Abstract
The WRKY family are transcription factors, involved in plant development, and response to biotic and abiotic stresses. Moso bamboo is an important bamboo that has high ecological, economic and cultural value and is widely distributed in the south of China. In this study, we performed a genome-wide identification of WRKY members in moso bamboo and identified 89 members. By comparative analysis in six grass genomes, we found the WRKY gene family may have experienced or be experiencing purifying selection. Based on relative expression levels among WRKY IIc members under three abiotic stresses, PeWRKY83 functioned as a transcription factor and was selected for detailed analysis. The transgenic Arabidopsis of PeWRKY83 showed superior physiological properties compared with the WT under salt stress. Overexpression plants were less sensitive to ABA at both germination and postgermination stages and accumulated more endogenous ABA under salt stress conditions. Further studies demonstrated that overexpression of PeWRKY83 could regulate the expression of some ABA biosynthesis genes (AtAAO3, AtNCED2, AtNCED3), signaling genes (AtABI1, AtPP2CA) and responsive genes (AtRD29A, AtRD29B, AtABF1) under salt stress. Together, these results suggested that PeWRKY83 functions as a novel WRKY-related TF which plays a positive role in salt tolerance by regulating stress-induced ABA synthesis.
Introduction
Grasses (Poaceae) are one of the most important plant families; they are not only the major source of food and potential renewable energy, but have huge economic and ecological value1. The grasses can be divided into several economically important subfamilies based on genomic analyses, such as the Bambusoideae (moso bamboo), Panicoideae (maize and sorghum), Ehrhartoideae (rice) and Pooideae (Brachypodium). Every year, the geographical distribution, growth, development and yield of many grass plants are limited by extreme environmental conditions, such as drought, high salinity or cold temperature2. Moso bamboo (Phyllostachys edulis), one of the most important woody bamboo, represents the only Bambusoideae plant. China is the most important area for moso bamboo, with many natural Phyllostachys edulis forests. However, in the south of China, unfavourable conditions (salinity and soil depletion) and extreme climate (drought and cold) limit the growth and distribution of moso bamboo.
Transcription factors (TFs) play a key role in plant growth and development, and respond to biotic and abiotic stresses through interaction with cis-acting elements in promoter regions, or with other TFs to regulate gene expression. Currently, there are more than 60 TF families identified with different functional roles3. The WRKY gene family is the largest TF family in plants. The WRKY TFs are named as such due to the conserved WRKYGQK sequence in the N-terminal, followed by a zinc finger motif4. In some WRKY genes, the WRKY domain is replaced by WKKY, WKRY, WSKY, WIKY, WRIC, WRMC, WRRY or WVKY5,6. There are three types of WRKY proteins in the WRKY gene family: group I-III. The classification is based on the number of WRKY domains and the pattern of the zinc finger motif. Proteins which contain two WRKY domains including a C2H2 (C-X4-5-C-X22-23-H-X-H) motif belong to group I. Group II and Group III proteins have one WRKY domain, but different finger motifs. Group II contains the same zinc-finger motif (C2H2) as group I. Group III has a C2-HC (C-X5-8-C-X25-28-H-X1-2-C) motif4. Most proteins with one WRKY domain belong to group II, and group II is further classified into five subgroups (IIa, IIb, IIc, IId, and IIe) based on their phylogenetic clades4. It is well known that WRKY proteins promote the expression of downstream target genes, since they can specifically interact with the W-box ((C/T)TGAC[T/C]) or the SURE (sugar-responsive cis-element) in the promoter region of many plant target genes7,8.
In 1994, the first WRKY protein (SPF1) was cloned from sweet potato by Ishiguro and Nakamura et al.9. Subsequently, many WRKY protein genes were cloned from different plant species especially in grass plants: rice, maize, Brachypodium distachyon, wheat and barley10–14. To date, only two WRKY homologues have been identified in non-plant species, Giardia lamblia and Dictyostelium discoideum 15,16. Moreover, the functions of WRKY proteins have been discovered. It has been shown that WRKY proteins play key roles in plant response to bacterial, fungal and viral pathogens in Arabidopsis, rice, tobacco, and parsley17–19. Furthermore, there is evidence that WRKY proteins are involved in response to various abiotic stresses, such as high temperature, low temperature, salt, drought, and ABA. In Arabidopsis, WRKY25, −26 and −33 play significant roles in response to heat tolerance20 and AtWRKY57 can improve drought stress21. There were at least nine WRKY genes in soybean which are differentially expressed under abiotic stresses22. The transgenic plants of GmWRKY21 and −13 were tolerant to cold and salt stresses, respectively. GmWRKY54 was involved in drought and salt tolerance22. In cotton, resistance to salt and drought of GhWRKY68 was reduced, when overexpressed in Nicotiana benthamiana 23. The function of WRKY proteins have been studied in grass plants. It is well documented that WRKY proteins are involved in the regulation of plant growth and developmental processes, including trichome development24, seed development and germination, embryogenesis and leaf senescence25–27.
The functional knowledge of WRKY proteins has been widely studied. The aim of this study was to identify whether PeWRKY proteins have a common function. The moso bamboo whole genome has been sequenced28; therefore a genome-wide analysis of the WRKY gene family in moso bamboo could be performed. The aim of our study was to characterize the WRKY protein family of moso bamboo, by identifying their protein sequence characteristics, phylogenetic relationships and gene structures. Moreover, PeWRKY83 was selected for detailed functional analysis.
Results
Identification and classification of PeWRKY genes
Based on the HHM profile of the WRKY domain (PF03106), 102 putative WRKY genes were identified in the moso bamboo genome (http://www.ncgr.ac.cn/bamboo, accessed February 2016). These candidates were subjected to Interproscan program to confirm the presence of the WRKY domains. Lastly, we obtained 89 protein-coding genes in moso bamboo. The 89 WRKY genes from moso bamboo are uniformly named as PeWRKY1-PeWRKY89, according to their physical locations (from top to bottom) on the chromosomes (Table S1).
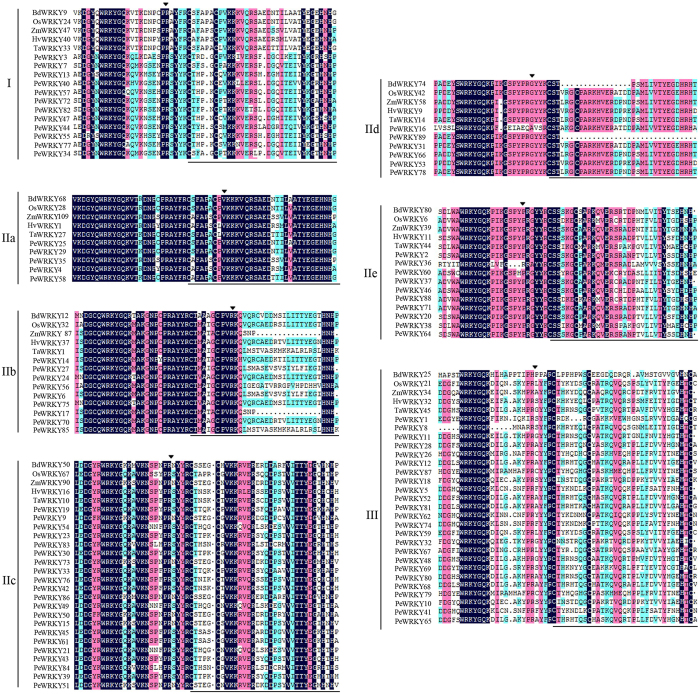
The phylogenetic relationship of the PeWRKY proteins was examined by multiple sequence alignment of their WRKY domains, which span approximately 60 amino acids (Fig. 1). The comparison analysis of the WRKY domains from six different grasses WRKY proteins, resulting in a better separation of the different groups and subgroups. For each of the groups or subgroups, I, IIa to IIe and III, one representative from Brachypodium distachyon, rice, maize, wheat and barley was chosen randomly. As shown in Fig. 1, the sequences of the WRKY domains were highly conserved. The highly conserved WRKY motif, including 80 WRKYGQK, seven WRKYGKK, one WRKYGQA and one WRKYGQQ existed in the 89 PeWRKY genes. In the 89 PeWRKYs, 13 of the WRKY proteins contained two complete WRKY domains and a C2-H2 type zinc finger motif. These proteins constituted group I. 76 WRKY proteins contained one WRKY domain, of which 51 (67%) have a C2H2-type (C-X4-5-C-X22-24-H-X1-2-H) zinc finger motif belonging to group II and the remaining 25 (33%) WRKY proteins belong to group III, which had a C2-HC type (C-X6-7-C-X23-33-H-X1-C) zinc finger motif. Furthermore, group II proteins can be divided into five distinct subgroups (2a-e), there were 5, 9, 21, 6 and 10 members in subgroups IIa-e, respectively.
Figure 1.
Alignment of multiple PeWRKY and selected other Gramineae WRKY domain amino acid sequences. Alignment was performed using the DNAMAN software. Residues that were highly conserved within each of the major groups are indark blue. The position of a conserved intron was indicated by an arrowhead. The black lines indicated the conserved zinc finger motifs.
The conserved motifs of PeWRKY proteins were further identified using MEME program. A total of 20 conserved motifs were identified. The different motifs were identified based on the biochemical properties of their amino acids and their specific location in the protein sequence (Figure S1)12. The conserved amino acids, the position of each residue in the WRKY sequence, and the residue varied greatly (Figure S1, Table S2). The conserved motifs 2, 4, 8 and 18 encoded the WRKY domains, which were broadly distributed in the PeWRKY protein sequences. From the 13 members of group I, two WRKY domains (motif 2 and motif 4) were identified, while in the other members from group II or III, there was only one WRKY domain. Most PeWRKY members from the same group, especially the closely related members, shared common motif compositions, while only seven motifs (motif 6, 8, 10, 15, 17, 18 and 19) were shared by different groups. These results suggest that the most prominent features of PeWRKY are highly conserved.
Phylogenetic analysis of PeWRKYs in six grass genomes
To examine the phylogenetic relationship among all 89 PeWRKY domains, a phylogenetic tree based on conserved WRKY domains containing Brachypodium distachyon, rice, maize, wheat and barley was constructed. Two different phylogenetic trees were created (Neighbour-Joining and Maximum Parsimony) using MEGA 6.0. The two kind of phylogenetics were largely comparable with only minor modifications at interior branches (data not shown). Therefore, the N-J phylogenetic tree was classified into three well-conserved subfamilies, group I to III (Fig. 2), as described previously and with significant statistical support10,11,13,14,29. This complex phylogenetic tree displayed the classification of PeWRKYs based on multiple sequence alignment of their WRKY domains, and the phylogenetic relationship of PeWRKYs was conserved (Figure S2).
Figure 2.
Phylogenetic tree of WRKY proteins of moso bamboo, Brachypodium distachyon, rice, maize, wheat and barley. The tree was generated using MEGA 6.0 by the NJ method with 1,000 bootstrap replicates. Moso bamboo, Brachypodium distachyon, rice, maize, wheat and barley proteins are indicated with different-coloured dots.
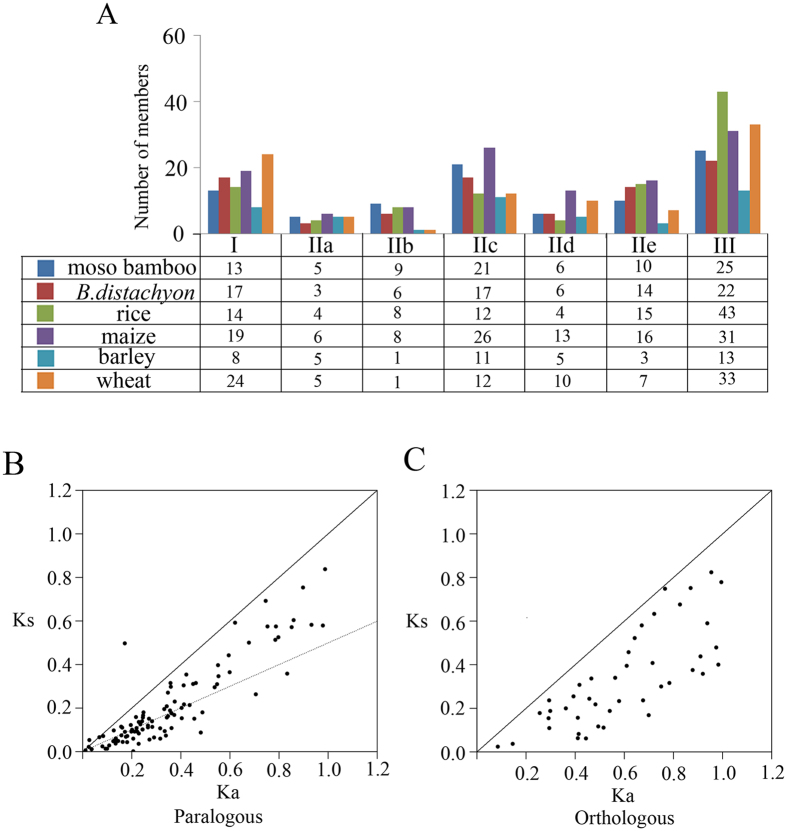
The number of WRKY proteins from each subfamily among six plant species was listed (Fig. 3A), revealing group II was the largest subfamily of the six-plant species. This was supported by that previously report-most proteins with one WRKY domain belong to group II. To clarify the paralogous and orthologous relationships among this family, we used two popular methods: phylogeny-based and bidirectional best-hit. Tables S3 details 104 putative paralogous pairs and 43 putative orthologous The Ka/Ks ratios of all the paralogous pairs ranged from 0.0078 to 0.974 and the mean value approximately 0.513 (Fig. 3B), while the Ka/Ks ratios of all 43 putative orthologous pairs ranged from 0.139 to 0.977 and the mean value approximately 0.517 (Fig. 3C). The Ka/Ks ratios of all the WRKY paralogous and orthologous pairs were less than 1, representing purifying selection on the WRKY genes.
Figure 3.
Comparison of WRKY family among six grass species and distribution of Ka and Ks values. (A) Comparison of group/subgroup size among moso bamboo, Brachypodium distachyon, Oryza sativa, Zea maize, Triticum aestivum and Hordeum vulgare WRKY family. (B) Above the black line, paralogous pairs with Ka/Ks ratio >1; between solid and dashed lines, pairs with Ka/Ks ratio 0.5–1. (C) Above the black line, orthologous pairs with Ka/Ks ratio >1; between solid and dashed lines, pairs with Ka/Ks ratio 0.5–1.
Expression profile of PeWRKY IIc genes
Numerous experiments in grasses have showed that WRKY IIc genes can be involved in various physiological processes under normal growth conditions and under various stresses. The sequence and evolutionary relationship analyses identified 21 PeWRKY genes in the group IIc. Thereafter, ten candidate PeWRKY IIc genes were selected and their expression levels in response to abiotic stress by quantitative real-time PCR (qRT-PCR) were performed.
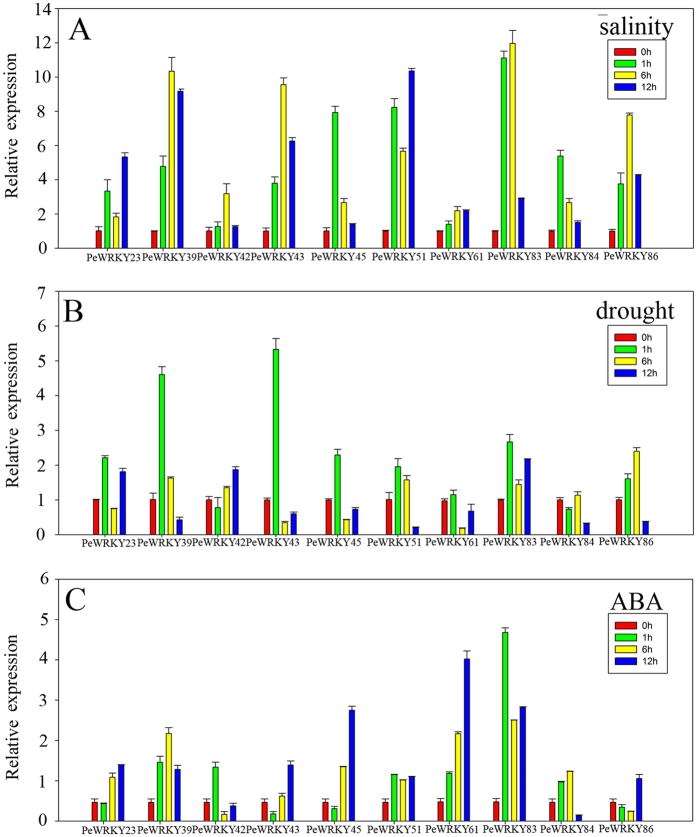
Expression of the 10 PeWRKY genes increased during the first 1 h of salt treatment. The expression of PeWRKY45, -51 and -83 were increased more than 8-fold (Fig. 4A). Two genes (PeWRKY42/84) were down-regulated rapidly following drought treatment. While the expression of the other eight PeWRKY genes peaked after 1 h. The expression of PeWRKY39 was higher 6-fold than the control (Fig. 4B). The expression of most PeWRKY genes changed significantly following ABA treatment, except for PeWRKY84 and PeWRKY86 (Fig. 4C). For example, PeWRKY61 was up-regulated more than 10-fold at 12 h. These results further support the hypothesis that WRKY genes can play important roles in regulating the response to abiotic stresses.
Figure 4.
Expression profiles of the WRKY IIc genes in 3-month-old moso bamboo seedlings subjected to salinity (A), dehydration (B) and ABA (C) stress treatments. The transcript levels of each PeWRKY in the stress-treated plants were plotted as the relative expression (fold) of the non-stressed control plants for 1, 6, and 12 h. The transcript level of Tonoplast intrinsic protein 41 gene (TIP41) was used as a reference. Mean values and standard errors (bar) were shown from three independent experiments.
From the qRT-PCR experimental results, PeWRKY83 was highly up-regulated following the three treatments and was therefore selected for further investigation (Fig. 4). The full-length cDNA of PeWRKY83 was comprised of 1074 bp and the deduced protein contained 357 amino acid residues with a predicted molecular mass of 37578.07 Da (Table S1).
Subcellular localization and DNA-binding assays
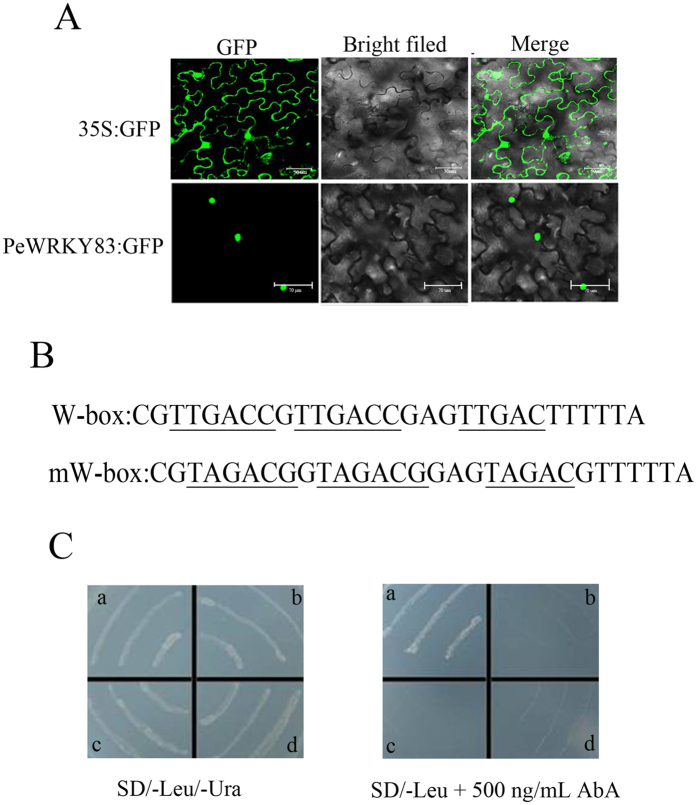
PeWRKY83 was located in the nucleus by TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/) server and WoLF PSORT (http://wolfpsort.org/). To verify this, pPeWRKY83-GFP gene expression vectors were constructed and transformed into N. tabacum. As depicted in Fig. 5A, PeWRKY83 was localized to the nucleus.
Figure 5.
Subcellular localization and DNA-binding assays. (A) Subcellular localization of PeWRKY83. (B) Sequence of the triple tandem repeats of the W-box and mW-box. (C) Yeast one-hybrid assay using the 3 × W-box or mW-box as bait. Yeast cells carrying pGAD-PeWRKY83 or pGAD7 were grown on SD/-Leu/-Ura or SD/-Leu containing 500 ng/ml AbA. (a) pAbAi-W-box/pGAD-PeWRKY83; (b) pAbAi-W-box/pGAD7; (c) pAbAi-mW-box/pGAD-PeWRKY83; and (d) pAbAi-mW-box/pGAD7.
Many studies have demonstrated that WRKY TFs can bind to the W-box [TTGAC(C/T)] to modulate protein expression. A yeast one-hybrid experiment was used to test whether PeWRKYs also have this binding characteristic. First, the two nucleotide chains (W-box/mW-box) were combined manually (Fig. 5B). Subsequently, they were inserted into vector pAbAi and fused to the yeast cells (Y1HGold), forming two strains: pAbAi-W-box and pAbAi-mW-box. Only pGAD-PeWRKY83/pAbAiW-box grew on SD/-Leu containing 500 ng/mL AbA (Fig. 5C).
Protein interaction analysis of PeWRKY83 with PeVQs
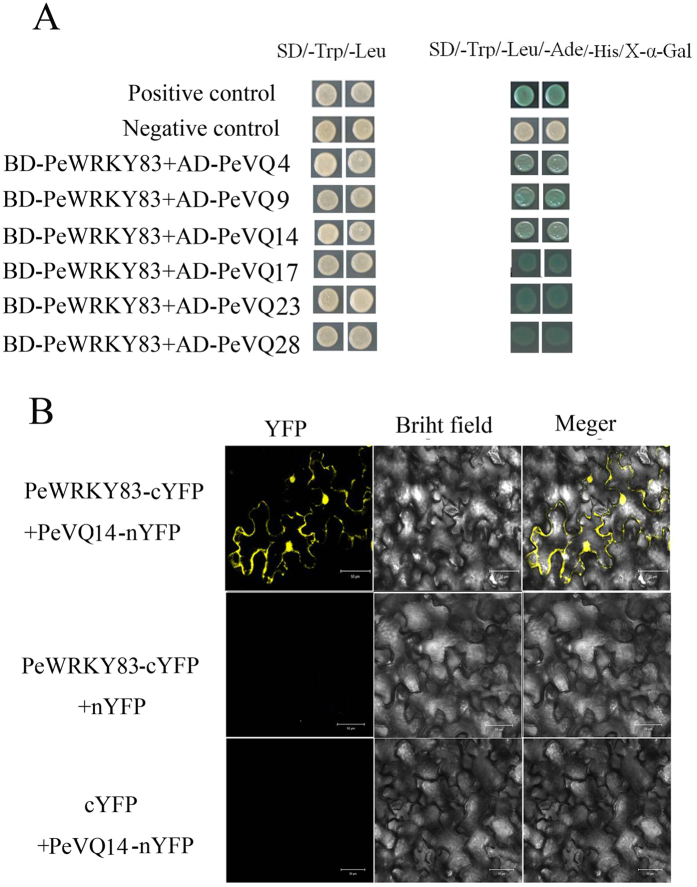
In general, the transcription factor cannot work without interaction with other proteins. Herein, PeWRKY83 showed no transactivation activity in yeast (Figure S3). WRKY proteins interact with other proteins, therefore we examined whether PeWRKY83 would interact with PeVQ proteins using a yeast two-hybrid assay. The PeWRKY83 was fused into pBD, while PeVQs were fused into pAD. The fused pBD and pAD vectors were then co-transformed into AH109 yeast cells and grown on SD/-Trp/-Leu/-Ade/-His/X-α-Gal plates for 3–5 days. The interaction ability was tested through ᾳ-galactosidase activity. As shown by the yeast two-hybrid assay (Fig. 6A), PeWRKY83 interacted with PeVQs. PeVQ14 is localized in the nucleus (data not shown), the same as PeWRKY83 (Fig. 5A). Figure 6B indicated that PeWRKY83 interacted with PeVQ14 in vivo by BiFC (bimolecular fluorescence complementation) assays in the leaves of tobacco, and the YFP signal was detected in the nuclear compartment of transformed cells.
Figure 6.
Interaction of PeWRKY83 with PeVQs. (A) Interaction of PeWRKY83 with PeVQs in yeast. The bait construct (pGBKT7-PeWRKY83) and the prey constructs (pGADT7-PeVQs) were co-transformed yeast strain AH109, then examined on SD/-Trp/-Leu and SD/-Trp/-Leu/-Ade/-His/X-α-Gal plates. Positive control, pGBKT7-53 + pGADT7-T; Negative control, pGBKT-53 + pGADT7-Lam. (B) BiFC assays of PeWRKY83 interaction with PeVQ14 in vivo. Yellow fluorescent protein (YFP) images were detected at an approximate frequency of 8.46% (1100 of 1300 tobacco leaf epidermal cells analysed exhibited BiFC events). Bars = 50 mm.
Salt tolerance of PeWRKY83 overexpressing Arabidopsis plants
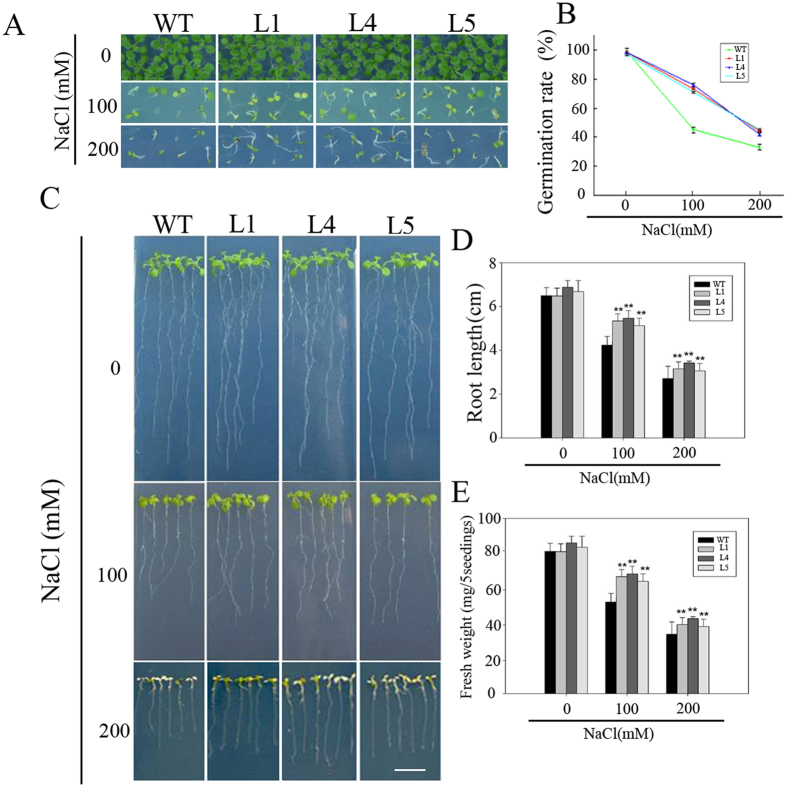
PeWRKY83, driven by CaMV 35 S promoter, was transformed into Arabidopsis plants. Three homozygous lines (L1, L4 and L5) with relatively high expression of the transgenosis were further analysed (Figure S4). We first carried out a seed germination experiment on MS plates, containing different concentrations of NaCl. No difference in germination was observed when the three transgenic lines and WT control seeds were grown on 0 mM NaCl MS plates. However, under high salt stress conditions caused by NaCl treatment, the germination rates of both WT and transgenic seeds were inhibited significantly, but the germination rate of WT seeds was significantly lower than that of transgenic seeds (Fig. 7A,B). Similarly, in the presence of 100 or 200 mM NaCl, the transgenic plants showed less growth inhibition than WT (Fig. 7C). The root length of overexpression lines (5.53–5.96 cm) showed significantly lesser suppression than WT (4.34 cm) and the fresh weight also lesser suppression (67.8%–72.57%) than WT (57.31%) under 100 mM NaCl treatment. Likewise, under 200 mM NaCl treatment, the root length of overexpression lines (3.03–3.29 cm) showed significantly lesser suppression than WT (2.75 cm) and the fresh weight also lesser suppression (39.83%–42.36%) than WT (36.43%) (Fig. 7D,E).
Figure 7.
Germination and phenotypes of PeWRKY83 in transgenic Arabidopsis under salt tolerance. (A) Germination performance of PeWRKY83-overexpression and WT seeds on 1/2 MS medium containing 0, 100, or 200 mM NaCl measured at 5 d after initiation. (B) Calculation of the germination rates of transgenic and WT seeds. (C and D) Effect of salt stress on root length and fresh weight of transgenic and WT plants. Values are means ± SE (n = 10). *P < 0.05, t-test; **P < 0.01, t-test.
The salt tolerance of transgenic plants grown in soil was also tested. After treatment with 200 mM NaCl for 20 d, most of the transgenic plants grew well, while most of WT plants wilted (Fig. 8A). Green leaf weight of the transgenic plants (0.452–0.506 g/plant) was more weighted than that of the WT plants (0.976 g/plant) (Fig. 8B). The physiological indices of WT and transgenic plants under salt stress were determined. After treatment with 200 mM NaCl, the levels of proline in the transgenic plants was higher (2.3-fold) than in the WT plants (Fig. 8C). As showed in Fig. 8D,E, REL (relative electrolyte leakage) and MDA was not significantly difference between the transgenic plants and WT before stress, yet was significantly lower than the WT plants following salt stress.
Figure 8.
Salt stress of PeWRKY83 in transgenic Arabidopsis plants. (A) Performance of WT and transgenic plants before and after salt treatment with 200 mM NaCl for 20d. (B–E) Green leaf weight of each plant, proline content, REL and MDA content were measured in WT and transgenic plants after salt treatment. Values are means ± SE (n = 3). *P < 0.05, t-test; **P < 0.01, t-test.
The transgenic Arabidopsis of PeWRKY83 was less sensitive to ABA
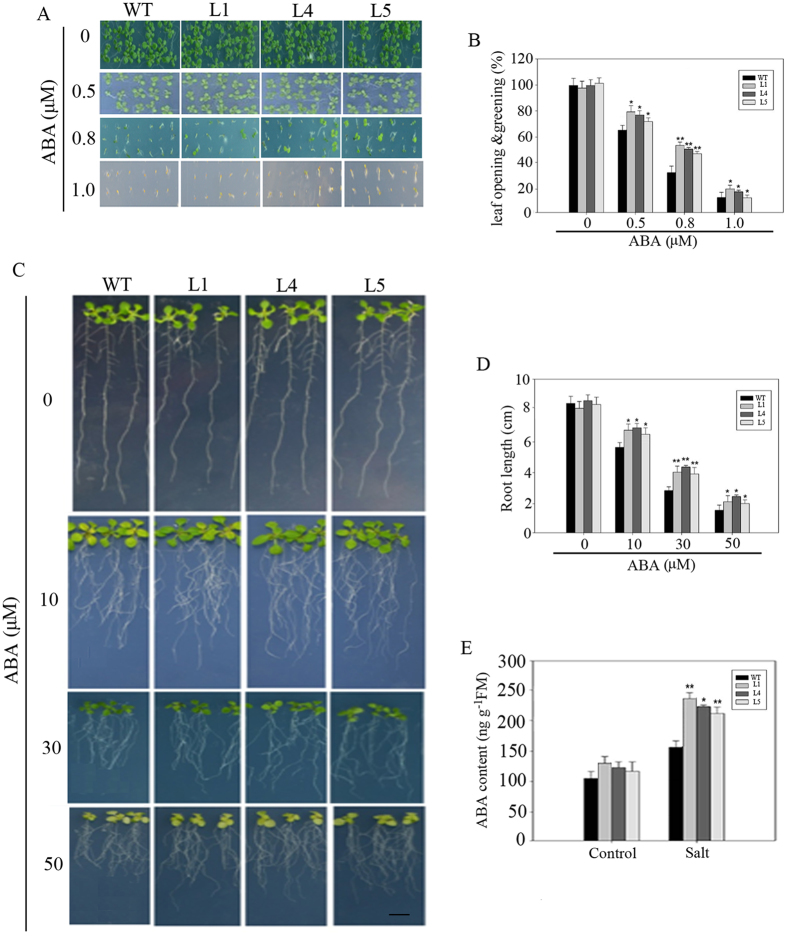
Since PeWRKY83 was strongly induced by ABA, we further tested if PeWRKY83 was involved in ABA sensitivity, an important aspect of the ABA-dependent regulation pathway. We tested ABA sensitivity of PeWRKY83 overexpressing plants at germination and seedling stage. As shown in Fig. 9A, in the normal medium, the germination rate was not significantly different from the WT and transgenic seeds. While in the ABA-supplemented medium, the germination rate of WT seeds was reduced significantly compared with the transgenic seeds. For example, under 0.8 μM ABA treatment, the leaf opening and greening rate of transgenic seeds was 56.76%–58.55%, while the leaf opening and greening rate of the WT seeds was 36% (Fig. 9B). Under different concentrations of ABA treatments, the WT plants were inhibited more severely than the PeWRKY83 overexpressing seedlings (Fig. 9C). In the medium containing 30 μM ABA, the root length of the transgenic plants (4.23–4.67 cm) was significantly greater than that of the WT plants (3.24 cm) (Fig. 9D). Figure 9E displayed that transgenic and WT plants showed non-significant differences for endogenous ABA content under normal growth conditions, however, it was significantly higher in transgenic seedlings (239–258 ng/g) than the WT (158 ng/g) under salt stress. These results suggest that overexpression of PeWRKY83 in plants can reduce sensitivity to ABA.
Figure 9.
Reduced sensitivity to ABA of PeWRKY83 in transgenic Arabidopsis. (A) Seed germination of transgenic and WT plants on 1/2 MS medium containing different ABA concentrations. (B) Statistical analysis of the leaf opening and greening rate in A. (C and D) Analysis of root length in WT and transgenic Arabidopsis under ABA stress. (E) ABA contents in normal or salt treated seedlings. Values are means ± SE (n = 10). *P < 0.05, t-test; **P < 0.01, t-test.
Expression of ABA-related genes in PeWRKY83 transgenic Arabidopsis
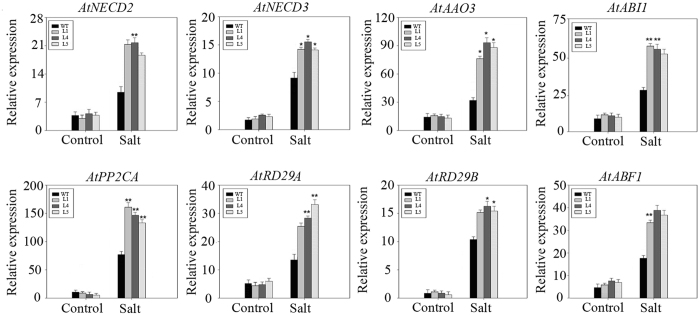
Since the transgenic plants were sensitive to exogenous ABA and accumulated more endogenous ABA under salt stress conditions, the transcription levels of some ABA biosynthesis and signaling genes were analyzed. As shown in Fig. 10, all the genes were strongly induced in the transgenic plants and wild type with salt treatment for 5 d. The transcript levels of three ABA biosynthesis genes (AtAAO3, AtNCED2 and AtNCED3) in both the transgenic plants and WT were clearly upregulated by salt stress, while the expression levels of these genes was significantly higher in transgenic plants as compared to WT. The two ABA signalling genes, AtABI1 and AtPP2CA, showed much higher expression levels in transgenic plants than that in WT under salt stress conditions. The expression levels of three well-documented ABA responsive genes, AtRD29A, AtRD29B and AtABF1, showed non-significant differences between the transgenic plants and WT under normal growth conditions. However, under salt stress conditions significant differences were observed for higher expression levels of these genes in transgenic plants as compared to WT were detected. These results suggested that PeWRKY83 can regulate the expression of ABA-related genes in transgenic Arabidopsis under salt treatment.
Figure 10.
Expression levels of ABA-related genes in PeWRKY83 transgenic plants. Leaves of WT and transgenic Arabidopsis plants were sampled at 5 d after salt stress. Values are means ± SE (n = 3). *P < 0.05, t-test; **P < 0.01, t-test.
Discussion
Moso bamboo is a fastest-growing plant and most important non-timber forest product in the world. WRKY proteins are members of a transcription factor family in higher plants and have a crucial role in plant growth and development. In this study, we identified 89 candidate WRKY proteins in Phyllostachys edulis. By multiple sequence alignment of their WRKY domains, the PeWRKY genes were classified into three groups (groups I, II, III). As illustrated in Fig. 1, the WRKYGQK signature was highly conserved among moso bamboo WRKY proteins, but a slight variation was identified in 9 genes (PeWRKY7, -21, -39, -43, -45, -51, -55, -61 and -84). Especially genes in group IIc had more variation (33.3%) than genes in other WRKY groups, which suggested that WRKY genes in group IIc are more active and variable. This phenomenon has also been reported in Arabidopsis thaliana, Oryza sativa, Hordeum vulgare, tobacco, canola, sunflower and soybean6,11,16,30–33. Based on sequence comparison and phylogenetic analysis, the PeWRKY genes were classified into three groups (groups I, II, III), and group II genes were further classified into IIa, IIb, IIc, IId and IIe. The phylogenetic analysis (Fig. 2) showed genes in group IIa clustered with group IIb and that group IId genes are closely related to IIe, which support the classifications of the three subgroups in group II: group IIa/group IIb, group IIc, and group IId/group IIe16,34. The Ka/Ks ratios of all WRKY paralogous and orthologous pairs were less than 1, suggesting purifying selection (Fig. 3).
The WRKY gene family play key roles in plant development and response to various abiotic stresses and, there is increasing evidence which suggest WRKY IIc proteins regulate response to abiotic stress. For example, Arabidopsis WRKY8 was highly up-regulated by salt treatment35. Transgenic Arabidopsis overexpressing TaWRKY19 displayed improved drought and salt stress36. A cotton WRKY IIc gene, GhWRKY68, responded to drought and salt stress23. In soybean, there were also two IIc genes (GmWRKY21/54) that conferred differential tolerance to abiotic stresses in transgenic Arabidopsis plants22. Hence, we randomly selected ten genes from group IIc to study the function of PeWRKY IIc genes. Expression profile analysis revealed that PeWRKY IIc genes were induced significantly by salt, drought and ABA stresses (Fig. 4). PeWRKY83 was highly expressed under the three treatments; therefore PeWRKY83 was selected for investigation.
To study the functions of PeWRKY83 in plants, we transformed PeWRKY83 into Arabidopsis. In many reports, Arabidopsis plants have been used in transgenic studies for stress-tolerant genes from species which are not easy to transform including wheat, soybean and moso bamboo37–39. Overexpression of PeWRKY83 improved stress tolerance in transgenic Arabidopsis plants based on phenotype and changes in physiological parameters such as proline, MDA and electrolyte leakage after stress treatments (Figs 7 and 8). The content of proline in PeWRKY83 transgenic plants was significantly higher than in WT plants under stress conditions. The plants retain water effectively, when the content of proline in plants is increased40. REL reflects membrane injury after stresses. MDA is a product of lipid peroxidation in biomembranes41. REL and MDA accumulation were reported as indicators of damage caused by abiotic stresses42,43. The transgenic plants of PeWRKY83 had lower MDA content than WT plants under salt stress, suggesting overexpression of PeWRKY83 in plants can lead to improved tolerance to oxidative stress caused by salt stress. In accordance with the lower content of MDA, overexpression of PeWRKY83 in Arabidopsis also led to a lower amount of REL compared with WT plants, suggesting that the degree of cell membrane damage in transgenic plants caused by abiotic stress was less than in WT plants.
Seed germination, seedling growth and plant development may be inhibited by ABA44. ABA accumulation is induced through ABA-dependent signalling pathways under various stresses45. Herein, seed germination and seedling root growth of PeWRKY83 transgenic plants were inhibited by exogenous ABA (Fig. 9). Moreover, the transgenic lines accumulated more ABA compared with the WT plants during salt stress (Fig. 9F). These results indicate that PeWRKY83 may increase salt tolerance by positively regulating ABA pathways. This was consistent with previous observations in other plant species22,23,46.
In previous studies, several experiments have demonstrated that overexpression of TFs regulate expression of stress/ABA-responsive genes and enhanced tolerance to various stresses by overexpression in plants47,48. In our study, expression of the eight marker genes was induced significantly in transgenic Arabidopsis plants compared with WT plants under salt treatment (Fig. 10), which indicated PeWRKY83 may affect expression in the upstream region. The expression of 9-cis epoxycarotenoid dioxygenase (NCED) and abscisic acid biosynthetic enzyme (AAO) were generally considered to be participated in ABA biosynthesis49–52. These three genes showed similar low expression levels in transgenic plants and WT under normal conditions. While under salt stress conditions, the expression levels of AtNCED2, AtNCED3 and AtAAO3 in transgenic plants was higher than that in the WT plants. Above results suggested that PeWRKY83 possible role in stress-induced ABA-biosynthesis along with additional factors. Furthermore, the expression levels of ABA signaling genes(ABI1 and AtPP2CA) and ABA-responsive genes(AtRD29A, AtRD29B and AtABF1) was examined in the transgenic plants and WT under normal and salt stress conditions. AtABI1 and AtPP2CA encode a protein phosphatase 2C, which plays a prime role in ABA-mediated signaling network related to stress responses53. Under salt stress conditions, ABI1 and AtPP2CA had higher expression levels in the transgenic plants than that in WT. AtRD29A, AtRD29B and AtABF1 have been reported to be marker genes in ABA-dependent stress response way54,55. We identified upon salt stress, the expression levels of these genes were significantly higher in the transgenic plants than WT. Overall the results suggested that the overexpression of PeWRKY83 had further induced ABA synthesis in transgenic plants under salt stress conditions, thus, leading to increased expression levels of ABA signaling and -related genes.
Most transcription factors interact via protein-protein and protein-DNA. The WRKY transcription factors can participate in protein-protein interactions with other transcriptional proteins. AtMEKK1 directly interacts with a senescence-related transcription factor (AtWRKY53) at the protein level56. Furthermore, Sun et al. showed that eleven Arabidopsis WRKY proteins (Group IId) can bind to a Ca2+-binding signalling protein (calmodulin)56. In addition, Cheng et al. used yeast two-hybrid assays to show that approximately half of the VQ proteins interact specifically with the WRKY domains of group Ic and group IIc WRKY proteins57. Recently, Chi et al. identified HDAC and histone proteins, which can interact with WRKY proteins58. In this study, we first examined the interaction between PeWRKY83 and PeVQs using yeast two-hybrid assay. In early reports, AtVQ9 interacted with AtWRKY8, and AtVQ1 interacted with AtWRKY33 3,35. In moso bamboo, there were 29 non-redundant VQ genes, named VQ1 to VQ29, according to their Sequence ID (Table S5). We randomly selected six PeVQs (PeVQ4, PeVQ9, PeVQ14, PeVQ17, PeVQ23 and PeVQ28) and found that PeWRKY83 (IIc) can interact with these six PeVQs by yeast two-hybrid assays (Fig. 6A); similar to findings of previous reports57. We further verified our result using BiFC assays (Fig. 6B), revealing that PeWRKY83 can interact with PeVQ14. By taking advantage of the results obtained from this study and previous observations, we conclude that the WRKY gene family in moso bamboo can take part in the interaction of protein and protein.
Experimental procedures
The WRKY genes in moso bamboo
To identify the non-redundant WRKY genes in the moso bamboo genome, we searched the National Centre for Gene Research database (http://www.ncgr.ac.cn/bamboo)28, using the Hidden Markov Model (HMM) profile of the WRKY domain (PF03106). All redundant sequences were discarded from further analysis based on Cluster W alignment results59. To confirm the WRKY domain in identified proteins, domain analysis was performed using Interproscan tool (http://www.ebi.ac.uk/Tools/pfa/iprscan5/)60. Furthermore, the WRKY domain and zinc-finger motif in the protein sequences was analysed by DNAMAN software and modified manually. Moso bamboo gene information, including chromosome locations, ORF lengths, molecular weight (MW), isoelectric point (pI) and the number of amino acids, were obtained from the Bamboo GDB server (http://www.bamboogdb.org). The WRKY proteins were analysed using MEME (Multiple Expectation Maximization for Motif Elicitation) program (http://meme.nbcr.net/meme/cgi-bin/meme.cgi)61,62. The ScanProsite database was used to annotate the identified protein motifs63.
Selective pressure analyses
A total of 415 WRKY protein sequences (85 Brachypodium distachyon, 102 Oryza sativa, 136 Zea maize, 92 Triticum aestivum) were obtained from Phytozome v11.1 (http://www.phytozome.net/), and 46 Hordeum vulgare WRKY proteins sequences were downloaded from NCBI databases. Phylogenetic trees were constructed based on the bootstrap neighbour-joining (NJ) method and bootstrap analysis (1,000 replicates) by MEGA 6.0 64. The putative orthologous and paralogous pairs were identified using the two popular methods: phylogeny-based and bidirectional best-hit, these criteria were described in previous studies65,66. The variations in selective pressures among these six species were evaluated using PAML software67.
Subcellular localization and DNA-binding assays
The coding region of PeWRKY83 was cloned and then constructed into pCAMBIAI1305 vector (Clontech, Beijing, country-region China), which contains a CaMV 35 S promoter and GFP gene, resulting in fusion PeWRKY83-GFP gene. The constructed pPeWRKY83-GFP vector was transformed into Agrobacterium tumefaciens GV3101 using a Gene Pulser Xcell (BIO-RAD, country-region USA). The suspensions were injected into the leaves of Nicotiana tabacum. The expression level of PeWRKY83-GFP was observed by confocal laser scanning microscopy (CarlZeiss LSM710, Germany).
The DNA-binding assays were tested by yeast one-hybrid assay as described by Jia et al.23. Briefly, W-box-specific reporter (pAbAi-Wbox) was used as bait. A yeast effectors vector, pGADT7-PeWRKY83, and an empty vector, pGADT7, were transformed into the yeast strain Y1H Gold carrying a pAbAiW-box or a pAbAi-mW-box plasmid. All of the transformed yeast cells were placed on SD/-Leu/-Ura medium and the yeast growth was restrained by Aureobasidin A (AbA). The mutant W-box and mW-box (TAGACG) were used as a negative control.
Yeast two-hybrid and BiFC assays
To confirm the interaction, the Matchmaker GAL4 two-hybrid system (Clontech, Palo Alto, CA) was used. pBD-PeWRKYs and pAD-PeVQs fusion constructs were generated from the full length of PeWRKY83 and PeVQ cDNAs, respectively. The prey and bait plasmids were transformed to yeast strain AH109. The transformed yeast cells were plated on selective SD/-Trp/-Leu, or SD/-Trp/-Leu/-Ade/-His plates, at 30 °C for 3–5 days to determine the protein-protein interaction.
The BiFC assays were carried out as previously described68. In short, the cDNAs of PeWRKY83 and PeVQ14 were cloned into pSPYN(C) E-35S. The resulting constructs (PeWRKY83-cYFP and PeVQ14-nYFP) were transformed into Agrobacterium strain GV3101 and infiltrated into the 3-week-old leaves of tobacco (Nicotiana tabacum) plants, and analysed after infected 48 h. Fluorescence was observed under a confocal laser scanning microscope (Olympus, http://www.olympus-global.com).
Plant growth conditions and abiotic stress treatments
Abiotic treatment experiments were performed on three-month-old moso bamboo seedlings, which were grown in an artificial growth chamber with a constant photoperiod (16 h light/8 h darkness) and kept at 22 °C. For dehydration, salinity and ABA treatments, the seedlings were poured into 25% PEG-6000, 200 mM NaCl and 10 μM ABA solutions, respectively. One whole plant was sampled as one replicate and in total four replicates was used in each RNA exaction. Each test was repeated at least three times. Samples for RNA extractions were collected at 0, 1, 6 and 12 h after treatment and stored at −80 °C for RNA isolation.
Salt tolerance of transgenic Arabidopsis plants
The coding sequence of PeWRKY83 was cloned into the transgenic vector pCAMBIA1301 under control of CaMV 35 S promoter, and then the vector was transferred into Agrobacterium tumefaciens strain GV3101 by freeze–thaw method. Positive transgenic Arabidopsis plants were selected on MS plates containing hygromycin (25 mg/L), homozygous T3 or T4 seeds were used.
For germination, the seeds of transgenic and WT were surface-sterilized and sown on 0, 100 and 200 mM NaCl MS plates, and kept at 4 °C for 3 d (for vernalization). The germination rate was determined after 3–5 d under normal conditions. Transgenic and WT seedlings were germinated normally on MS plates using the same method, then moved to MS plates containing 0, 100 and 200 mM NaCl under normal conditions for 7 d, root length and fresh weight were recorded. These seedlings were further transferred to pots under normal conditions. For salt stress, three-week plants were irrigated with 200 mM NaCl solution for 20 d and phenotypic changes were recorded for both treated and control plants.
ABA sensitivity test of transgenic plants
First, the seeds of transgenic and WT were germinated on 1/2 MS medium plates, or containing 0.5 μM, 0.8 μM and 1 μM ABA to test germination rate. After 3–5 d, the leaf opening and greening was recorded. The geminated Arabidopsis plantlets of the transgenic seed and WT at the same stage were transferred to 1/2 MS medium with different concentrations of ABA (0, 10, 30 and 50 μM). After 7 days in the greenhouse at 22 °C, root length was measured. ABA was extracted as described previously by Olivella et al. (1988), and the ABA content was measured using an ELISA kit (Fangcheng, Beijing, China) according to the manufacturer’s instructions.
RNA isolation, qRT-PCR analysis
Total RNA was extracted according to the manufacturer’s instructions of the RNAprep Pure Plant Kit (Tiangen). First-strand cDNA was synthesised using a PrimeScriptTM RT Reagent Kit (TaKaRa). Gene-specific primers of PeWRKYs and ABA-related genes were designed using Primer Express 3.0 (Table S4). Moso bamboo TIP41 (Genbank: gil242384689) or Arabidopsis Tublin was used as internal controls for normalization of the template cDNA. Real-time quantitative RT-PCR (qRT-PCR) was performed on an ABI 7300 Real-Time system (Applied Biosystems). Each reaction was performed in 20 μl (total volume) and consisted of 10 μl SYBR Green Master mix (Applied Biosystems, USA), 1 pmol of each primer, 2 μl cDNA templates and sterile H2O. The steps performed during real-time PCR were as follows: step (1) 50 °C, 2 min; step (2) 95 °C, 7 min; step (3) (95 °C, 5 s; 60 °C, 30 s) × 40 cycles. The data from real-time PCR amplification was estimated in terms of comparative fold expression following 2−∆∆ct method and three biological replicates were carried out.
Measurement of relative electrolyte leakage (REL), proline and malondialdehyde (MDA) contents
Seedlings of transgenic and control plants were cultured in an artificial climate chamber (22 °C) for three weeks. The seedlings were then subjected to salt treatment with 200 mM NaCl solution for 20 d, the levels of relative electrolyte leakage (REL), proline and malondialdehyde (MDA) in WT and transgenic lines without or with salt treatment were measured by the methods described previously69,70.
Statistical analysis
Data analysis was conducted using SPSS v10.0, and significant differences were determined by Student’s t-test at significance levels of P < 0.01 (**) and P < 0.05 (*).
Electronic supplementary material
Acknowledgements
This work was supported by the National Nature Science Foundation of China (gant No. 31670672).
Author Contributions
M.W. and R.C. conceived and designed the study, performed the experiments, and drafted the manuscript. H.L. participated in the design of the experiments. G.H. and F.L. implemented the software. Y.X. conceived and directed the study, participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10795-z
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bevan MW, Garvin DF, Vogel JP. Brachypodium distachyon genomics for sustainable food and fuel production. Curr Opin Biotechnol. 2010;21:211–217. doi: 10.1016/j.copbio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Chittaranjan K, et al. Application of genomics-assisted breeding for generation of climate resilient crops: progress and prospects. Front. Plant Sci. 2015;6:563. doi: 10.3389/fpls.2015.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai Z, et al. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell. 2011;23:3824. doi: 10.1105/tpc.111.090571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eulgem T, Rushton PJ, Robatzek S, Somssich I, Somssich IE. WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 5.Rushton DL, et al. WRKY transcription factors: key components in abscisic acid signalling. Plant Biotechnol. J. 2012;10:2–11. doi: 10.1111/j.1467-7652.2011.00634.x. [DOI] [PubMed] [Google Scholar]
- 6.Xie Z, et al. Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant physiol. 2005;137:176–189. doi: 10.1104/pp.104.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R. Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol. Biol. 1995;29:691–702. doi: 10.1007/BF00041160. [DOI] [PubMed] [Google Scholar]
- 8.Sun C, et al. A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 2003;15:2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. MGG. 1994;244:563–571. doi: 10.1007/BF00282746. [DOI] [PubMed] [Google Scholar]
- 10.Wen F, et al. Genome-wide evolutionary characterization and expression analyses of WRKY family genes in Brachypodium distachyon. DNA Res. 2014;21:327. doi: 10.1093/dnares/dst060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangelsen E, et al. Phylogenetic and comparative gene expression analysis of barley (Hordeum vulgare) WRKY transcription factor family reveals putatively retained functions between monocots and dicots. BMC Genom. 2008;9:194. doi: 10.1186/1471-2164-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross CA, Liu Y, Shen QJ. The WRKY gene family in rice (Oryza sativa) Journal of Integrative Plant Biology. 2007;49:827–842. doi: 10.1111/j.1744-7909.2007.00504.x. [DOI] [Google Scholar]
- 13.Wei K-F, Chen J, Chen Y-F, Wu L-J, Xie D-X. Molecular phylogenetic and expression analysis of the complete WRKY transcription factor family in maize. DNA Res. 2012;19:153–164. doi: 10.1093/dnares/dsr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu X, et al. WRKY transcription factors in wheat and their induction by biotic and abiotic stress. Plant Mol. Biol. 2013;31:1053–1067. doi: 10.1007/s11105-013-0565-4. [DOI] [Google Scholar]
- 15.Glöckner G, et al. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature. 2002;418:79–85. doi: 10.1038/nature00847. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 2005;5:1. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 2003;51:21–37. doi: 10.1023/A:1020780022549. [DOI] [PubMed] [Google Scholar]
- 18.Kalde M, Barth M, Somssich IE, Lippok B. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol Plant Microbe Interact. 2003;16:295–305. doi: 10.1094/MPMI.2003.16.4.295. [DOI] [PubMed] [Google Scholar]
- 19.Oh S-K, et al. CaWRKY2, a chili pepper transcription factor, is rapidly induced by incompatible plant pathogens. Molecules & Cells. 2006;22:58. [PubMed] [Google Scholar]
- 20.Li S, Fu Q, Chen L, Huang W, Yu D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta. 2011;233:1237–1252. doi: 10.1007/s00425-011-1375-2. [DOI] [PubMed] [Google Scholar]
- 21.Jiang Y, Liang G, Yu D. Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Molecular Plant. 2012;5:1375–1388. doi: 10.1093/mp/sss080. [DOI] [PubMed] [Google Scholar]
- 22.Zhou QY, et al. Soybean WRKY‐type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 2008;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- 23.Jia H, et al. GhWRKY68 reduces resistance to salt and drought in transgenic Nicotiana benthamiana. PloS one. 2015;10:e0120646. doi: 10.1371/journal.pone.0120646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang W, Yu D. Arabidopsis WRKY2 transcription factor mediates seed germination and postgermination arrest of development by abscisic acid. BMC Plant Biol. 2009;9:96. doi: 10.1186/1471-2229-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao Y, Laun T, Zimmermann P, Zentgraf U. Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 2004;55:853–867. doi: 10.1007/s11103-005-2142-1. [DOI] [PubMed] [Google Scholar]
- 26.Ueda M, Zhang Z, Laux T. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell. 2011;20:264–270. doi: 10.1016/j.devcel.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Xie Z, Zhang Z-L, Hanzlik S, Cook E, Shen QJ. Salicylic acid inhibits gibberellin-induced alpha-amylase expression and seed germination via a pathway involving an abscisic-acid-inducible WRKY gene. Plant Mol. Biol. 2007;64:293–303. doi: 10.1007/s11103-007-9152-0. [DOI] [PubMed] [Google Scholar]
- 28.Peng Z, et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla) Nature Genet. 2013;45:456–461. doi: 10.1038/ng.2569. [DOI] [PubMed] [Google Scholar]
- 29.Narusaka Y, et al. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003;34:137–148. doi: 10.1046/j.1365-313X.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 30.Bencke-Malato M, et al. Genome-wide annotation of the soybean WRKY family and functional characterization of genes involved in response to Phakopsora pachyrhizi infection. BMC Plant Biol. 2014;14:236. doi: 10.1186/s12870-014-0236-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang B, Jiang Y, Rahman MH, Deyholos MK, Kav NN. Identification and expression analysis of WRKY transcription factor genes in canola (Brassica napus L.) in response to fungal pathogens and hormone treatments. BMC Plant Biol. 2009;9:68. doi: 10.1186/1471-2229-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giacomelli JI, Ribichich KF, Dezar CA, Chan RL. Expression analyses indicate the involvement of sunflower WRKY transcription factors in stress responses, and phylogenetic reconstructions reveal the existence of a novel clade in the Asteraceae. Plant Sci. 2010;178:398–410. doi: 10.1016/j.plantsci.2010.02.008. [DOI] [Google Scholar]
- 33.van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJ. A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol. 2008;146:1983–1995. doi: 10.1104/pp.107.112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends Plant Sci. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Hu Y, et al. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. 2013;74:730–745. doi: 10.1111/tpj.12159. [DOI] [PubMed] [Google Scholar]
- 36.NIU CF, et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 2012;35:1156–1170. doi: 10.1111/j.1365-3040.2012.02480.x. [DOI] [PubMed] [Google Scholar]
- 37.Ge W, et al. Main regulatory pathways, key genes and microRNAs involved in flower formation and development of moso bamboo (Phyllostachys edulis) Plant Biotechnol. J. 2017;15:82–96. doi: 10.1111/pbi.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao YJ, et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011;68:302–313. doi: 10.1111/j.1365-313X.2011.04687.x. [DOI] [PubMed] [Google Scholar]
- 39.He X, Hou X, Shen Y, Huang Z. TaSRG, a wheat transcription factor, significantly affects salt tolerance in transgenic rice and Arabidopsis. FEBS letters. 2011;585:1231–1237. doi: 10.1016/j.febslet.2011.03.055. [DOI] [PubMed] [Google Scholar]
- 40.Song S, Chen Y, Zhao M, Zhang W-H. A novel Medicago truncatula HD-Zip gene, MtHB2, is involved in abiotic stress responses. Environ Exp Bot. 2012;80:1–9. doi: 10.1016/j.envexpbot.2012.02.001. [DOI] [Google Scholar]
- 41.Heath RL, Packer L. Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys. 1968;125:189–198. doi: 10.1016/0003-9861(68)90654-1. [DOI] [PubMed] [Google Scholar]
- 42.Dong C-H, et al. A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Molecular and Cellular Biology. 2006;26:9533–9543. doi: 10.1128/MCB.01063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taulavuori E, Hellström EK, Taulavuori K, Laine K. Comparison of two methods used to analyse lipid peroxidation from Vaccinium myrtillus (L.) during snow removal, reacclimation and cold acclimation. J. Exp. Bot. 2001;52:2375–2380. doi: 10.1093/jexbot/52.365.2375. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, et al. Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 2014;14:103. doi: 10.1186/1471-2229-14-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiong L, Wang R-G, Mao G, Koczan JM. Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant physiol. 2006;142:1065–1074. doi: 10.1104/pp.106.084632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Q-L, et al. Functional analysis of a novel chrysanthemum WRKY transcription factor gene involved in salt tolerance. Plant Mol. Biol. 2014;32:282–289. doi: 10.1007/s11105-013-0639-3. [DOI] [Google Scholar]
- 47.Peng Y, et al. Overexpression of a PLDα1 gene from Setaria italica enhances the sensitivity of Arabidopsis to abscisic acid and improves its drought tolerance. Plant Cell Rep. 2010;29:793–802. doi: 10.1007/s00299-010-0865-1. [DOI] [PubMed] [Google Scholar]
- 48.Xu G-Y, et al. A novel rice calmodulin-like gene, OsMSR2, enhances drought and salt tolerance and increases ABA sensitivity in Arabidopsis. Planta. 2011;234:47–59. doi: 10.1007/s00425-011-1386-z. [DOI] [PubMed] [Google Scholar]
- 49.Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005;56:165–185. doi: 10.1146/annurev.arplant.56.032604.144046. [DOI] [PubMed] [Google Scholar]
- 50.Iuchi S, et al. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 51.Tan BC, et al. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003;35:44–56. doi: 10.1046/j.1365-313X.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 52.Koiwai H, et al. Tissue-specific localization of an abscisic acid biosynthetic enzyme, AAO3, in Arabidopsis. Plant Physiol. 2004;134:1697–1707. doi: 10.1104/pp.103.036970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007;12:343–351. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 54.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol. 2003;6:410–417. doi: 10.1016/S1369-5266(03)00092-X. [DOI] [PubMed] [Google Scholar]
- 55.Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun J, et al. The Brachypodium distachyon BdWRKY36 gene confers tolerance to drought stress in transgenic tobacco plants. Plant Cell Rep. 2015;34:23–35. doi: 10.1007/s00299-014-1684-6. [DOI] [PubMed] [Google Scholar]
- 57.Cheng Y, et al. Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant physiol. 2012;159:810–825. doi: 10.1104/pp.112.196816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chi Y, et al. Protein–protein interactions in the regulation of WRKY transcription factors. Molecular plant. 2013;6:287–300. doi: 10.1093/mp/sst026. [DOI] [PubMed] [Google Scholar]
- 59.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chu Z, et al. Genome-wide identification, classification, and analysis of two-component signal system genes in maize. Genet Mol Res. 2011;10:3316–3330. doi: 10.4238/2011.December.8.3. [DOI] [PubMed] [Google Scholar]
- 63.De Castro E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang S, Zhang X, Yue J-X, Tian D, Chen J-Q. Recent duplications dominate NBS-encoding gene expansion in two woody species. MGG. 2008;280:187–198. doi: 10.1007/s00438-008-0355-0. [DOI] [PubMed] [Google Scholar]
- 66.Gu Z, Cavalcanti A, Chen F-C, Bouman P, Li W-H. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol Biol Evol. 2002;19:256–262. doi: 10.1093/oxfordjournals.molbev.a004079. [DOI] [PubMed] [Google Scholar]
- 67.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Bioinformatics. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 68.Walter M, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 69.Pennycooke JC, et al. The low temperature-responsive, SolanumCBF1 genes maintain high identity in their upstream regions in a genomic environment undergoing gene duplications, deletions, and rearrangements. Plant Mol. Biol. 2008;67:483. doi: 10.1007/s11103-008-9333-5. [DOI] [PubMed] [Google Scholar]
- 70.Wang N, et al. Adult intussusception: a retrospective review of 41 cases. World J Gastroenterol. 2009;15:3303–3308. doi: 10.3748/wjg.15.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.