM, Liang L, Gunn SK, Darlington G, Ellis KJ. 2005. A combination of prebiotic short- and long-chain inulin-type fructans enhances calcium absorption and bone mineralization in young adolescents. Am J Clin Nutr 82:471–476. [DOI] [PubMed] [Google Scholar]
- 34.Costabile A, Kolida S, Klinder A, Gietl E, Bäuerlein M, Frohberg C, Landschütze V, Gibson GR. 2010. A double-blind, placebo-controlled, crossover study to establish the bifidogenic effect of a very-long-chain inulin extracted from globe artichoke (Cynara scolymus) in healthy human subjects. Br J Nutr 104:1007–1017. doi: 10.1017/S0007114510001571. [DOI] [PubMed] [Google Scholar]
- 35.Nickerson KP, Chanin R, McDonald C. 2015. Deregulation of intestinal antimicrobial defense by the dietary additive, maltodextrin. Gut Microbes 6:78–83. doi: 10.1080/19490976.2015.1005477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kredier RB, Earnest CP, Lundberg J, Rasmussen C, Greenwood M, Cowan P, Almada AL. 2007. Effects of ingesting protein with various forms of carbohydrate following resistance-exercise on substrate availability and markers of anabolism, catabolism, and immunity. J Int Soc Sports Nutr 4:18. doi: 10.1186/1550-2783-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, Calignano A, Khan AA, Gilbert JA, Nagler CR. 2016. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J 10:742–750. doi: 10.1038/ismej.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magoc T, Salzberg SL. 2011. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, GonzalezPeña A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McDonald D, Price MN, Goodrich J, Nawrocki EP, De Santis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Filippis F, Vannini L, La Storia A, Laghi L, Piombino P, Stellato G, Serrazanetti DI, Gozzi G, Turroni S, Ferrocino I, Lazzi C, Di Cagno R, Gobbetti M, Ercolini D. 2014. The same microbiota and a potentially discriminant metabolome in the saliva of omnivore, ovo-lacto-vegetarian and vegan individuals. PLoS One 9:e112373. doi: 10.1371/journal.pone.0112373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, Serrazanetti DI, Di Cagno R, Ferrocino I, Lazzi C, Turroni S, Cocolin L, Brigidi P, Neviani E, Gobbetti M, O'Toole PW, Ercolini D. 2016. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 65:1812–1821. doi: 10.1136/gutjnl-2015-309957. [DOI] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- 48.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. 2013. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.