Abstract
We expanded on a series of pyrido[2,1-f]purine-2,4-dione derivatives as human adenosine A3 receptor (hA3R) antagonists to determine their kinetic profiles and affinities. Many compounds showed high affinities and a diverse range of kinetic profiles. We found hA3R antagonists with very short residence time (RT) at the receptor (2.2 min for 5) and much longer RTs (e.g., 376 min for 27 or 391 min for 31). Two representative antagonists (5 and 27) were tested in [35S]GTPγS binding assays, and their RTs appeared correlated to their (in)surmountable antagonism. From a kon–koff–KD kinetic map, we divided the antagonists into three subgroups, providing a possible direction for the further development of hA3R antagonists. Additionally, we performed a computational modeling study that sheds light on the crucial receptor interactions, dictating the compounds’ binding kinetics. Knowledge of target binding kinetics appears useful for developing and triaging new hA3R antagonists in the early phase of drug discovery.
Introduction
The adenosine A3 receptor is the youngest member discovered in the family of adenosine receptors,1 all of which belong to class A G-protein coupled receptors (GPCRs) and fall into four distinct subtypes (A1, A2A, A2B, and A3). Although all subtypes are activated by the endogenous ligand adenosine, these purinergic receptors differ from each other in their distribution and to which G protein they are coupled. Following agonist activation, the A1 and A3 adenosine receptors cause a decrease in cAMP levels as they primarily couple to Gi proteins. The A2A and A2B adenosine receptors, on the other hand, are primarily linked to Gs proteins, and this leads to increased levels of cAMP upon receptor activation.2
Although the pharmacological characterization of adenosine receptors has been well documented,3 the human adenosine A3 receptor (hA3R) is less well characterized because of its “dichotomy” in different therapeutic applications.4 Moreover, certain ligands have been described as cytoprotective or cytotoxic merely depending on the concentration employed, highlighting the difficulties that arise when characterizing novel hA3R compounds.5 Nevertheless, there is no doubt that the hA3R has therapeutic potential in clinical indications (i.e., cardiovascular diseases,6,7 cancer,7,8 and respiratory diseases7,9−11) due to its overexpression on cancer and inflammatory cells.3,12−15
Traditional drug screening methods, and those employed in previous hA3R drug discovery attempts, revolve around the use of a ligand’s affinity as the selection criterion for further optimization in a so-called structure–affinity relationships (SAFIRs) approach. In recent years, however, there has been emerging the realization that selecting ligands based on their affinity, an equilibrium parameter, does not necessarily predict in vivo efficacy. This is due to the dynamic conditions in vivo that often are in contrast to the equilibrium conditions applied in in vitro assays.16 In fact, a ligand’s kinetic properties may provide a better indication of how a ligand will perform in vivo.17 Specifically, the parameter of residence time (RT) has been proposed as a more relevant selecting criterion. The RT reflects the lifetime of the ligand–receptor complex and can be calculated as the reciprocal of the ligand’s dissociation constant (RT = 1/koff).18,19
While the binding kinetics of some (labeled) hA3R agonists have been studied,20 this parameter has not been part of medicinal chemistry efforts for antagonists, i.e., yielding structure–kinetics relationships (SKRs), next to SAFIRs.21 Therefore, to provide the first SKR analysis on the hA3R, a highly potent and selective hA3R antagonist scaffold was chosen. The pyrido[2,1-f]purine-2,4-dione template has been previously characterized with respect to affinity alone. In a Topliss approach,22 we had synthesized and characterized a number of highly potent and selective hA3R antagonists.23,24 One of the reference antagonists (1) with good affinity and selectivity over other adenosine receptors is represented in Table 1. Using this compound as the starting point, we further selected and synthesized compounds to add to the library of pyrido[2,1-f]purine-2,4-dione derivatives. Using radioligand displacement assays and competition association assays, we obtained affinity (Ki) and kinetic parameters (kon, koff, and RTs). This allowed a full SKR study alongside a more traditional SAFIR analysis. The findings provide information on the structural requirements for a favorable kinetic profile at the hA3R and consequently may improve the in vitro to in vivo translation for hA3R antagonists.
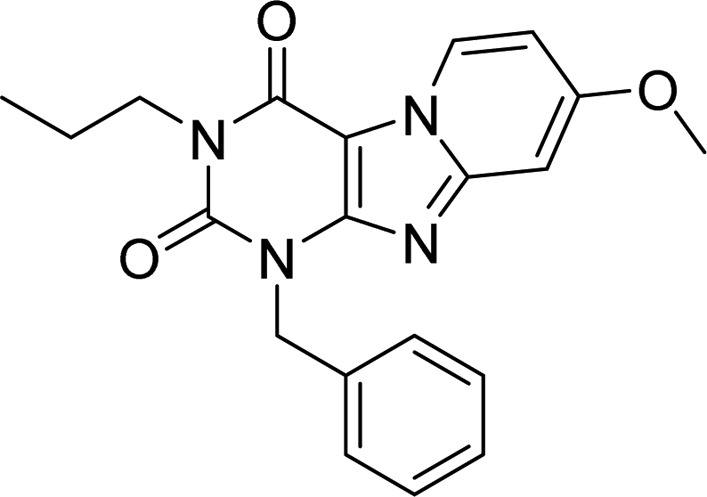
Table 1. Binding Affinity and Kinetic Parameters of 1-Benzyl-8-methoxy-3-propylpyrido[2,1-f]purine-2,4(1H,3H)-dione23,24.
| compd | pKia ± SEM (mean Ki in nM) | KRIb | konc (M–1 s–1) | koffd (s–1) | RTe (min) |
|---|---|---|---|---|---|
| 1 | 8.5 ± 0.02 (3.2) | 0.99 (0.97, 1.0) | (8.5 ± 1.2) × 105 | (3.2 ± 0.02) × 10–4 | 52 ± 0.3 |
pKi ± SEM (n ≥ 3, average Ki value in nM), obtained at 25 °C from radioligand binding assays with [3H]34 on human aenosine A3 receptors stably expressed on CHO cell membranes.
KRI (n = 2, individual estimates in parentheses), obtained at 10 °C from dual-point competition association assays with [3H]34 on human aenosine A3 receptors stably expressed on CHO cell membranes.
kon ± SEM (n ≥ 3), obtained at 10 °C from competition association assays with [3H]34 on human aenosine A3 receptors stably expressed on CHO cell membranes.
koff ± SEM (n ≥ 3), obtained at 10 °C from competition association assays with [3H]34 on human aenosine A3 receptors stably expressed on CHO cell membranes.
RT (min) = 1/(60 × koff).
Results and Discussion
Chemistry
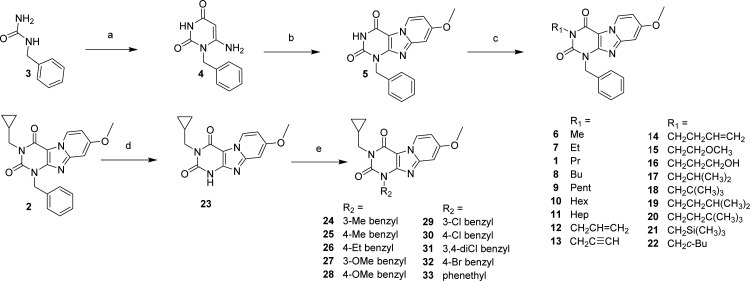
The synthesis approach shown in Scheme 1 was adapted from Priego et al.23,24 Starting from the commercially available materials benzylurea (3), ethyl cyanoacetate, and sodium methoxide. 1-benzyl-6-amino-uracil (4) was synthesized in an 88% yield.25 In situ dibromination of uracil 4 at the C5 position by N-bromosuccinimide, followed by cyclization with 4-methoxypyridine, gave the pyrido[2,1-f]purine-2,4-dione (5) in a one-pot reaction. Final compounds 1, 2, and 6–22 (as depicted in Table 1) were obtained, with yields varying in the range of 3–86%, by alkylating the N3 position of 5 using a variety of alkyl, alkenyl, and alkynyl bromides in acetonitrile and 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) as a base. Second, to be able to diversify on the N1 (R2) position, building block 23 had to be obtained. Full conversion of methylcyclopropyl compound 2 into the desired debenzylated 23 was realized by multiple additions of ammonium formate and Pd(OH)2 at 80 °C in ethanol overnight. Because of poor solubility, 23 was extracted with hot DMF and Pd(OH)2 was removed by filtration, resulting in a quantitative yield. Finally, various N1 substituted benzyl (24–32) and phenethyl (33) derivatives (Scheme 1) were made starting from the respective benzyl- or phenethyl bromides in DMF with K2CO3 used as base.
Scheme 1. Synthesis of 1,3-Disubstituted-1H,3H-pyrido[2,1-f]purine-2,4-dione Derivatives.
(a) ethyl cyanoacetate, NaOEt, EtOH, reflux, overnight; (b) (i) NBS, CH3CN, 80 °C, 1 h, (ii) 4-methoxypyridine, 80 °C, overnight; (c) R1-Br, DBU, CH3CN, 80 °C, overnight; (d) 20% Pd(OH)2, ammonium formate, EtOH, reflux, overnight; (e) R2-Br, K2CO3, DMF, 40 °C, overnight.
Biological Evaluation
All binding affinities of the pyrido[2,1-f]purine-2,4-dione derivatives were determined at 25 °C in a 2 h incubation protocol. All compounds were able to concentration-dependently inhibit specific [3H]8-ethyl-4-methyl-2-phenyl-(8R)-4,5,7,8-tetrahydro-1H- imidazo[2,1-i]-purin-5-one26 ([3H]PSB-11, 34) binding to the human adenosine A3 receptor, and their affinities are listed in Tables 1, 2 and 3. All compounds had (sub)nanomolar binding affinities ranging from 0.38 nM for compound 27 to 108 nM for compound 5.
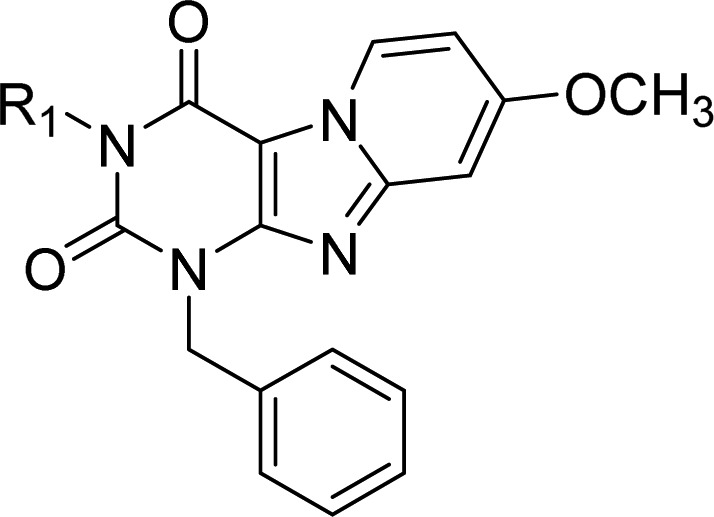
Table 2. Binding Affinities and Kinetic Parameters of Pyrido[2,1-f]purine-2,4-dione Derivatives with Modification on N-3 Position (R1 Group).
| compd | R1 | pKia ± SEM (mean Ki in nM) | KRIb | konc (M–1 s–1) | koffd (s–1) | RTe (min) |
|---|---|---|---|---|---|---|
| 5 | H | 7.0 ± 0.02 (108) | 0.38 ± 0.12 | (5.3 ± 1.5) × 105 | (1.4 ± 0.5) × 10–2 | 2.2 ± 1.4 |
| 6 | CH3 | 7.7 ± 0.1 (20.8) | 0.54 (0.52, 0.55) | ndf | nd | nd |
| 7 | CH2CH3 | 8.0 ± 0.1 (10.7) | 0.80 (0.85, 0.75) | nd | nd | nd |
| 8 | CH2CH2CH2CH3 | 8.8 ± 0.1 (1.5) | 1.29 (1.27, 1.31) | nd | nd | nd |
| 9 | CH2CH2CH2CH2CH3 | 8.5 ± 0.02 (3.5) | 1.11 (0.98, 1.24) | (1.1 ± 0.1) × 106 | (6.0 ± 0.5) × 10–4 | 28 ± 2.2 |
| 10 | CH2CH2CH2CH2CH2CH3 | 8.6 ± 0.1 (2.8) | 2.18 (2.15, 2.21) | (2.3 ± 1.0) × 105 | (8.2 ± 1.3) × 10–5 | 213 ± 35 |
| 11 | CH2CH2CH2CH2CH2CH2CH3 | 8.2 ± 0.2 (6.8) | 4.06 (3.66, 4.46) | (4.2 ± 0.3) × 105 | (6.2 ± 0.2) × 10–5 | 278 ± 45 |
| 12 | CH2CH=CH2 | 8.3 ± 0.1 (5.9) | 0.72 (0.46, 0.99) | nd | nd | nd |
| 13 | CH2C≡CH | 8.4 ± 0.02 (4.3) | 1.20 (1.16, 1.23) | nd | nd | nd |
| 14 | CH2CH2CH=CH2 | 8.9 ± 0.1 (1.4) | 1.23 (1.04, 1.41) | nd | nd | nd |
| 15 | CH2CH2OCH3 | 7.7 ± 0.2 (23) | 0.70 (0.70, 0.70) | (4.3 ± 0.8) × 105 | (6.3 ± 0.7) × 10–4 | 27 ± 2.6 |
| 16 | CH2CH2CH2OH | 7.1 ± 0.1 (81) | 1.04 ± 0.11 | nd | nd | nd |
| 17 | CH2CH(CH3)2 | 8.9 ± 0.02 (1.2) | 1.64 ± 0.24 | (7.8 ± 2.7) × 105 | (2.0 ± 0.8) × 10–4 | 148 ± 102 |
| 18 | CH2C(CH3)3 | 8.5 ± 0.1 (3.5) | 1.73 ± 0.28 | (5.5 ± 1.3) × 105 | (1.1 ± 0.4) × 10–4 | 250 ± 147 |
| 19 | CH2CH2CH(CH3)2 | 8.5 ± 0.04 (3.5) | 1.39 (1.23; 1.55) | nd | nd | nd |
| 20 | CH2CH2C(CH3)3 | 8.1 ± 0.02 (8.0) | 0.95 (1.02, 0.87) | nd | nd | nd |
| 21 | CH2Si(CH3)3 | 8.6 ± 0.03 (2.7) | 1.36 (1.26, 1.45) | nd | nd | nd |
| 2 | CH2C3H5 | 9.0 ± 0.02 (1.0) | 2.68 ± 0.48 | (2.8 ± 0.5) × 106 | (6.0 ± 1.7) × 10–5 | 315 ± 105 |
| 22 | CH2C4H7 | 8.6 ± 0.03 (2.7) | 1.48 (1.66, 1.30) | nd | nd | nd |
pKi ± SEM (n ≥ 3, average Ki value in nM), obtained at 25 °C from radioligand binding assays with [3H]34 on human adenosine A3 receptors stably expressed on CHO cell membranes.
KRI ± SEM (n = 3) or KRI (n = 2, individual estimates in parentheses), obtained at 10 °C from dual-point competition association assays with [3H]34 on human adenosine A3 receptors stably expressed on CHO cell membranes.
kon ± SEM (n ≥ 3), obtained at 10 °C from competition association assays with [3H]34 on human adenosine A3 receptors stably expressed on CHO cell membranes.
koff ± SEM (n ≥ 3), obtained at 10 °C from competition association assays with [3H]34 on human adenosine A3 receptors stably expressed on CHO cell membranes.
RT (min) = 1/(60 × koff).
nd = not determined.
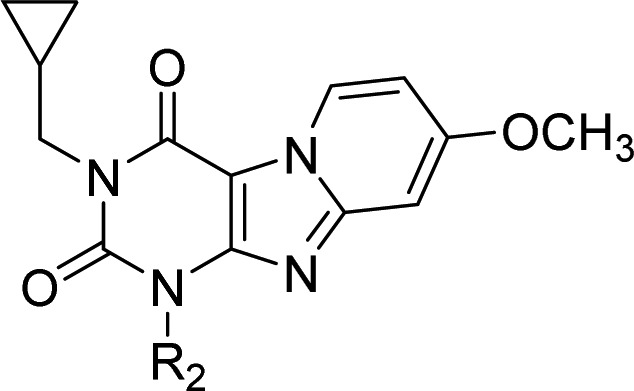
Table 3. Binding Affinities and Kinetic Parameters of Pyrido[2,1-f]purine-2,4-dione Derivatives with Modification at R2.
| compd | R2 | pKia ± SEM (mean Ki in nM) | KRIb | konc (M–1 s–1) | koffd (s–1) | RTe (min) |
|---|---|---|---|---|---|---|
| 2 | benzyl | 9.0 ± 0.02 (1.0) | 2.68 ± 0.48 | (2.8 ± 0.5) × 106 | (6.0 ± 1.7) × 10–5 | 315 ± 105 |
| 24 | 3-CH3-benzyl | 8.8 ± 0.02 (1.5) | 1.18 (1.18, 1.17) | ndf | nd | nd |
| 25 | 4-CH3-benzyl | 9.0 ± 0.1 (0.92) | 1.15 (1.03, 1.27) | nd | nd | nd |
| 26 | 4-CH2CH3-benzyl | 9.2 ± 0.04 (0.71) | 0.81 (0.82, 0.79) | nd | nd | nd |
| 27 | 3-OCH3-benzyl | 9.4 ± 0.03 (0.38) | 2.24 (2.32, 2.15) | (4.8 ± 0.2) × 105 | (4.7 ± 0.7) × 10–5 | 376 ± 58 |
| 28 | 4-OCH3-benzyl | 8.9 ± 0.01 (1.4) | 1.39 (1.22, 1.55) | (4.8 ± 0.1) × 105 | (7.8 ± 2.0) × 10–5 | 250 ± 72 |
| 29 | 3-Cl-benzyl | 8.3 ± 0.02 (4.9) | 0.89 (1.06, 0.72) | (8.2 ± 1.3) × 105 | (4.7 ± 0.7) × 10–4 | 36 ± 5.5 |
| 30 | 4-Cl-benzyl | 8.9 ± 0.01 (1.2) | 1.11 (1.02, 1.20) | (3.0 ± 0.3) × 106 | (8.2 ± 0.2) × 10–4 | 20 ± 0.5 |
| 31 | 3,4-dichlorobenzyl | 8.3 ± 0.01 (5.3) | 3.12 (3.49, 2.75) | (1.0 ± 0.1) × 105 | (5.3 ± 1.5) × 10–5 | 391 ± 137 |
| 32 | 4-Br-benzyl | 8.9 ± 0.1 (1.2) | 1.19 (1.30, 1.08) | nd | nd | nd |
| 33 | phenethyl | 8.1 ± 0.04 (7.7) | 1.09 (1.21, 0.97) | nd | nd | nd |
pKi ± SEM (n ≥ 3, average Ki value in nM), obtained at 25 °C from radioligand binding assays with [3H]34 on human adenosine A3 receptors stably expressed on CHO cell membranes.
KRI ± SEM (n = 3) or KRI (n = 2, individual estimates in parentheses), obtained at 10 °C from dual-point competition association assays with [3H]34 on human adenosine A3 receptors stably expressed on CHO cell membranes.
kon ± SEM (n ≥ 3), obtained at 10 °C from competition association assays with [3H]34 on human adenosine A3 receptors stably expressed on CHO cell membranes.
koff ± SEM (n ≥ 3), obtained at 10 °C from competition association assays with [3H]34 on human adenosine A3 receptors stably expressed on CHO cell membranes.
RT (min) = 1/(60 × koff).
n.d. = not determined.
Subsequently, the human adenosine A3 receptor ligands were screened in a so-called “dual-point” competition association assay,27 allowing for the semiquantitative estimation of the compounds’ dissociation rates and therefore the compounds’ RTs. The specific binding of [3H]34 was measured after 20 and 240 min in the absence and presence of a single concentration (i.e., 1 × IC50) of unlabeled human adenosine A3 receptor antagonists, which yielded their kinetic rate index (KRI). A long RT compound shows a characteristic “overshoot” followed by a steady decrease in specific binding until a new equilibrium is reached; in such a case. the KRI value is greater than unity. Conversely, a ligand with a fast dissociation rate is represented by a more shallow curve, yielding a KRI value smaller than one when dividing the binding at t1 by the binding at t2. The KRI values in the series ranged from 0.38 to 4.06 (Table 1, 2, and 3).
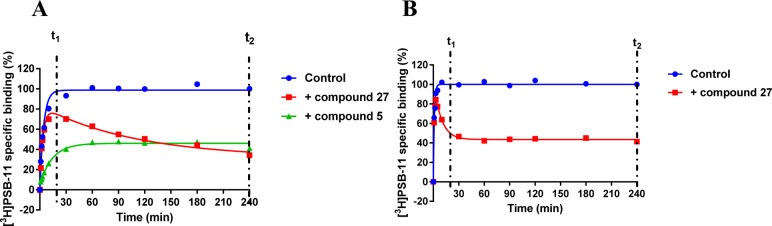
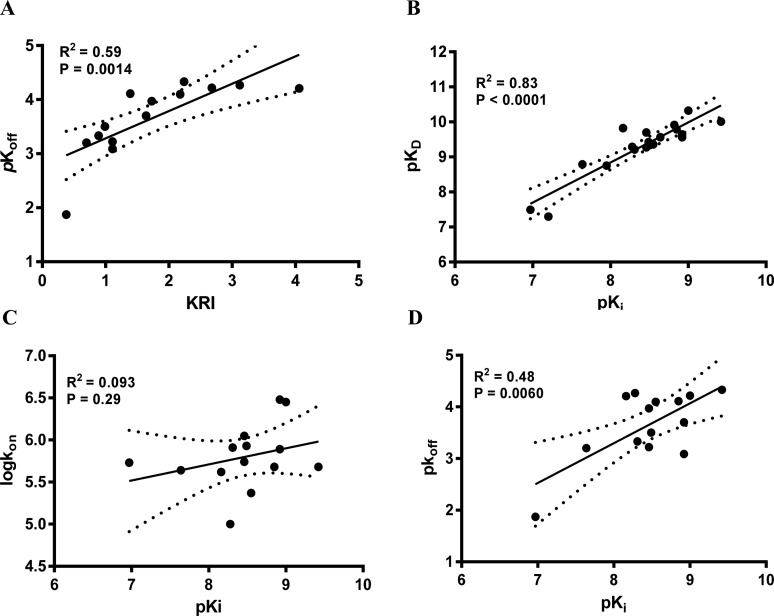
Compounds with a KRI value less than 0.7 or greater than 1.5 were selected for complete kinetic characterization through the use of a competition association assay with [3H]34 (Figure 1A). To obtain extensive structure–kinetics relationships (SKRs), close structural analogues (9, 28, 29, and 30) of 1 were also tested to obtain their association (kon) and dissociation (koff) rate constants. Association rate constants varied by 30-fold, ranging from (1.0 ± 0.1) × 105 M–1 s–1 for antagonist 31 to (3.0 ± 0.3) × 106 M–1 s–1 for antagonist 30 (Table 3). Interestingly, there was an approximately 290-fold difference in dissociation rate constants, reflecting the divergent KRI values. Antagonist 5 had the fastest dissociation rate constant of (1.4 ± 0.5) × 10–2 s–1 and thus the shortest RT of 2.2 min, while both antagonist 27 and 31 had the slowest dissociation rate constants of (4.7 ± 0.7) × 10–5 s–1 and (5.3 ± 1.5) × 10–5 s–1, respectively, and thus the longest RTs of 376 and 391 min, respectively. Notably, the long RT antagonist 27 (Figure 1A) displayed a typical “overshoot” in the competition association curve, indicative of a slower dissociation than the radiolabeled probe [3H]34, while the short RT antagonists, exemplified by antagonist 5 (Figure 1A), presented more shallow, gradually ascending curves. There was a good correlation between the negative logarithm of the antagonists’ dissociation rate constants and their KRI values derived from the kinetic screen (Figure 2A), which confirmed that a compound’s KRI value is a good predictor for its dissociation rate constant. Notably, the experimental temperatures in the kinetic assays were lower than in the equilibrium displacement assays (25 °C vs 10 °C) because kinetic studies performed at 25 °C were compromised by the nature of the compounds tested. This is shown in Figure 1B, where the “overshoot” of long RT antagonist 27 happened before the t1 checkpoint of 20 min, which did not happen at 10 °C. A significant correlation was also observed between the antagonist affinities (Ki values) determined in equilibrium displacement experiments and their kinetic KD values derived from competition association experiments (Figure 2B), despite the differences in assay temperature (25 °C vs 10 °C). Interestingly, the kinetic association rate constants (kon) did not show any significant correlation with affinity (Figure 2C), while the dissociation rate constants (koff) had a fair correlation with affinity (Figure 2D).
Figure 1.
(A) Representative competition association assay curves of [3H]34 in the absence (control) or presence of a long residence time compound 27 and a short residence time compound 5. Experiments were performed at 10 °C using the compound’s respective IC50 value at the hA3R. (B) Competition association curves of [3H]34 in the absence (control) or presence of long residence time compound 27. Experiments were performed at 25 °C using the compound’s respective IC50 value at the hA3R. t1 is the radioligand binding at 20 min, while t2 is the radioligand binding at 240 min.
Figure 2.
Correlations between the negative logarithm of the human adenosine A3 receptor antagonists’ dissociation rates (pkoff) and their kinetic rate index (KRI) (A), the human adenosine A3 receptor antagonists’ affinity (pKi) and their “kinetic KD” (pKD) (B), association rate constants (log kon) (C), and dissociation rate constants (pkoff) (D). The central line corresponds to the linear regression of the data, the dotted lines represent the 95% confidence intervals for the regression. Data used in these plots are detailed in Tables 1–3. Data are expressed as mean from at least three independent experiments.
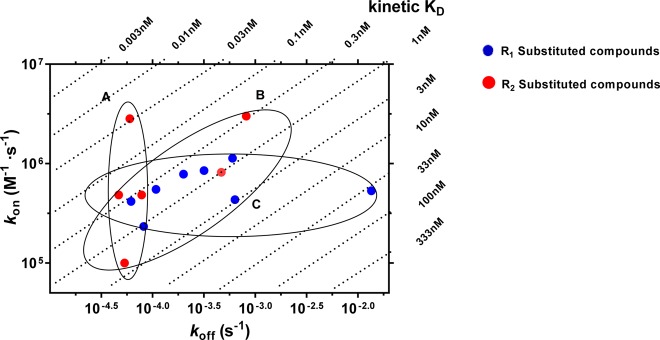
The representative long RT and short RT antagonists (27 and 5) were selective for the hA3 receptor when compared to other adenosine receptors (i.e., human adenosine A1 and A2A receptor, Supporting Information, Table S1). These two antagonists (27 and 5) with comparable association rate constants but distinct dissociation rate constants (or RTs) were further analyzed in a [35S]GTPγS binding assay in which we studied the (in)surmountable antagonism induced by the two compounds (Figure 3). Moreover, a kon–koff–KD “kinetic map” (Figure 4) was constructed based on the compounds’ divergent affinities (expressed as kinetic KD values) and kinetics parameters, yielding a division of these antagonists into three different subcategories: antagonists that show similar koff values (<2-fold) but due to differing kon values (>28-fold) have different KD values (∼100-fold, group A), antagonists that display similar KD values (<10-fold) despite showing divergent koff and kon values (17-fold and 30-fold, group B), and antagonists with similar kon values (<5-fold) but due to differing koff values (∼290-fold) have different KD values (>110-fold, group C). Additionally, we applied molecular modeling to compare the binding behavior in some molecular detail of several antagonists with similar affinities (2 vs 10; 31 vs 29 or 30) (Figure 5).
Figure 3.
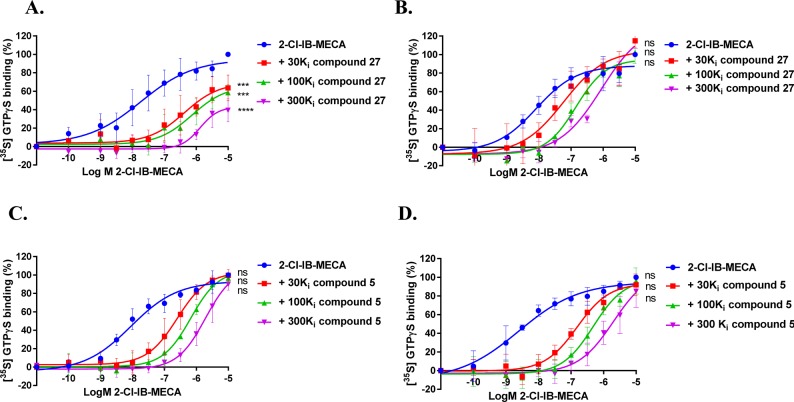
2-Cl-IB-MECA-stimulated [35S] GTPγS binding to hA3R stably expressed on CHO cell membranes (25 °C) in the absence or presence of long-residence-time antagonist 27 (A and B, normalized and combined, n ≥ 3) or short-residence-time antagonist 5 (C and D, normalized and combined, n ≥ 3). Antagonist 27 (A) and 5 (C) were incubated for 60 min prior to the challenge of the hA3R agonist 2-Cl-IB-MECA, at a concentration ranging from 0.1 nM to 10 μM, for another 30 min. Antagonist 27 (B) and 5 (D) were coincubated with 2-Cl-IB-MECA, at the same concentration range, for 30 min. The agonist curves were generated in the presence of increasing concentrations of antagonists, namely 30-, 100-, and 300-fold their respective Ki values. Curves were fitted to a four parameter logistic dose–response equation. Data is from at least three independent experiments performed in duplicate, normalized according to the maximal response (100%) produced by 2-Cl-IB-MECA alone. The shift in agonist EC50 values was determined to perform Schild analyses. Two-way ANOVA with Dunnett’s post-test was applied for the comparison of Emax by agonist control, * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, **** p < 0.0001, ns for not significant.
Figure 4.
Kinetic map (y axis, kon in M–1 s–1; x axis, koff in s–1) of all compounds that were kinetically characterized in this study. kon and koff values were obtained through competition association assays performed at the hA3R. The kinetically derived affinity (KD = koff/kon) is represented through diagonal parallel lines. Group A: compounds that show similar koff values but due to differing kon values have different KD values. Group B: compounds that display similar KD values despite showing divergent koff and kon values. Group C: compounds with similar kon values, but due to differing koff values have different KD values.
Figure 5.
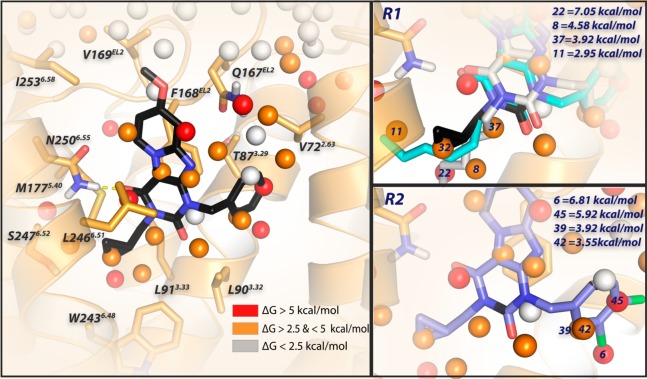
Docking of antagonist 2 into the binding site of the homology model of the adenosine A3 receptor based on the crystal structure of the adenosine A2A receptor (PDB 4EIY).41 Antagonist 2 is represented by black sticks, and residues within 5 Å of 2 are visualized as orange sticks. The protein is represented by orange ribbons. Ligand and residues atoms color code: red = oxygen, blue = nitrogen, white = hydrogen. The overlay of consecutively numbered hydration sites (colored spheres; for color code, see below) were calculated by WaterMap (left). Hydration sites shown as red and orange spheres represent positions were “unstable” water molecules can be found, which should be displaced by antagonist 2. White spheres symbolize “stable” water molecules, which are in exchange with the bulk solvent. Two different binding modes are represented for antagonist 10 (cyan and gray sticks), which shows that the flexible hexyl chain can displace different hydration sites (8 for gray and 11 for cyan). For the key hydration sites (8, 11, 22, 32, 37) surrounding the lipophilic “tails”, calculated ΔG values (in kcal/mol) with respect to bulk solvent are shown (upper right). Hydration sites 6, 39, 42, and 45 are proposed to be displaced by the 3,4 dichloro substituents of 31; calculated ΔG values (in kcal/mol) with respect to bulk solvent are shown (lower right).
Structure–Affinity Relationships (SAFIRs) and Structure–Kinetics Relationships (SKRs)
According to previous studies from our group,23,24 methoxy-substitution at the C8 position (Table 1) of the pyrido[2,1-f]purine-2,4-dione scaffold yielded selective hA3R antagonists with good affinity (3.2 nM for 1 as a reference compound). From our preliminary studies, this methoxy-group appeared important for slow dissociation (1 vs compound S2 from Supporting Information, Figure S1, Table S2). Because of the nanomolar affinity and close-to-unity KRI value of 1, it was treated as the starting point of this SAFIR and SKR study, having, on further analysis, an association rate constant of (8.5 ± 1.2) × 105 M–1 s–1 and a dissociation rate constant of (3.2 ± 0.02) × 10–4 s–1 (RT = 52 min). Next, we decided to investigate R1 substitutions (Table 2), beginning with antagonist 5 (R1 = H).
The Substitutions at R1 (Table 2)
First, an increase in alkyl chain length was investigated, indicating an elongated carbon chain had a cumulative effect on KRI (5, 6, 7, 8, 10, 11), with the exception of antagonist 9 (KRI values from 0.38 to 4.06). One could point to a possible correlation between lipophilicities and dissociation rate constants (and consequently RTs) to explain this trend (Supporting Information, Figure S2A). However, with all of the antagonists kinetically characterized, no such correlation was observed (Supporting Information, Figure S2B). Therefore, other reasons should be taken into account as to why elongating the carbon chain has such a profound effect on the ligand’s dissociation rate. The role of membrane–drug interactions in determining the pharmacological profile is a possible reason, especially the role long carbon tails have in such interactions.28 It is interesting to point out that the affinity of antagonist 11, which had a traditional lead selection process taken place, would most likely have resulted in the elimination of this compound due to the more favorable affinity and hydrophilic properties of its shorter carbon chain counterparts (8 or 9) (affinities, 6.8 nM vs 1.5 nM or 3.5 nM; KRI values, 4.06 vs 1.29 or 1.11). This would have overlooked the efficacy this compound could offer due to its longer residence time.
Second, the presence of a more rigid substitution of the R1 group of saturated equivalents (antagonists 1 and 8) led to antagonists 12 and 14, with similar improvement in the affinity pairs (12 and 14, 5.9 and 1.4 nM; 1 and 8, 3.2 and 1.5 nM) and KRI values (12 and 14, 0.72 and 1.23; 1 and 8, 0.99 and 1.29). Further rigidification with alkyne (13) rather than alkene (12) maintained affinities (4.3 vs 5.9 nM) and increased KRI values (1.20 vs 0.72). This alkyne could be the starting point for a further study on “click-chemistry” for introducing, e.g., fluorescent tags.29−31
Third, the introduction of a polar atom or group in antagonist 15 or 16, respectively, led to a decrease in affinity compared to their nonpolar counterpart 1 (23 or 81 nM vs 3.2 nM). The changes in KRI values between antagonist 1 and its polar counterparts 15 and 16 can be considered minor (0.99 vs 0.70 and 1.04). Of note, by comparing affinities and kinetic profiles of polar antagonist 15 with its nonpolar equivalent 1, we found the polarity at the “lipophilic carbon chain” resulted in slower association (kon of (4.3 ± 0.8) × 105 M–1 s–1 vs (8.5 ± 1.2) × 105 M–1 s–1) but faster dissociation (koff of (6.3 ± 0.7) × 10–4 s–1 vs (3.2 ± 0.02) × 10–4 s–1), with a concomitant decrease in affinity (23 vs 3.2 nM).
Moreover, the bulkiness of the substituents was studied with branched carbon side chains (17, 18, 19, 20, and 21) or aliphatic rings (2 and 22). As to the branched carbon side chains, compound affinities remained in the nanomolar range, while in terms of KRI values, 2-carbon-linker branched side chains (17 and 18) caused larger KRI values than those of either their linear counterparts (8 and 9) or 3-carbon-linear branched side chains (19 and 20) (17, 1.64 vs 1.29 or 1.39; 18, 1.73 vs 1.11 or 0.95). Although the association rate constants of 17 and 18 were similar to other antagonists in Table 2, the dissociation rate constants suggest their branched side chains have an extra “anchoring” effect compared with the linear counterparts. For example, the koff of 18 with a 5-carbon branched side chain was quite similar to 10 or 11, having a 6 or 7-carbon linear side chain ((1.1 ± 0.4) × 10–4 s–1 vs (8.2 ± 1.3) × 10–5 s–1 or (6.2 ± 0.2) × 10–5 s–1). The presence of a slightly less polar but larger silicon atom (21) instead of carbon (18) made the KRI value decrease (1.36 vs 1.73), although the affinity remained virtually the same (2.7 vs 3.5 nM).
Interestingly, another reported analogue (2)23 of compound 1, with cyclopropylmethyl substitution at the R1 group, led to unique kinetic parameters, i.e., a combination of a fast association rate constant ((2.8 ± 0.5) × 106 M–1 s–1 vs (8.5 ± 1.2) × 105 M–1 s–1) and a slow dissociation rate constant ((6.0 ± 1.7) × 10–5 s–1 vs (3.2 ± 0.02) × 10–4 s–1), although the affinities of 2 and 1 were similar (1.0 ± 0.03 nM vs 3.2 ± 0.1 nM, respectively). The RT of compound 2 was the longest in Table 2 with 315 min. For the antagonist with cyclobutylmethyl (22), affinity (2.7 vs 1.0 nM) and KRI value (1.48 vs 2.68) were lower than for compound 2.
Although the dissociation rate constants of the antagonists in Table 2 varied greatly depending on the R1 substituent, the association rate constants were more similar (within 5-fold). Association rate constants are often reasoned to be caused by a diffusion limited process whereby the collision rate of ligand and receptor determines the rate of ligand–receptor complex formation.32 When no conformational changes are required for the receptor and ligand to bind and when taking into account the proportion of the receptor responsible for binding, this sets the association rate constant at observed limits of around 107 M–1 s–1.33 As the association rate constants for all R1 substituted compounds were slower than the diffusion limit by at least 3.5 fold (2), we hypothesize target engagement for R1 substituted antagonists is more hampered than imposed by the diffusion limit.
The Substitutions at R2 Group (Table 3)
From Table 2, we learned that cyclopropylmethyl-substituted antagonist 2 exhibited a kinetic profile as a long RT compound while showing the affinity previously reported.23 As a result, this compound became the starting point for our exploration of the substitutions (R2 group) on the aromatic ring.
Introduction of a nonpolar alkyl substituent on antagonist 2’s benzyl ring (24, 25, 26), resulted in a decrease in KRI values (from 2.68 to 0.81), while slight variations in affinity were observed.
Then, introduction of a polar methoxy substituent on antagonist 2’s benzyl ring led to mixed results with a small decrease in RT at para-position and a slight increase in RT at meta-position in 28 (250 vs 315 min) and 27 (376 vs 315 min), respectively. In particular, the long residence time for 27 in combination with its subnanomolar affinity (0.38 nM) made this compound stand out in the series.
Next, halogen substitutions on antagonist 2’s benzyl ring were examined. Apparently, the position of halogen substitution is important for affinity as para-substitution in antagonist 30 and 32 yielded similar affinity compared to 2 (1.2 vs 1.2 vs 1.0 nM). The one compound with meta-substitution, 29, showed a 5-fold decrease in affinity compared to 2 (4.9 vs 1.0 nM). Dichloro-substituted compound 31 had the largest KRI value (3.12) among the halogen-substituted antagonists; the para-bromo substituted compound 32 was similar in this respect to para-chloro substituted 30 (1.19 vs 1.11). In a full competition association experiment, we determined the rate constants for 31 and learned it had the longest RT of all compounds kinetically characterized (391 min), concomitant with the slowest association rate constant of the compounds kinetically characterized ((1.0 ± 0.1) × 105 M–1 s–1). Previous theoretical studies have indicated the strength of halogen bonding can be increased through the introduction of electron withdrawing groups onto halobenzenes.34 Such would be the case for 31, where the additional chloro substituent forms a stronger halogen bonding interaction with the R2 binding pocket. Introducing a phenethyl (33) rather than benzyl substituent (2) led to a decrease in affinity (7.7 vs 1.0 nM), while the KRI value was also strongly affected (1.09 vs 2.68). This observation parallels our previous findings that the binding pocket for the R2 substituent is of limited size.23
Functional Assay
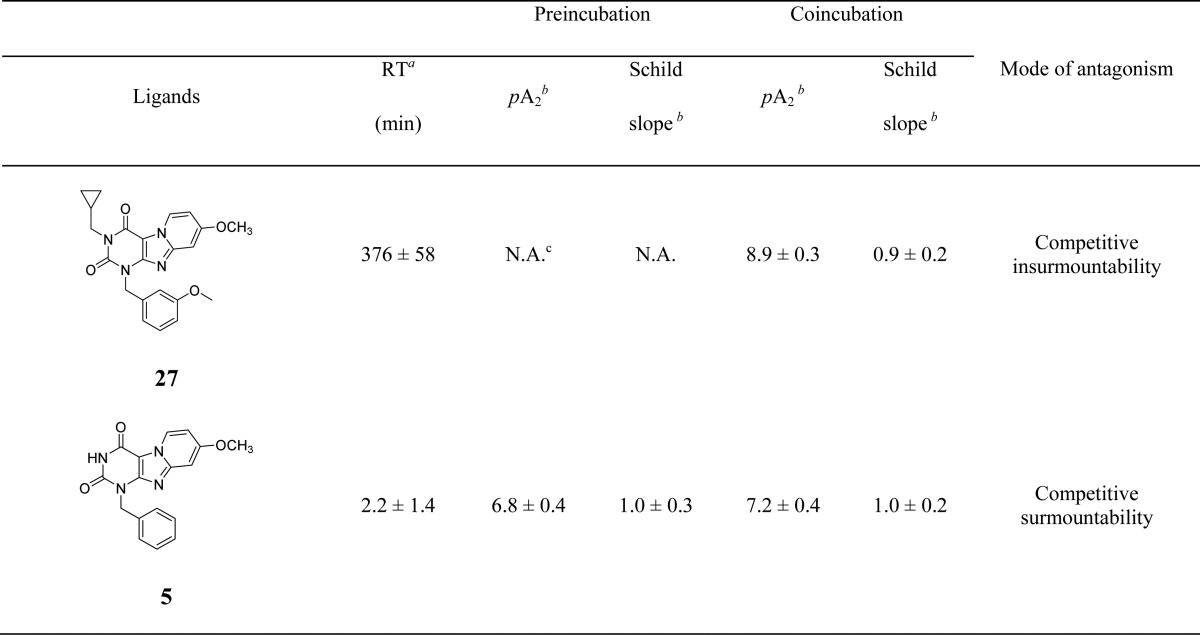
Following kinetic characterization, a long (27) and a short (5) RT compound were chosen for functional characterization in a [35S]GTPγS binding assay, also because for these two compounds the kon values were similar (4.8 ± 0.2) × 105 M–1 s–1 vs (5.3 ± 1.5) × 105 M–1 s–1). This difference allowed a possible link to be made between RTs and efficacies. Pretreatment of hA3 receptor membranes with increasing concentrations of the long RT antagonist 27, before stimulation by the A3 receptor agonist 2-Cl-IB-MECA, induced insurmountable antagonism. In other words, the 2-Cl-IB-MECA concentration–effect curves were shifted to the right with a concomitant decrease in the maximal response (Figure 3A). Conversely, the short RT antagonist 5 displayed surmountable antagonism, shifting 2-Cl-IB-MECA’s curves to the right without affecting its maximum effect (Figure 3B). In this experimental setup, the Schild-slope of 5 generated from Schild-plots was close to unity (Table 4), and the compound’s pA2 value was comparable with its pKi value (6.8 ± 0.4 vs 7.0 ± 0.02). We also performed coincubation experiments with these antagonists in the presence of 2-Cl-IB-MECA. In this experimental setup, all antagonists produced a rightward shift of the 2-Cl-IB-MECA concentration–effect curves without a suppression of the maximal response (Figure 3C,D). Notably, the Schild-slopes of both long and short RT antagonists (27 and 5) were close to unity (0.9 ± 0.2 for 27, 1.0 ± 0.2 for 5, Table 4). In addition, the pA2 value of 5 was comparable with the result from the preincubation condition (7.2 ± 0.4 vs 6.8 ± 0.4, Table 4), and the pA2 value of 27 was also in agreement with its pKi value (8.9 ± 0.3 vs 9.4 ± 0.03).
Table 4. Functional Activity of hA3 Receptor Antagonists from [35S]GTPγS Binding Assays.
Kinetic Map
Using the association (kon) and dissociation (koff) rate constants obtained from competition association experiments (Tables 1–3), a kinetic map (Figure 4) was constructed by plotting these values on the y-axis and x-axis, respectively. The dashed diagonal parallel lines represent the kinetically derived KD values (KD = koff/kon). Out of this map, three subgroups emerged. Group A represents compounds that exhibit similar koff values but with vastly different kon values. As a consequence, a diverse range of KD values was observed. Previous SKR studies have primarily focused on optimizing dissociation rates and RTs for predicting in vivo efficacy and creating a kinetically favorable ligand. Yet recently, there has been greater acknowledgment of the important role that the association rate constants may play in determining the efficacy of a drug as the result of increased rebinding or increased drug–target selectivity.19 A kinetic map would thus allow for the selection of compounds with appropriate RTs while exploring the role of association rate constants in determining efficacy by choosing a rapidly or slowly associating compound, i.e., 2 or 31 ((2.8 ± 0.5) × 106 M–1 s–1 vs (1.0 ± 0.1) × 105 M–1 s–1). Group B displays ligands that exhibit a narrow range of affinity (KD: 0.1–1 nM) yet a wide range of koff values that result in RTs ranging from 20 to 391 min. This information would have gone unnoticed in a traditional SAFIR hit-to-lead approach and would most likely have led to the selection of high affinity compounds not in possession of a potentially efficacy promoting long residence time. Thus, combining SAFIR with SKR aspects in lead optimization would allow the selection of not only potent but also long RT compounds through the drug development pipeline. Lastly, group C represents compounds that present similar kon values but due to differing koff values show considerable differences in affinities (KD). This illustrates the differences that were observed in the binding kinetics of the R1 and R2 substituents, as group C mainly consists of R1 substituents (noncyclopropylmethyl substituents), while group A mainly consists of R2 substituents (cyclopropylmethyl substituents). This difference also suggests a different mode of receptor–ligand interaction during the binding process of the two ligand groups.
Altogether, the construction of a kinetic map allows for a more detailed categorization of compounds’ affinities as dictated by their kinetic rate constants. In previous studies, such a separation has explained the different therapeutic effects molecules exhibit highlighting the benefits of such an in-depth analysis.35,36
Given the putative link between RT and clinical efficacy, it may be postulated that the lack of hA3R antagonists progressing from preclinical trials is due to insufficient selection criteria employed in these initial phases of hA3R drug screening. As previously reported, hA3R antagonists are reasoned to be beneficial in the treatment of chronic obstructive pulmonary disease (COPD).37 For this indication, a number of antagonists are available that act at the muscarinic M3 receptor.38 For these therapeutics, their dosing regime and thus duration of action have been linked to their RT. For example, aclidinium, which requires a twice daily dosing regimen, exhibits a much shorter RT than tiotropium that in turn requires only once daily dosing.16 This extended duration of action that enables long-lasting efficacy and practical dosing regimens at the muscarinic M3 receptor is thought to be a beneficial feature in the treatment of chronic illnesses.39,40 As hA3R antagonists can be used to treat chronic COPD but also a number of other chronic disorders, we could imagine that considering the ligand’s kinetic profile early in the drug screening process would reduce the likelihood of failure due to insufficient efficacy in future clinical trials. Perhaps when selecting hA3R antagonists with a favorable long RT, i.e., group A in the kinetic map, will we see the therapeutic potential of the hA3R fulfilled.
Computational Studies
Finally, we decided to further investigate the ligand–receptor interactions using a homology model of the adenosine A3 receptor, based on the crystal structure of the adenosine A2A receptor (PDB 4EIY).41 WaterMap calculations were applied to try and explain the variance in kinetic profiles of different ligands by unfavorable hydration.42,43
Antagonist 2 (in black stick representation) was docked in the homology model. As a first step, it was placed inside the transmembrane bundle, with the tricyclic ring system surrounded by TM3, TM6, and EL2. Hydrogen bonding was constrained between the amide-hydrogen (−NH2, δ+) from Asn2506.55 and the carbonyl-oxygen (−C=O, δ–) at the C4-position of the pyrido[2,1-f]purine-2,4-dione scaffold (Figure 5, left). To compare differences between the ligands, an “apo” WaterMap of the hA3 receptor was generated. Hydration sites shown as red and orange spheres represent positions where “unstable” water molecules are found. Antagonist 10 (hexyl-substitution), with comparable koff ((8.2 ± 1.3) × 10–5 s–1 vs (6.0 ± 1.7) × 10–5 s–1) to 2 but 10-fold slower kon ((2.3 ± 1.0) × 105 M–1 s–1 vs (2.8 ± 0.5) × 106 M–1 s–1), was docked with two different binding modes in the same binding site (Figure 5 upper right, cyan and gray sticks). We found additional unstable waters (8, 11, 22 in Figure 5 upper right) surrounding the lipophilic substituents of the compounds, which could be explained as hindrance when the antagonist is associating with the binding site.
The same WaterMap was used to investigate the kinetic profile of antagonist 31. Indeed, hydration sites 6, 39, 42, and 45 are proposed to be displaced by the 3,4-dichloro substituent. Thus, both the association and dissociation of 31 were slowed down by these unstable waters. For the association process, the lipophilic 3,4-dichloro moiety has difficulty in approaching the occupied unstable hydration sites ((1.0 ± 0.1) × 105 M–1 s–1, slowest kon in the whole series); the same lipophilic 3,4-dichloro substituent seems to provide more stabilization to the receptor–ligand complex, thus hampering the dissociation process ((5.3 ± 1.5) × 10–5 s–1, slowest koff in the whole series). Interestingly, by removing a single chloro atom at either the 3- or 4- position on the benzyl-ring (30 or 29), association and dissociation rate constants became faster by approximately 10-fold. Although the differences in their kon and koff values were modest (2–3 fold), the unstable hydration sites may prevent the 4-chloro-substituted antagonist 30 from reaching the hydration sites 6, 39, and 42 that interact with the 3-Cl substituent; consequently, both its association and dissociation rate constants were faster than of the 3-chloro-substituted counterpart 29 (kon, (3.0 ± 0.3) × 106 M–1 s–1 vs (8.2 ± 1.3) × 105 M–1 s–1; koff, (8.2 ± 0.2) × 10–4 s–1 vs (4.7 ± 0.7) × 10–4 s–1).
Conclusions
We have demonstrated that, next to affinity, additional knowledge of target binding kinetics is useful for selecting and developing new hA3R antagonists in the early phase of drug discovery. By introducing proper substituents at the N3 position or the N1 benzyl ring of a series of pyridopurinediones, divergences in kinetic profiles were observed, while almost all compounds had high and often similar affinity. Two representative ligands (5 and 27) were tested in [35S]GTPγS binding assays, confirming the link between their RTs and their (in)surmountable antagonism. According to these findings, a kon–koff–KD kinetic map was constructed and subsequently the antagonists were divided into three subgroups. Additionally, we also performed a computational modeling study that sheds light on the crucial interactions (including with water molecules) for both the association and dissociation kinetics of this family of antagonists. It should be mentioned that the kinetic parameters were derived at the hA3R, which may be different in, e.g., rodents used in advanced animal models. Still, this study suggests that favorable long RTs would be a proper indicator in the development of hA3R antagonists for chronic inflammatory conditions, e.g., COPD.
Experimental Section
Chemistry
All solvents and reagents were purchased from commercial sources and were of analytical grade. Distilled water will be referred to as H2O. TLC analysis was performed to monitor the reactions, using Merck silica gel F254 plates. Grace Davison Davisil silica column material (LC60A, 30–200 μm) was used to perform column chromatography. Microwave reactions were performed in an Emrys Optimizer (Biotage AB, formerly Personal Chemistry). 1H and 13C NMR spectra were recorded on a Bruker DMX-400 (400 MHz) spectrometer, using tetramethylsilane as internal standard. Chemical shifts are reported in δ (ppm) and the following abbreviations are used: s, singlet; d, doublet; dd, double doublet; t, triplet; m, multiplet. The analytical purity of the final compounds is 95% or higher and was determined by high-performance liquid chromatography (HPLC) with a Phenomenex Gemini 3 μm C18 110A column (50 mm × 4.6 mm, 3 μm), measuring UV absorbance at 254 nm. The sample preparation and HPLC method was as follows: 0.3–0.6 mg of compound was dissolved in 1 mL of a 1:1:1 mixture of CH3CN/H2O/t-BuOH and eluted from the column within 15 min at a flow rate of 1.3 mL/min. The elution method was set up as follows: 1–4 min isocratic system of H2O/CH3CN/1% TFA in H2O, 80:10:10; from the fourth min, a gradient was applied from 80:10:10 to 0:90:10 within 9 min, followed by 1 min of equilibration at 0:90:10 and 1 min at 80:10:10. Liquid chromatography–mass spectrometry (LC–MS) analyses were performed using a Thermo Finnigan Surveyor—LCQ Advantage Max LC–MS system and a Gemini C18 Phenomenex column (50 mm × 4.6 mm, 3 μm). The elution method was set up as follows: 1–4 min isocratic system of H2O/CH3CN/1% TFA in H2O, 80:10:10; from the fourth min, a gradient was applied from 80:10:10 to 0:90:10 within 9 min, followed by 1 min of equilibration at 0:90:10 and 1 min at 80:10:10.
1-Benzyl-8-methoxy-1H,3H-pyrido[2,1-f]purine-2,4-dione (5)24
6-Amino-1-benzyluracil (4)25 (10.8 g, 49.7 mmol, 1.00 equiv) was suspended in CH3CN (370 mL). N-Bromosuccinimide (17.7 g, 99.4 mmol, 2.00 equiv) was added to the suspension, and the mixture was heated at 80 °C for 1 h, after which full conversion was shown by TLC (1:9 CH3OH/CH2Cl2 + 3% triethylamine). Subsequently, 4-methoxypyridine (15.1 mL, 149.2 mL, 3.00 equiv) was added and the mixture was heated at 80 °C during 10 h. Full consumption of the bromo intermediate was shown by TLC (1% CH3OH/CH2Cl2). A precipitate was formed overnight at RT, which was collected by filtration and washed with diethyl ether. This yielded the desired compound as a white solid (10.2 g, 31.6 mmol, 64%). 1H NMR (400 MHz, DMSO-d6) δ: 11.11 (s br, 1H), 8.72 (d, J = 7.2 Hz, 1H), 7.39–7.29 (m, 4H), 7.28–7.22 (m, 2H), 6.91 (dd, J = 7.2, 2.0 Hz, 1H), 5.19 (s, 2H), 3.89 (s, 3H) ppm. NMR was according to literature data.24
General Procedure for the Preparation of N3-Substituted 1-Benzyl-8-methoxy-1H,3H-pyrido[2,1-f]purine-2,4-diones (1, 2, 6–22).24
The compounds were synthesized using the procedure described by Priego et al.,24 but 5 equiv of the alkyl halide was used instead of 1.5 equiv and the reaction mixture was heated to 80 °C overnight in all cases. The pure compounds were obtained by silica column chromatography using a mixture of petroleum ether/ethyl acetate (2:1) as eluent, if not otherwise stated.
1-Benzyl-8-methoxy-3-propyl-1H,3H-pyrido[2,1-f]purine-2,4-dione (1).24
The pure product was obtained by column chromatography using petroleum ether/ethyl acetate (1:1) as eluent, yielding a white solid 113 mg, 0.31 mmol, 60%. 1H NMR (400 MHz, CDCl3) δ: 8.83 (d, J = 7.2 Hz, 1H), 7.54 (d, J = 7.2 Hz, 2H), 7.34–7.25 (m, 3H), 6.98 (d, J = 2.0 Hz, 1H), 6.74 (dd, J = 7.6, 2.8 Hz, 1H), 5.37 (s, 2H), 4.02 (t, J = 7.6 Hz, 2H), 3.92 (s, 3H), 1.74 (sextet, J = 7.6 Hz, 2H), 0.99 (t, J = 7.6 Hz, 3H) ppm.24 MS [ESI + H]+: calcd for C20H20N4O3, 364.15; found, 365.0.
1-Benzyl-3-(cyclopropylmethyl)-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (2).23
The pure product was obtained by column chromatography using a mixture of 2% CH3OH/CH2Cl2 as eluent, yielding a white solid, 1.51 g, 3.98 mmol, 86%. 1H NMR (400 MHz, CDCl3) δ: 8.82 (d, J = 7.2 Hz, 1H), 7.54 (d, J = 7.2 Hz, 2H), 7.33–7.23 (m, 3H), 6.97 (d, J = 2.4 Hz, 1H), 6.73 (dd, J = 8.4, 2.4 Hz, 1H), 5.37 (s, 2H), 3.94 (d, J = 7.4 Hz, 2H), 3.92 (s, 3H), 1.35–1.25 (m, 1H), 0.47–0.44 (m, 4H) ppm. MS [ESI + H]+: calcd for C21H20N4O3, 376.15; found, 376.9.
1-Benzyl-8-methoxy-3-methylpyrido[2,1-f]purine-2,4(1H,3H)-dione (6)
Reaction was performed in a sealed tube, and 50 equiv of methyl iodide was used. The residue was purified by silica column chromatography eluting with a petroleum ether/ethyl acetate (1:1) mixture, yielding a white solid, 25 mg, 0.074 mmol, 15%. 1H NMR (400 MHz, CDCl3) δ: 8.83 (d, J = 7.6 Hz, 1H), 7.54 (d, J = 7.2 Hz, 2H), 7.33–7.24 (m, 3H), 6.99 (d, J = 2.4 Hz, 1H), 6.75 (dd, J = 7.6, 2.4 Hz, 1H), 5.37 (s, 2H), 3.93 (s, 3H), 3.45 (s, 3H) ppm. MS [ESI + H]+: calcd for C18H16N4O3, 336.12; found, 337.2.
1-Benzyl-3-ethyl-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (7)
The pure product was obtained by column chromatography using petroleum ether/ethyl acetate (1:1) as eluent, yielding a white solid, 58 mg, 0.17 mmol, 33%. 1H NMR (400 MHz, CDCl3) δ: 8.83 (d, J = 7.2 Hz, 1H), 7.54 (d, J = 7.2 Hz, 2H), 7.33–7.24 (m, 3H), 6.98 (d, J = 2.0 Hz, 1H), 6.74 (dd, J = 7.6, 2.8 Hz, 1H), 5.36 (s, 2H), 4.12 (q, J = 7.2 Hz, 2H), 3.92 (s, 3H), 1.28 (t, J = 7.2 Hz, 3H) ppm. MS [ESI + H]+: calcd for C19H18N4O3, 350.14; found, 351.0.
1-Benzyl-3-butyl-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (8)
Purified by column chromatography using petroleum ether/EtOAc (3:1), yielding a white solid 10 mg, 0.026 mmol, 5%. 1H NMR (400 MHz, CDCl3) δ: 8.83 (d, J = 7.6 Hz, 1H), 7.54 (d, J = 6.8 Hz, 2H), 7.24–7.33 (m, 3H), 6.98 (d, J = 2.4 Hz, 1H), 6.74 (dd, J = 7.4, 2.6 Hz, 1H), 5.36 (s, 2H), 4.04 (t, J = 7.6 Hz, 2H), 3.93 (s, 3H), 1.70–1.64 (m, 2H), 1.40 (sextet, J = 3.6 Hz, 2H), 0.95 (t, J = 7.2 Hz, 3H) ppm. MS [ESI + H]+: calcd for C21H22N4O3, 378.17; found, 378.9.
1-Benzyl-8-methoxy-3-pentyl-1H,3H-pyrido[2,1-f]purine-2,4-dione (9)
White solid, 110 mg, 0.28 mmol, yield 56%. 1H NMR (400 MHz, CDCl3) δ: 8.83 (d, J = 7.6 Hz, 1H), 7.56 (d, J = 7.2 Hz, 2H), 7.34–7.25 (m, 3H), 6.97 (d, J = 2.0 Hz, 1H), 6.74 (dd, J = 7.6, 2.8 Hz, 1H), 5.37 (s, 2H), 4.05 (t, J = 7.6 Hz, 2H), 3.93 (s, 3H), 1.72–1.66 (m, 2H), 1.39–1.37 (m, 4H), 0.91 (t, J = 7.2 Hz, 3H) ppm. MS [ESI + H]+: calcd for C22H24N4O3, 392.18; found, 393.1.
1-Benzyl-3-hexyl-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (10)
White solid, 90 mg, 0.22 mmol, yield 44%. 1H NMR (400 MHz, CDCl3) δ: 8.83 (d, J = 7.2 Hz, 1H), 7.55 (d, J = 6.8 Hz, 2H), 7.34–7.25 (m, 3H), 6.98 (d, J = 2.0 Hz, 1H), 6.74 (dd, J = 7.6, 2.4 Hz, 1H), 5.37 (s, 2H), 4.05 (t, J = 7.6 Hz, 2H), 3.92 (s, 3H), 1.69 (pentet, J = 7.6 Hz, 2H), 1.40–1.26 (m, 6H), 0.89 (t, J = 7.2 Hz, 3H) ppm. MS [ESI + H]+: calcd for C23H26N4O3, 406.20; found, 407.1.
1-Benzyl-3-heptyl-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (11)
Column chromatography using a mixture of petroleum ether/ethyl acetate (3:1) as eluent yielded 131 mg of the pure product as white solid, 0.31 mmol, 62%. 1H NMR (400 MHz, CDCl3) 1H NMR (400 MHz, CDCl3) δ: 8.80 (d, J = 7.6 Hz, 1H), 7.54 (d, J = 6.8 Hz, 2H), 7.32–7.25 (m, 3H), 6.95 (d, J = 2.4 Hz, 1H), 6.71 (dd, J = 7.2, 2.4 Hz, 1H), 5.35 (s, 2H), 4.03 (t, J = 7.6, 2H), 3.90 (s, 3H), 1.67 (pentet, J = 7.6 Hz, 2H), 1.38–1.26 (m, 8H), 0.87 (t, J = 7.2 Hz, 3H) ppm. MS [ESI + H]+: calcd for C24H48N4O3, 420.22; found, 421.2.
3-Allyl-1-benzyl-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (12)
Purified by column chromatography using petroleum ether/ethyl acetate (1:1) as eluent, yielding a white solid, 73 mg, 0.20 mmol, 40%. 1H NMR (400 MHz, CDCl3) δ: 8.82 (d, J = 7.6 Hz, 1H), 7.55 (d, J = 7.2 Hz, 2H), 7.34–7.25 (m, 3H), 6.99 (d, J = 2.0 Hz, 1H), 6.75 (dd, J = 7.6, 2.4 Hz, 1H), 6.02–5.92 (m, 1H), 5.38 (s, 2H), 5.29 (dd, J = 17.2, 1.2 Hz, 1H), 5.20 (d, J = 10.0 Hz, 1H), 4.68 (d, J = 5.6 Hz, 2H), 3.93 (s, 3H) ppm. MS [ESI + H]+: calcd for C20H18N4O3, 362.14; found, 363.0.
1-Benzyl-8-methoxy-3-(prop-2-yn-1-yl)pyrido[2,1-f]purine-2,4(1H,3H)-dione (13)
The pure product was obtained by column chromatography using petroleum ether/ethyl acetate (1:1) as eluent, yielding a white solid, 60 mg, 0.17 mmol, 3%. 1H NMR (400 MHz, CDCl3) δ: 8.82 (d, J = 7.2 Hz, 1H), 7.57 (d, J = 6.8 Hz, 2H), 7.36–7.26 (m, 3H), 6.99 (d, J = 2.4 Hz, 1H), 6.76 (dd, J = 7.2, 2.4 Hz, 1H), 5.38 (s, 2H), 4.83 (d, J = 2.4 Hz, 2H), 3.94 (s, 3H), 2.19 (t, J = 2.4 Hz, 1H) ppm. MS [ESI + H]+: calcd for C20H16N4O3, 360.12; found, 361.1.
1-Benzyl-3-(but-3-en-1-yl)-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (14)
White solid, 18 mg, 0.05 mmol, yield 10%. 1H NMR (400 MHz, CDCl3) δ: 8.84 (d, J = 7.6 Hz, 1H), 7.54 (d, J = 7.2 Hz, 2H), 7.35–7.28 (m, 3H), 7.00 (d, J = 2.4 Hz, 1H), 6.77 (dd, J = 7.6, 2.4 Hz, 1H), 5.91–5.84 (m, 1H), 5.39 (s, 2H), 5.08 (dd, J = 16.8, 1.2 Hz, 1H), 5.02 (d, J = 10.4, 1H), 4.15 (t, J = 7.2 Hz, 2H), 3.95 (s, 3H), 2.48 (q, J = 7.2 Hz, 2H) ppm. MS [ESI + H]+: calcd for C21H20N4O3, 376.15; found, 376.9.
1-Benzyl-8-methoxy-3-(2-methoxyethyl)-1H,3H-pyrido[2,1-f]purine-2,4-dione (15)
Purified by column chromatography using a mixture of 2% CH3OH in CH2Cl2 yielded the product as a white solid, 70 mg, 0.18 mmol, 37%. 1H NMR (400 MHz, CDCl3) δ: 8.83 (d, J = 7.2 Hz, 1H), 7.54 (d, J = 6.8 Hz, 2H), 7.33–7.24 (m, 3H), 6.98 (d, J = 2.4 Hz, 1H), 6.74 (dd, J = 7.6, 2.4 Hz, 1H), 5.36 (s, 2H), 4.30 (t, J = 6.0 Hz, 2H), 3.93 (s, 3H), 3.68 (t, J = 6.0 Hz, 2H), 3.36 (s, 3H) ppm. MS [ESI + H]+: calcd for C20H20N4O3, 380.15; found, 380.8.
1-Benzyl-3-(3-hydroxypropyl)-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (16)
Purified by column chromatography using a mixture of petroleum ether/EtOAc (1:2), followed by recrystallization from CH3OH/EtOAc. Yield: white solid, 106 mg, 0.28 mmol, 28%. 1H NMR (400 MHz, CDCl3) δ: 8.80 (d, J = 7.2 Hz, 1H), 7.53 (d, J = 7.6 Hz, 2H), 7.33–7.24 (m, 3H), 6.99 (d, J = 2.4 Hz, 1H), 6.76 (dd, J = 7.6, 2.4 Hz, 1H), 5.37 (s, 2H), 4.22 (t, J = 6.0 Hz, 2H), 3.93 (s, 3H), 3.53 (t, J = 5.6 Hz, 2H), 1.92 (pentet, J = 5.6 Hz, 2H) ppm. MS [ESI + H]+: calcd for C20H20N4O3, 380.15; found, 380.9.
1-Benzyl-3-isobutyl-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (17)
White solid, 0.08 mmol, 29 mg, yield 15%. 1H NMR (400 MHz, CDCl3) δ: 8.86 (d, J = 7.2 Hz, 1H), 7.55 (d, J = 7.2 Hz, 2H), 7.33–7.28 (m, 3H), 7.00 (d, J = 2.0 Hz, 1H), 6.76 (dd, J = 7.6, 2.4 Hz, 1H), 5.39 (s, 2H), 3.95 (s, 3H), 3.92 (d, J = 7.6 Hz, 2H), 2.22 (nonet, J = 7.2 Hz, 1H), 0.97 (d, J = 6.8 Hz, 6H) ppm. MS [ESI + H]+: calcd for C21H22N4O3, 378.17; found, 379.0.
1-Benzyl-8-methoxy-3-(3-neopentyl)-1H,3H-pyrido[2,1-f]purine-2,4-dione (18)
Purified by column chromatography using a mixture of 1% CH3OH in CH2Cl2 as eluent, yielding the product as a white solid, 0.025 mmol, 20 mg, 5%. 1H NMR (400 MHz, CDCl3) δ: 8.84 (d, J = 7.2 Hz, 1H), 7.52 (d, J = 7.2 Hz, 2H), 7.32–7.25 (m, 3H), 6.98 (d, J = 2.4 Hz, 1H), 6.73 (dd, J = 7.2, 2.4 Hz, 1H), 5.37 (s, 2H), 3.99 (s, 2H), 3.92 (s, 3H), 0.98 (s, 9H) ppm. MS [ESI + H]+: calcd for C22H24N4O3, 392.18; found, 393.0.
1-Benzyl-3-isopentyl-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (19)
Yield: white solid, 104 mg, 0.26 mmol, 53%. 1H NMR (400 MHz, CDCl3) δ: 8.83 (d, J = 7.2 Hz, 1H), 7.53 (d, J = 7.2 Hz, 2H), 7.33–7.24 (m, 3H), 6.98 (d, J = 2.4 Hz, 1H), 6.74 (dd, J = 7.6, 2.4 Hz, 1H), 5.36 (s, 2H), 4.08–4.04 (m, 2H), 3.93 (s, 3H), 1.69 (nonet, J = 6.8 Hz, 1H), 1.59–1.53 (m, 2H), 0.98 (d, J = 6.4 Hz, 6H) ppm. MS [ESI + H]+: calcd for C22H24N4O3, 392.18; found, 393.1.
1-Benzyl-3-(3,3-dimethylbutyl)-8-methoxy-pyrido[2,1-f]purine-2,4(1H,3H)-dione (20)
Yield: white solid, 81 mg, 0.20 mmol, 40%. 1NMR (400 MHz, CDCl3) δ: 8.84 (d, J = 7.6 Hz, 1H), 7.55 (d, J = 7.2, 2H), 7.35–7.27 (m, 3H), 6.98 (d, J = 2.4 Hz, 1H), 6.74 (dd, J = 7.2, 2.4 Hz, 1H), 5.38 (s, 2H), 4.12–4.08 (m, 2H), 3.93 (s, 3H), 1.61–1.57 (m, 2H), 1.04 (s, 9H) ppm. MS [ESI + H]+: calcd for C23H26N4O3, 406.20; found, 407.1.
1-Benzyl-8-methoxy-3-((trimethylsilyl)methyl)pyrido[2,1-f]purine-2,4(1H,3H)-dione (21)
White solid, 137 mg, 0.34 mmol, yield 67%. 1H NMR (400 MHz, CDCl3) δ: 8.84 (d, J = 7.2 Hz, 1H), 7.50 (d, J = 6.8 Hz, 2H), 7.32–7.24 (m, 3H), 6.98 (d, J = 2.4 Hz, 1H), 6.73 (d, J = 7.2, 2.4 Hz, 1H), 5.39 (s, 2H), 3.93 (s, 3H), 3.64 (s, 2H), 0.08 (s, 9H) ppm. MS [ESI + H]+: calcd for C21H24N4O3Si, 408.16; found, 409.2.
1-Benzyl-3-(cyclobutylmethyl)-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (22)
White solid, 90 mg, 0.23 mmol, yield 46%. 1H NMR (400 MHz, CDCl3) δ: 8.82 (d, J = 7.2 Hz, 1H), 7.53 (d, J = 7.2 Hz, 2H), 7.33–7.13 (m, 3H), 6.69 (d, J = 1.8 Hz, 1H), 6.73 (dd, J = 7.2, 2.4 Hz, 1H), 5.35 (s, 2H), 4.11 (d, J = 7.6 Hz, 2H), 3.92 (s, 3H), 2.83–2.72 (m, 1H), 2.05–1.95 (m, 2H), 1.90–1.79 (m, 4H) ppm. MS [ESI + H]+: calcd for C22H22N4O3, 390.17; found, 391.2.
3-(Cyclopropylmethyl)-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (23).23
Prepared following a slightly modified procedure described by Priego et al. In total, four portions of 8 equiv of ammonium formate (after 0, 2, 4, and 6 h) and three portions of 0.15 equiv of 20% Pd(OH)2 (0, 4, and 6 h) were added, after which full conversion was reached after overnight reflux visualized by TLC (3% CH3OH/CH2Cl2). The reaction mixture was filtered over Celite and the residue extracted 5 times with hot DMF. The combined organic layer was concentrated in vacuo, which resulted in a quantitative yield. 1H NMR in accordance to data in literature.23
General Procedure for the Preparation of N1-Substituted-3-cyclopropylmethyl-8-methoxy-1H,3H-pyrido[2,1-f]purine-2,4-diones (24–33).23
The compounds were synthesized according to the procedure described by Priego et al.2 Purification by silica column chromatography using an eluent mixture of petroleum ether/ethyl acetate (3:1) yielded the pure final products.
3-(Cyclopropylmethyl)-8-methoxy-1-(3-methylbenzyl)pyrido[2,1-f]purine-2,4(1H,3H)-dione (24)
White solid, 79 mg, 0.20 mmol, yield 57%. 1H NMR (400 MHz, CDCl3) δ: 8.84 (d, J = 7.2 Hz, 1H), 7.36 (s, 2H), 7.22 (t, J = 7.6 Hz, 1H), 7.09 (d, J = 7.6 Hz, 1H), 6.99 (d, J = 2.4 Hz, 1H), 6.75 (dd, J = 7.2, 2.4 Hz, 1H), 5.36 (s, 2H), 3.97 (d, J = 7.2 Hz, 2H), 3.94 (s, 3H), 2.34 (s, 3H), 1.37–1.31 (m, 1H), 0.52–0.45 (m, 4H) ppm. MS [ESI + H]+: calcd for C22H22N4O3, 390.17; found, 391.0.
3-(Cyclopropylmethyl)-8-methoxy-1-(4-methylbenzyl)pyrido[2,1-f]purine-2,4(1H,3H)-dione (25)
See ref (23).
3-(Cyclopropylmethyl)-1-(4-ethylbenzyl)-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (26)
White solid, 43 mg, 0.11 mmol, yield 21%. 1H NMR (400 MHz, CDCl3) δ: 8.82 (d, J = 7.6 Hz, 1H), 7.48 (d, J = 8.0 Hz, 2H), 7.14 (d, J = 8.0 Hz, 2H), 6.98 (d, J = 2.0 Hz, 1H), 6.73 (dd, J = 7.2, 2.4 Hz, 1H), 5.34 (s, 2H), 3.95–3.93 (m, 5H), 2.60 (q, J = 7.6 Hz, 2H), 1.34–1.28 (m, 1H), 1.19 (t, J = 7.6 Hz, 3H), 0.50–0.42 (m, 4H) ppm. MS [ESI + H]+: calcd for C23H24N4O3, 404.18; found, 405.2.
3-(Cyclopropylmethyl)-8-methoxy-1-(3-methoxybenzyl)pyrido[2,1-f]purine-2,4(1H,3H)-dione (27)
White solid, 20 mg, 0.049 mmol, yield 14%. 1H NMR (400 MHz, CDCl3) δ: 8.83 (d, J = 7.6 Hz, 1H), 7.23 (t, J = 8.0 Hz, 1H), 7.15–7.10 (m, 2H), 6.97 (d, J = 1.6 Hz, 1H), 6.80 (dd, J = 8.4, 2.4 Hz, 1H), 6.74 (dd, J = 7.6, 2.4 Hz, 1H), 5.35 (s, 2H), 3.96–3.92 (m, 5H), 3.77 (s, 3H), 1.35–1.28 (m, 1H), 0.50–0.45 (m, 4H) ppm. MS [ESI + H]+: calcd for C22H22N4O4, 406.16; found, 406.9.
3-(Cyclopropylmethyl)-8-methoxy-1-(4-methoxybenzyl)pyrido[2,1-f]purine-2,4(1H,3H)-dione (28)
See ref (23).
1-(3-Chlorobenzyl)-3-(cyclopropylmethyl)-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (29)
See ref (23).
1-(4-Chlorobenzyl)-3-(cyclopropylmethyl)-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (30)
See ref (23).
3-(Cyclopropylmethyl)-1-(3,4-dichlorobenzyl)-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (31)
See ref (23).
1-(4-Bromobenzyl)-3-(cyclopropylmethyl)-8-methoxypyrido[2,1-f]purine-2,4(1H,3H)-dione (32)
White solid, 22 mg, 0.048 mmol, yield 14%. 1H NMR (400 MHz, CDCl3) δ: 8.85 (d, J = 7.2 Hz, 1H), 7.46 (s, 4H), 6.99 (d, J = 1.6 Hz, 1H), 6.87 (dd, J = 7.2, 2.0 Hz, 1H), 5.33 (s, 2H), 3.99–3.85 (m, 5H), 1.34–1.24 (m, 1H), 0.50–0.44 (m, 4H) ppm. MS [ESI + H]+: calcd for C21H19N4O3, 454.06; found, 455.0.
3-(Cyclopropylmethyl)-8-methoxy-1-phenylethylpyrido[2,1-f]purine-2,4(1H,3H)-dione (33)
White solid, 112 mg, 0.29 mmol, yield 82%. 1H NMR (400 MHz, CDCl3) δ: 8.81 (d, J = 7.2 Hz, 1H), 7.35–7.25 (m, 4H), 7.20 (t, J = 7.2 Hz, 1H), 6.96 (d, J = 2.4 Hz, 1H), 6.73 (dd, J = 7.2, 2.4 Hz, 1H), 4.40 (t, J = 8.0 Hz, 2H), 3.95–3.90 (m, 5H), 3.12 (t, J = 8.0 Hz, 2H), 1.32–1.23 (m, 1H), 0.49–0.40 (m, 4H) ppm. MS [ESI + H]+: calcd for C22H22N4O3, 390.17; found, 391.0.
Biology
Chemicals and Reagents
[3H]8-Ethyl-4-methyl-2-phenyl-(8R)-4,5,7,8-tetrahydro-1H-imidazo[2,1-i]-purin-5-one26 ([3H]34, specific activity 56 Ci·mmol–1) was a gift from Prof. C. E. Müller (University of Bonn, Germany). Unlabeled 34 was purchased from Tocris Ltd. (Abingdon, UK). 5′-N-Ethylcarboxamidoadenosine (NECA) was purchased from Sigma-Aldrich (Steinheim, Germany). Adenosine deaminase (ADA) was purchased from Boehringer Mannheim (Mannheim, Germany). Bicinchoninic acid (BCA) and BCA protein assay reagents were purchased from Pierce Chemical Company (Rockford, IL, USA). Chinese hamster ovary cells stably expressing the human adenosine A3 receptor (CHOhA3) were a gift from Dr. K.-N. Klotz (University of Würzburg, Germany). All other chemicals were obtained from standard commercial sources and were of analytical grade.
Cell Culture and Membrane Preparation
Chinese hamster ovary (CHO) cells, stably expressing the human adenosine A3 receptor (CHOhA3), were cultured and membranes were prepared and stored as previously described.44 Protein determination was done through use of the bicinchoninic acid (BCA) method.45
Radioligand Displacement Assay
Membrane aliquots containing ∼15 μg of CHOhA3 protein were incubated in a total volume of 100 μL of assay buffer (50 mM Tris-HCl, 5 mM MgCl2, supplemented with 0.01% CHAPS and 1 mM EDTA, pH 7.4) at 25 °C for 120 min. Displacement experiments were performed using six concentrations of competing antagonist in the presence of a final concentration of ∼10 nM [3H] 34. At this concentration, total radioligand binding did not exceed 10% of that added to prevent ligand depletion. Nonspecific binding (NSB) was determined in the presence of 100 μM NECA. Incubation was terminated by rapid filtration performed on 96-well GF/B filter plates (PerkinElmer, Groningen, The Netherlands), using a PerkinElmer Filtermate harvester (PerkinElmer, Groningen, The Netherlands). After drying the filter plate at 50 °C for 30 min, the filter-bound radioactivity was determined by scintillation spectrometry using the 2450 MicroBeta2 plate counter (PerkinElmer, Boston, MA).
Radioligand Association and Dissociation Assays
Association experiments were performed by incubating membrane aliquots containing ∼15 μg of CHOhA3 membrane in a total volume of 100 μL of assay buffer at 10 or 25 °C with ∼10 nM [3H] 34. The amount of radioligand bound to the receptor was measured at different time intervals during a total incubation of 120 min. Dissociation experiments were performed by preincubating membrane aliquots containing ∼15 μg of protein in a total volume of 100 μL of assay buffer at 10 or 25 °C for 60 min. After the preincubation, radioligand dissociation was initiated by the addition of 5 μL of 100 μM unlabeled NECA. The amount of radioligand still bound to the receptor was measured at various time intervals for a total of 120 min to ensure that full dissociation from hA3 receptor was reached. Incubations were terminated and samples were obtained as described under Radioligand Displacement Assay.
Radioligand Competition Association Assay
The binding kinetics of unlabeled ligands were quantified using the competition association assay based on the theoretical framework by Motulsky and Mahan.46 The competition association assay was initiated by adding membrane aliquots (15 μg/well) at different time points for a total of 240 min to a total volume of 100 μL of assay buffer at 10 or 25 °C with ∼10 nM [3H] 34 in the absence or presence of a single concentration of competing hA3R antagonists (i.e., at their IC50 value). Incubations were terminated and samples were obtained as described under Radioligand Displacement Assay. The “dual-point” competition association assays were designed as described previously,27 where in this case the two time points were selected at 20 (t1) and 240 min (t2).
[35S] GTPγS Binding Assay
The assays were performed by incubating 15 μg of homogenized CHOhA3 membranes in a total volume of 80 μL of assay buffer (50 mM Tris-HCl buffer, 5 mM MgCl2, 1 mM EDTA, 0.05% BSA, and 1 mM DTT, pH 7.4) supplemented with 1 μM GDP and 5 μg of saponin. The assays were performed in a 96-well plate format, where DMSO stock solutions of the compounds were added using a HP D300 Digital Dispenser (Tecan, Männedorf, Switserland). The final concentration of organic solvent per assay point was ≤0.1%. In all cases, the basal level of [35S] GTPγS binding was measured in untreated membrane samples, whereas the maximal level of [35S] GTPγS binding was measured by treatment of the membranes with 10 μM 2-Cl-IBMECA. For the insurmountability experiments, membrane preparations were preincubated with or without antagonists (30-, 100-, 300-fold Ki values) for 60 min at 25 °C, prior to the addition of 2-Cl-IBMECA (10 μM to 0.1 nM) and 20 μL of [35S] GTPγS (final concentration ∼0.3 nM), after which incubation continued for another 30 min at 25 °C. For the surmountability (control) experiments, antagonists and 2-Cl-IBMECA were coincubated with [35S] GTPγS for 30 min at 25 °C. For all experiments, incubations were terminated and samples were obtained as described under Radioligand Displacement Assay by using GF/B filters (Whatman International, Maidstone, UK).
Data Analysis
All experimental data were analyzed using the nonlinear regression curve fitting program GraphPad Prism 6.0 (GraphPad Software, Inc., San Diego, CA). From displacement assays, IC50 values were obtained by nonlinear regression analysis of the displacement curves. The obtained IC50 values were converted into Ki values using the Cheng–Prusoff equation to determine the affinity of the ligands.47 The observed association rates (kobs) derived from both assays were obtained by fitting association data using one phase exponential association. The dissociation rates were obtained by fitting dissociation data to a one phase exponential decay model. The kobs values were converted into association rate constants (kon) using the equation kon = (kobs – koff)/[L], where [L] is the amount of radioligand used for the association experiments. The association and dissociation rates were used to calculate the kinetic KD using the equation KD = koff/kon. Association and dissociation rate constants for unlabeled compounds were calculated by fitting the data into the competition association model using “kinetics of competitive binding”:46
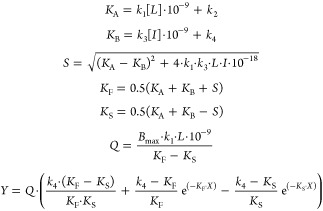 |
where k1 is the kon of the radioligand (M–1 s–1), k2 is the koff of the radioligand (s–1), L is the radioligand concentration (nM), I is the concentration of the unlabeled competitor (nM), X is the time (s), and Y is the specific binding of the radioligand (DPM). The control curve (without competitor) from competition association assays generates the k1 value, and the k2 value was obtained from Radioligand Association and Dissociation Assays. With that, the k3, k4 and Bmax can be calculated, where k3 represents the kon (M–1 s–1) of the unlabeled ligand, k4 stands for the koff (s–1) of the unlabeled ligand, and Bmax equals the total binding (DPM). All competition association data were globally fitted. The residence time (RT, in min) was calculated using the equation RT = 1/(60 × koff), as koff values are expressed in s–1. [35S] GTPγS binding curves were analyzed by nonlinear regression using “log (agonist) vs response-variable slope” to obtain potency, inhibitory potency, or efficacy values of agonists and inverse agonists/antagonists (EC50, IC50 or Emax, respectively). In the (in)surmountability assays, Gaddum/Schild EC50 shift equations were used to obtain Schild-slopes and pA2 values; statistical analysis of two-way ANOVA with Tukey’s post-test was applied. All experimental values obtained are means of at least three independent experiments performed in duplicate, unless stated otherwise. R2 and P values were calculated using the GraphPad Prism linear regression analysis function. Log P (log partition coefficient) values were calculated using Chemdraw Professional 15.0 (Cambridge Soft, PerkinElmer, Waltham Mass).
Computational Studies
A ligand optimized homology model of the hA3R was generated by following a similar approach as has been used before48 and using the Maestro software package (Schroedinger Inc., New York). In short: first, different homology models were constructed based on the high resolution crystal structure of the adenosine A2A receptor (PDB 4EIY)41 and using a sequence alignment from GPCRDB.49,50 In the subsequent steps, we iteratively optimized the model using Prime.51−53 During every step, the best model was selected based on enrichment (BEDROC-160.9 and ROC). For this, we used a set of 100 diverse antagonists from ChEMBL54 obtained by “Cluster Molecules”.55 We matched 50 decoys to every ligand ionization state, using the DUD-e web service.56 The final model used here showed excellent enrichment (BEDROC-160.9, 0.55; ROC, 0.80). We introduced a long residence time ligand, 2, in the putative ligand binding site using Induced fit docking,57 with H-bond constraints on Asn2506.55. On the basis of this, we generated a WaterMap42,43 of the apo state of the receptor. Other ligands were docked using core-constrained docking (using the core of 2 as constraints). Figures were rendered using PyMol.58
Acknowledgments
The research in this study has been performed in the “Kinetics for Drug Discovery (K4DD)” consortium. The K4DD project is supported by the Innovative Medicines Initiative Joint Undertaking (IMI JU) under grant agreement no. 115366, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA companies’ in kind contribution. We thank Prof C. E. Mueller (Bonn University, Germany) for her kind help in obtaining [3H] PSB-11, the radiolabeled probe used in this study. Lizi Xia thanks Dr. Daan van der Es for his assistance in preparing the chemistry section.
Glossary
Abbreviations Used
- ADA
adenosine deaminase
- BCA
bicinchoninic acid
- BSA
bovine serum albumin
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- CHO
Chinese hamster ovary
- COPD
chronic obstructive pulmonary disease
- GTPγS
guanosine 5′-O-[γ-thio]triphosphate
- hA3R
human adenosine A3 receptor
- KD
equilibrium dissociation constant of ligand
- koff
dissociation rate constant
- kon
association rate constant
- KRI
kinetic rate index
- NECA
5′-(N-ethylcarboxamide)adenosine
- RT
residence time
- SA(FI)R
structure–affinity relationships
- SKR
structure–kinetics relationships
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jmedchem.7b00950.
The binding affinities of short RT antagonist (5) and long RT antagonist (27) at human adenosine A1 and A2A receptors, the KRI values of pyrido[2,1-f]purine-2,4-dione derivatives without methoxy-substitution at C-8 position, the comparison with their methoxy-substituted counterparts, and the correlation between pkoff and Log P of the compounds (PDF)
Molecular formula strings (CSV)
Author Contributions
L.X., A.P.IJ., and L.H.H. conceived the study. A.P.IJ. and L.H.H. supervised the project. The chemical synthesis was designed and supervised by J.P.D.V and performed by B.J.K. and J.P.D.V. The bioassays were supervised by L.H.H. and performed by W.A.C.B., T.T.D., E.P., and L.X. The computational work was performed by E.B.L. The manuscript was written by L.X., W.A.C.B., J.P.D.V., and A.P.IJ.
The authors declare no competing financial interest.
Supplementary Material
References
- Meyerhof W.; Müller-Brechlin R.; Richter D. Molecular cloning of a novel putative G-protein coupled receptor expressed during rat spermiogenesis. FEBS Lett. 1991, 284, 155–160. 10.1016/0014-5793(91)80674-R. [DOI] [PubMed] [Google Scholar]
- Fredholm B. B.; IJzerman A. P.; Jacobson K. A.; Klotz K.-N.; Linden J. International union of pharmacology. XXV. Nomenclature and classification of adenosine receptors. Pharmacol. Rev. 2001, 53, 527–552. [PMC free article] [PubMed] [Google Scholar]
- Jacobson K. A.; Gao Z.-G. Adenosine receptors as therapeutic targets. Nat. Rev. Drug Discovery 2006, 5, 247–264. 10.1038/nrd1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessi S.; Merighi S.; Varani K.; Leung E.; Mac Lennan S.; Borea P. A. The A3 adenosine receptor: an enigmatic player in cell biology. Pharmacol. Ther. 2008, 117, 123–140. 10.1016/j.pharmthera.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Jacobson K. A.; Moro S.; Kim Y.-C.; Li A.-H. A3 adenosine receptors: protective vs. damaging effects identified using novel agonists and antagonists. Drug Dev. Res. 1998, 45, 113–124. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishat S.; Khan L. A.; Ansari Z. M.; Basir S. F. Adenosine A3 receptor: a promising therapeutic target in cardiovascular disease. Curr. Cardiol. Rev. 2016, 12, 18–26. 10.2174/1573403X12666160111125116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscsó B.; Csóka B.; Pacher P.; Haskó G. Investigational A3 adenosine receptor targeting agents. Expert Opin. Invest. Drugs 2011, 20, 757–768. 10.1517/13543784.2011.573785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman P.; Bar-Yehuda S.; Synowitz M.; Powell J. D.; Klotz K. N.; Gessi S.; Borea P. A. Adenosine receptors and cancer. Handb. Exp. Pharmacol. 2009, 193, 399–441. 10.1007/978-3-540-89615-9_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Schneider D. J.; Blackburn M. R. Adenosine signaling and the regulation of chronic lung disease. Pharmacol. Ther. 2009, 123, 105–116. 10.1016/j.pharmthera.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. N.; Nadeem A.; Spina D.; Brown R.; Page C. P.; Mustafa S. J.. Adenosine receptors and asthma In Adenosine Receptors in Health and Disease; Handbook of Experimental Pharmacology, Vol. 193; Springer: Berlin, Heidelberg, 2009; Vol. 193, pp 329–362, DOI 10.1007/978-3-540-89615-9_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. N. Adenosine receptors and asthma in humans. Br. J. Pharmacol. 2008, 155, 475–486. 10.1038/bjp.2008.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borea P. A.; Varani K.; Vincenzi F.; Baraldi P. G.; Tabrizi M. A.; Merighi S.; Gessi S. The A3 adenosine receptor: history and perspectives. Pharmacol. Rev. 2015, 67, 74–102. 10.1124/pr.113.008540. [DOI] [PubMed] [Google Scholar]
- Baraldi P. G.; Cacciari B.; Romagnoli R.; Merighi S.; Varani K.; Borea P. A.; Spalluto G. A3 adenosine receptor ligands: history and perspectives. Med. Res. Rev. 2000, 20, 103–128. . [DOI] [PubMed] [Google Scholar]
- Baraldi P. G.; Preti D.; Borea P. A.; Varani K. Medicinal chemistry of A3 adenosine receptor modulators: pharmacological activities and therapeutic implications. J. Med. Chem. 2012, 55, 5676–5703. 10.1021/jm300087j. [DOI] [PubMed] [Google Scholar]
- Hasko G.; Linden J.; Cronstein B.; Pacher P. Adenosine receptors: therapeutic aspects for inflammatory and immune diseases. Nat. Rev. Drug Discovery 2008, 7, 759–770. 10.1038/nrd2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D.; Hillger J. M.; Ijzerman A. P.; Heitman L. H. Drug-target residence time—a case for G protein-coupled receptors. Med. Res. Rev. 2014, 34, 856–892. 10.1002/med.21307. [DOI] [PubMed] [Google Scholar]
- Cusack K. P.; Wang Y.; Hoemann M. Z.; Marjanovic J.; Heym R. G.; Vasudevan A. Design strategies to address kinetics of drug binding and residence time. Bioorg. Med. Chem. Lett. 2015, 25, 2019–2027. 10.1016/j.bmcl.2015.02.027. [DOI] [PubMed] [Google Scholar]
- Copeland R. A.; Pompliano D. L.; Meek T. D. Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discovery 2006, 5, 730–739. 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- Copeland R. A. The drug-target residence time model: a 10-year retrospective. Nat. Rev. Drug Discovery 2016, 15, 87–95. 10.1038/nrd.2015.18. [DOI] [PubMed] [Google Scholar]
- Zeilinger M.; Pichler F.; Nics L.; Wadsak W.; Spreitzer H.; Hacker M.; Mitterhauser M. New approaches for the reliable in vitro assessment of binding affinity based on high-resolution real-time data acquisition of radioligand-receptor binding kinetics. EJNMMI Res. 2017, 7, 22. 10.1186/s13550-016-0249-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D.; Heitman L. H.; Ijzerman A. P. Kinetic aspects of the interaction between ligand and G protein-coupled receptor: the case of the adenosine receptors. Chem. Rev. 2017, 117, 38–66. 10.1021/acs.chemrev.6b00025. [DOI] [PubMed] [Google Scholar]
- Topliss J. G. A manual method for applying the Hansch approach to drug design. J. Med. Chem. 1977, 20, 463–469. 10.1021/jm00214a001. [DOI] [PubMed] [Google Scholar]
- Priego E.-M.; Pérez-Pérez M.-J.; von Frijtag Drabbe Kuenzel J. K.; de Vries H.; Ijzerman A. P.; Camarasa M.-J.; Martín-Santamaría S. Selective human adenosine A3 antagonists based on pyrido[2,1-f]purine-2,4-diones: novel features of hA3 antagonist binding. ChemMedChem 2008, 3, 111–119. 10.1002/cmdc.200700173. [DOI] [PubMed] [Google Scholar]
- Priego E.-M.; von Frijtag Drabbe Kuenzel J.; Ijzerman A. P.; Camarasa M.-J.; Pérez-Pérez M.-J. Pyrido[2,1-f]purine-2,4-dione derivatives as a novel class of highly potent human A3 adenosine receptor antagonists. J. Med. Chem. 2002, 45, 3337–3344. 10.1021/jm0208469. [DOI] [PubMed] [Google Scholar]
- Kalla R.; Perry T.; Elzein E.; Varkhedkar V.; Li X.; Ibrahim P.; Palle V.; Xiao D.; Zablocki J.. Xanthine Derivatives as A2B Adenosine Receptor Antagonists. WO2004106337 A1, 2004.
- Müller C. E.; Diekmann M.; Thorand M.; Ozola V. [3H]8-Ethyl-4-methyl-2-phenyl-(8R)-4,5,7,8-tetrahydro-1H-imidazo[2,1-i]-purin-5-one ([3H]PSB-11), a novel high-affinity antagonist radioligand for human A3 adenosine receptors. Bioorg. Med. Chem. Lett. 2002, 12, 501–503. 10.1016/S0960-894X(01)00785-5. [DOI] [PubMed] [Google Scholar]
- Guo D.; van Dorp E. J. H.; Mulder-Krieger T.; van Veldhoven J. P. D.; Brussee J.; IJzerman A. P.; Heitman L. H. Dual-point competition association assay: a fast and high-throughput kinetic screening method for assessing ligand-receptor binding kinetics. J. Biomol. Screening 2013, 18, 309–320. 10.1177/1087057112464776. [DOI] [PubMed] [Google Scholar]
- Vauquelin G. Cell membranes··· and how long drugs may exert beneficial pharmacological activity in vivo. Br. J. Clin. Pharmacol. 2016, 82, 673–682. 10.1111/bcp.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M.-H.; Chen X.; Chen S.-B.; Ou T.-M.; Yao M.; Gu L.-Q.; Huang Z.-S.; Tan J.-H. A new application of click chemistry in situ: development of fluorescent probe for specific G-quadruplex topology. Sci. Rep. 2015, 5, 17202. 10.1038/srep17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb H. C.; Sharpless K. B. The growing impact of click chemistry on drug discovery. Drug Discovery Today 2003, 8, 1128–1137. 10.1016/S1359-6446(03)02933-7. [DOI] [PubMed] [Google Scholar]
- Tian H.; Fürstenberg A.; Huber T. Labeling and single-molecule methods yo monitor G protein-coupled receptor dynamics. Chem. Rev. 2017, 117, 186–245. 10.1021/acs.chemrev.6b00084. [DOI] [PubMed] [Google Scholar]
- Tummino P. J.; Copeland R. A. Residence time of receptor–ligand complexes and Its Effect on Biological Function. Biochemistry 2008, 47, 5481–5492. 10.1021/bi8002023. [DOI] [PubMed] [Google Scholar]
- Smith G. F.Medicinal chemistry by the numbers: the physicochemistry, thermodynamics and kinetics of modern drug design. In Progress in Medicinal Chemistry; Lawton G., Witty D. R., Eds.; Elsevier BV: Burlington, 2009; Vol. 48, p 1. [DOI] [PubMed] [Google Scholar]
- Wilcken R.; Zimmermann M. O.; Lange A.; Joerger A. C.; Boeckler F. M. Principles and applications of halogen bonding in medicinal chemistry and chemical biology. J. Med. Chem. 2013, 56, 1363–1388. 10.1021/jm3012068. [DOI] [PubMed] [Google Scholar]
- Markgren P.-O.; Schaal W.; Hämäläinen M.; Karlén A.; Hallberg A.; Samuelsson B.; Danielson U. H. Relationships between structure and interaction kinetics for HIV-1 protease inhibitors. J. Med. Chem. 2002, 45, 5430–5439. 10.1021/jm0208370. [DOI] [PubMed] [Google Scholar]
- Yu Z.; van Veldhoven J. P. D.; Louvel J.; ’t Hart I. M. E.; Rook M. B.; van der Heyden M. A. G.; Heitman L. H.; Ijzerman A. P. Structure–affinity relationships (SARs) and structure–kinetics relationships (SKRs) of Kv11.1 blockers. J. Med. Chem. 2015, 58, 5916–5929. 10.1021/acs.jmedchem.5b00518. [DOI] [PubMed] [Google Scholar]
- Polosa R.; Blackburn M. R. Adenosine receptors as targets for therapeutic intervention in asthma and chronic obstructive pulmonary disease. Trends Pharmacol. Sci. 2009, 30, 528–535. 10.1016/j.tips.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P. J. The role of anticholinergics in chronic obstructive pulmonary disease. Am. J. Med. Supplements 2004, 117, 24–32. 10.1016/j.amjmed.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Tashkin D. P. Is a long-acting inhaled bronchodilator the first agent to use in stable chronic obstructive pulmonary disease?. Curr. Opin. Pulm. Med. 2005, 11, 121–128. 10.1097/00063198-200503000-00004. [DOI] [PubMed] [Google Scholar]
- Cazzola M.; Page C. Long-acting bronchodilators in COPD: where are we now and where are we going?. Breathe 2014, 10, 110–120. 10.1183/20734735.014813. [DOI] [Google Scholar]
- Liu W.; Chun E.; Thompson A. A.; Chubukov P.; Xu F.; Katritch V.; Han G. W.; Roth C. B.; Heitman L. H.; IJzerman A. P.; Cherezov V.; Stevens R. C. Structural basis for allosteric regulation of GPCRs by sodium ions. Science 2012, 337, 232–236. 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel R.; Young T.; Farid R.; Berne B. J.; Friesner R. A. Role of the active-site solvent in the thermodynamics of factor Xa ligand binding. J. Am. Chem. Soc. 2008, 130, 2817–2831. 10.1021/ja0771033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young T.; Abel R.; Kim B.; Berne B. J.; Friesner R. A. Motifs for molecular recognition exploiting hydrophobic enclosure in protein–ligand binding. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 808–813. 10.1073/pnas.0610202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman L. H.; Göblyös A.; Zweemer A. M.; Bakker R.; Mulder-Krieger T.; van Veldhoven J. P. D.; de Vries H.; Brussee J.; Ijzerman A. P. A Series of 2,4-disubstituted quinolines as a new class of allosteric enhancers of the adenosine A3 receptor. J. Med. Chem. 2009, 52, 926–931. 10.1021/jm8014052. [DOI] [PubMed] [Google Scholar]
- Smith P. K.; Krohn R. I.; Hermanson G. T.; Mallia A. K.; Gartner F. H.; Provenzano M. D.; Fujimoto E. K.; Goeke N. M.; Olson B. J.; Klenk D. C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Motulsky H. J.; Mahan L. C. The kinetics of competitive radioligand binding predicted by the law of mass action. Mol. Pharmacol. 1984, 25, 1–9. [PubMed] [Google Scholar]
- Cheng Y.-C.; Prusoff W. H. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50% inhibition (IC50) of an enzymatic reaction. Biochem. Pharmacol. 1973, 22, 3099–3108. 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- Louvel J.; Guo D.; Soethoudt M.; Mocking T. A.; Lenselink E. B.; Mulder-Krieger T.; Heitman L. H.; IJzerman A. P. Structure-kinetics relationships of Capadenoson derivatives as adenosine A 1 receptor agonists. Eur. J. Med. Chem. 2015, 101, 681–691. 10.1016/j.ejmech.2015.07.023. [DOI] [PubMed] [Google Scholar]
- Isberg V.; Mordalski S.; Munk C.; Rataj K.; Harpsøe K.; Hauser A. S.; Vroling B.; Bojarski A. J.; Vriend G.; Gloriam D. E. GPCRdb: an information system for G protein-coupled receptors. Nucleic Acids Res. 2016, 44, D356–D364. 10.1093/nar/gkv1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munk C.; Isberg V.; Mordalski S.; Harpsøe K.; Rataj K.; Hauser A.; Kolb P.; Bojarski A.; Vriend G.; Gloriam D. GPCRdb: the G protein-coupled receptor database–an introduction. Br. J. Pharmacol. 2016, 173, 2195–2207. 10.1111/bph.13509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Release 2016-3 Prime; Schrödinger LLC: New York, 2016.
- Jacobson M. P.; Friesner R. A.; Xiang Z.; Honig B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 2002, 320, 597–608. 10.1016/S0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- Jacobson M. P.; Pincus D. L.; Rapp C. S.; Day T. J.; Honig B.; Shaw D. E.; Friesner R. A. A hierarchical approach to all-atom protein loop prediction. Proteins: Struct., Funct., Genet. 2004, 55, 351–367. 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Gaulton A.; Bellis L. J.; Bento A. P.; Chambers J.; Davies M.; Hersey A.; Light Y.; McGlinchey S.; Michalovich D.; Al-Lazikani B.; Overington J. P. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012, 40, D1100–D1107. 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipeline Pilot, version 9.2; Scitegic Accelrys Software Inc.: San Diego, 2016. [Google Scholar]
- Mysinger M. M.; Carchia M.; Irwin J. J.; Shoichet B. K. Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J. Med. Chem. 2012, 55, 6582–6594. 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman W.; Day T.; Jacobson M. P.; Friesner R. A.; Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006, 49, 534–553. 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, version 1.8; Schrödinger, LLC: New York, 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.