Abstract
As a well-known long non-coding RNA, HOTAIR has been demonstrated to be involved in carcinogenesis and progression of various human cancers. Previous studies have investigated the potential association between HOTAIR polymorphisms and cancer risk in Chinese population. However, the results remain conflicting. Therefore, for the first time, we conducted a meta-analysis to derive a more precise estimation of these associations for Chinese. PubMed, Embase, CNKI and Wanfang databases were systematically searched. Odds ratios with 95% confidence intervals were applied to assess the association between rs920778, rs4759314, rs7958904, rs874945 and rs1899663 polymorphisms of HOTAIR and cancer susceptibility. Heterogeneity, sensitivity analysis and publication bias were conducted to measure the robustness of our findings. A total of 21 eligible studies comprising 12,278 cases and 14,532 controls were analyzed. The pooled data showed that rs920778 polymorphism was significantly associated with an increased cancer risk in all five genetic models in Chinese population. As for rs4759314 and rs874945 polymorphisms, similarly increased risks were found in specific genetic models and stratified groups. However, significant decreases in cancer risk were observed for rs7958904 in the total population, as well as in subgroup analyses. In addition, lack of association was detected between rs1899663 polymorphism and cancer susceptibility. In summary, our meta-analysis implicates possible relationship between HOTAIR polymorphisms and cancer risk in Chinese population.
Keywords: HOTAIR, polymorphism, cancer risk, Chinese population, meta-analysis
INTRODUCTION
Long non-coding RNAs (lncRNAs), as a type of transcribed RNAs which are longer than 200 nucleotides with no protein-coding capacity, were initially claimed to be a fake transcriptional noise [1, 2]. Nowadays, it is becoming clear that lncRNAs are involved in a wide range of biological regulation in the carcinogenesis and progression of various human cancers [3–6]. HOTAIR, an lncRNA located on chromosome 12q13.13, is coded from the homebox C gene (HOXC) locus [7]. It has been demonstrated that HOTAIR could specifically interact with polycomb repressive complex 2 (PRC2) and LSD1/CoREST/REST complex, in turn induce its relating methylation of histone H3K27 and demethylation of histone H3K4 respectively, and consequently result in the alteration of genes expression profile [8, 9]. The aberrant expression of HOTAIR has been reported in a variety of human cancers such as breast cancer, gastric cancer, colorectal cancer and liver cancer [10–13]. In addition, HOTAIR was also shown to be involved in the progression of multiple types of cancers, indicating that HOTAIR might serve as a useful biomarker for tumorigenesis and progression [8, 14–16].
Several single nucleotide polymorphisms (SNPs) located in HOTAIR locus have been identified [17, 18]. Among them, the rs920778, rs4759314, rs7958904, rs874945 and rs1899663 polymorphisms are common and widely studied. In 2014, Zhang et al. firstly reported the association between three HOTAIR polymorphisms and cancer risk in Chinese population [19]. From then on, increasing epidemiologic studies from Chinese population explored the association of the common polymorphisms in HOTAIR with the risk of cancers including gastrointestinal cancers [18–22], estrogen-dependent cancers (cervical cancer, ovarian cancer and breast cancer) [23–27], thyroid carcinoma [28] and osteosarcoma [29]. However, the results are inconsistent. Also, as individual studies with limited sample sizes are difficult to obtain reliable conclusions; further validation of the results is needed. Thus, to get a more precise conclusion, we conducted a meta-analysis involving all eligible studies published to date to estimate the association between HOTAIR polymorphisms and cancer risk in Chinese population. To our knowledge, this is the first meta-analysis which investigates the association for Chinese.
RESULTS
Study characteristics
The screening process of the studies was shown in Figure 1. A total of 51 relevant articles were initially retrieved by using our search strategy. After reviewing the titles and abstracts, 27 obviously irrelevant or duplicate articles were first excluded and 24 potential articles were left for further evaluation. Among these 24 articles, 8 reviews, letters or meta-analyses, 1 studies not on focus polymorphism locus [30], 1 study unavailable for data extraction [31] and 2 studies not relating to Chinese population [32, 33] were excluded. Finally, 12 eligible articles (21 studies) published from 2014 to 2016 were included in our meta-analysis. There are 13 studies available for rs920778 C>T polymorphism [19, 20, 25–28], 12 studies for rs4759314 A>G polymorphism [18–25, 28, 29], 6 studies for rs7958904 G>C polymorphism [18, 21, 24, 29], 5 studies for rs874945 G>A polymorphism [18, 21, 24, 29] and 5 studies for rs1899663 G>T polymorphism [19, 20, 23, 25, 28], respectively. The main characteristics and genotype distributions of all included studies were summarized in Supplementary Table 1.
Figure 1. Flow diagram of the study selection process.
Quantitative analysis
Meta-analysis for HOTAIR rs920778 C>T polymorphism
Thirteen eligible studies including 6,854 cases and 8,477 controls were recruited in the meta-analysis. The results for the association between HOTAIR rs920778 polymorphism and cancer risk are presented in Table 1. The pooled analyses indicated that rs920778 polymorphism was significantly associated with an increased susceptibility of overall cancer in allelic, recessive, dominant, homozygous and heterozygous genetic models (Figure 2). The similar associations were observed both in estrogen-dependent cancers and gastrointestinal cancers when subsequently stratified by cancer type (Table 1). Analyses accounting for the source of controls in all five genetic models showed that rs920778 was remarkably associated with increased cancer risk in both population and hospital based groups in Chinese population (Table 1). And subgroup analyses based on the genotyping method also revealed similar results in RFLP and Taqman groups. In addition, we excluded four studies not satisfied with Hardy-Weinberg equilibrium (HWE) to reanalyze, and elevated risk was still found. Sensitivity analysis showed that the pooled ORs were not qualitatively changed by any single study in all five genetic models, which indicated that the results of our meta-analysis remained robust in Chinese population (Supplementary Figure 1). Visual inspection of funnel plot and Egger's test were performed to assess the publication bias. As shown in Supplementary Figure 2, the visual inspection of funnel plot revealed no obvious asymmetry but under allelic, dominant and heterozygous models. However, the results of Egger's test showed a publication bias only except homozygous model (T vs. C: P = 0.001; TT vs. TC+CC: P = 0.018; TC+TT vs. CC: P = 0.013; TT vs. CC: P = 0.215; TC vs. CC: P = 0.041).
Table 1. Summary ORs and 95% CIs of HOTAIR rs920778 polymorphism and cancer risk.
| Locus | N* | Allele (T vs. C) | Recessive (TT vs. TC+CC) | Dominant (TC+TT vs. CC) | Homozygote (TT vs. CC) | Heterozygote (TC vs. CC) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) P | I2 (%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2 (%) | OR (95%CI) P | I2 (%) | OR (95%CI) P | I2 (%) | ||
| Total | 13 | 1.47 (1.39–1.55) < 0.01 |
49.0 | 1.50 (1.33–1.68) < 0.01 |
39.3 | 1.47 (1.38–1.58)< 0.01 |
38.6 | 2.55 (2.21–2.95) < 0.01 |
7.9 | 1.34 (1.25–1.44)< 0.01 |
28.1 |
| Source of controls | |||||||||||
| Population | 5 | 1.42 (1.30–1.54) < 0.01 |
0 | 1.76 (1.23–2.51) 0.002 |
70.8 | 1.40 (1.25–1.56) < 0.01 |
0 | 2.70 (2.12–3.42) < 0.01 |
0 | 1.26 (1.12–1.41) < 0.01 |
0 |
| Hospital | 4 | 1.69 (1.50–1.91) < 0.01 |
44.8 | 1.71 (1.25–2.34) 0.001 |
0 | 1.71 (1.47–1.99) < 0.01 |
44.1 | 2.96 (2.18–4.02) < 0.01 |
0 | 1.64 (1.27–2.12) < 0.01 |
53.9 |
| Method | |||||||||||
| RFLP | 9 | 1.43 (1.34–1.51) < 0.01 |
36.1 | 1.69 (1.36–2.10) < 0.01 |
54.5 | 1.43 (1.33–1.54) < 0.01 |
22.6 | 2.54 (2.15–3.01) < 0.01 |
33.6 | 1.31 (1.21–1.42) < 0.01 |
0 |
| Taqman | 3 | 1.92 (1.46–2.53) < 0.01 |
51.3 | 1.35 (0.93–1.94) 0.114 |
0 | 1.89 (1.50–2.38) < 0.01 |
47.7 | 2.40 (1.67–3.45) < 0.01 |
0 | 1.81 (1.17–2.80) 0.008 |
63.1 |
| MOLDI-TOF-MS | 1 | 1.55 (1.28–1.86) < 0.01 |
NA | 1.91 (1.18–3.09) 0.009 |
NA | 1.52 (1.20–1.91) < 0.01 |
NA | 2.89 (1.80–4.64) < 0.01 |
NA | 1.34 (1.05–1.71) 0.019 |
NA |
| Type of cancer | |||||||||||
| Estrogen-dependent | 5 | 1.66 (1.40–1.97) < 0.01 |
51.5 | 1.25 (1.06–1.48) 0.008 |
17.6 | 1.68 (1.43–1.97) < 0.01 |
30.8 | 2.44 (1.86–3.19) < 0.01 |
0 | 1.48 (1.24–1.75) < 0.01 |
46.5 |
| Gastrointestinal | 5 | 1.45 (1.34–1.58) < 0.01 |
18.3 | 2.00 (1.59–2.53) < 0.01 |
0 | 1.43 (1.30–1.58) < 0.01 |
19.0 | 2.89 (2.30–3.64) < 0.01 |
0 | 1.30 (1.17–1.44) < 0.01 |
5.6 |
| Controls in HWE | 9 | 1.45 (1.37–1.54) < 0.01 |
33.7 | 1.66 (1.35–2.05) < 0.01 |
51.4 | 1.46 (1.35–1.57) < 0.01 |
12.2 | 2.56 (2.17–3.02) < 0.01 |
34.1 | 1.33 (1.23–1.44) < 0.01 |
0 |
* Numbers of comparisons. RFLP: Restriction Fragment Length Polymorphism. MOLDI-TOF-MS: Matrix-Assisted Laser Desorption/ Ionization Time of Flight Mass Spectrometry. HWE: Hardy-Weinberg Equilibrium. NA: not available.
Figure 2. Forests for HOTAIR rs920778 polymorphism and cancer.
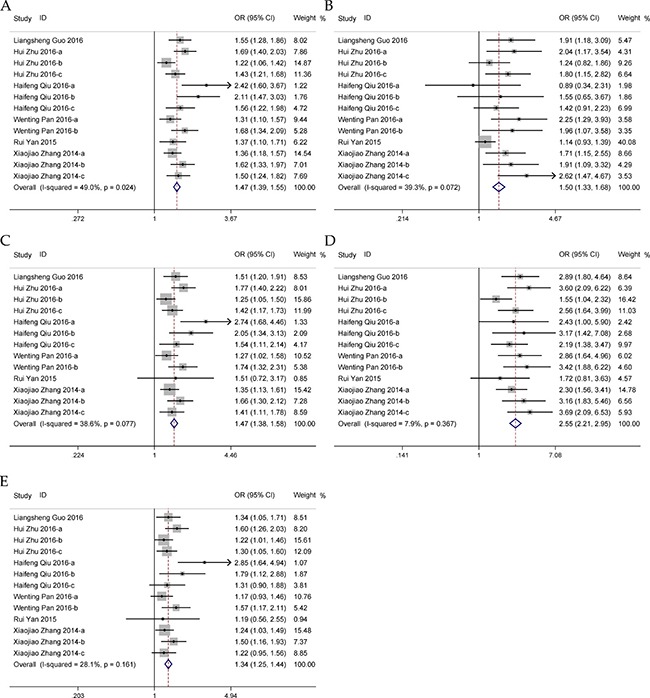
(A) allele model (T vs. C); (B) recessive model (TT vs. TC + CC); (C) dominant model (TC + TT vs. CC); (D) homozygous model (TT vs. CC); (E) heterozygous model (TC vs. CC).
Meta-analysis for HOTAIR rs4759314 A>G polymorphism
Twelve studies comprising 8,136 cases and 9,472 controls reported the association of rs4759314 polymorphism with cancer susceptibility. As shown in Table 2, a significant association between HOTAIR rs4759314 polymorphism and cancer risk was observed in dominant and heterozygous models for Chinese (Figure 3). When different cancer types were considered, increased susceptibility was found only in estrogen-dependent cancer group (Table 2). Subsequent subgroup analyses also revealed significant association in the genotyping method of MALDI-TOF-MS group as well as age and sex matched group (Table 2). Publication bias was evaluated by the visual inspection of funnel plot and Egger's test, and there was no publication bias detected (G vs. A: P = 0.827; GG vs. GA+AA: P = 0.099; GA+GG vs. AA: P = 0.615; GG vs. AA: P = 0.069; GA vs. AA: P = 0.488; Supplementary Figure 3). However, further sensitivity analysis revealed that omission of each study made some significant differences on the findings (Supplementary Figure 4).
Table 2. Summary ORs and 95% CIs of HOTAIR rs4759314 polymorphism and cancer risk.
| Locus | N* | Allele (G vs. A) | Recessive (GG vs. GA+AA) | Dominant (GA+GG vs. AA) | Homozygote (GG vs. AA) | Heterozygote (GA vs. AA) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | ||
| Total | 12 | 1.11 (0.99–1.26) 0.087 |
50.4 | 1.11 (0.80–1.55) 0.532 |
0 | 1.11 (1.01–1.21) 0.026 |
44.4 | 1.31 (0.95–1.80) 0.096 |
0 | 1.09 (1.00–1.20) 0.058 |
38.1 |
| Source of controls | |||||||||||
| Population | 4 | 1.00 (0.84–1.19) 0.962 |
0 | 0.93 (0.29–3.00) 0.898 |
0 | 1.00 (0.83–1.20) 0.977 |
0 | 0.93 (0.29–2.99) 0.901 |
0 | 1.00 (0.83–1.20) 0.990 |
0 |
| Hospital | 6 | 1.13 (0.92–1.38) 0.259 |
68.7 | 0.96 (0.60–1.53) 0.862 |
17.2 | 1.13 (0.91–1.41) 0.256 |
67.3 | 1.07 (0.68–1.67) 0.782 |
25.1 | 1.13 (0.91–1.40) 0.273 |
65.4 |
| Method | |||||||||||
| RFLP | 5 | 1.04 (0.89–1.22) 0.629 |
0 | 0.89 (0.32–2.45) 0.817 |
0 | 1.04 (0.89–1.23) 0.604 |
0 | 0.95 (0.35–2.60) 0.925 |
0 | 1.05 (0.89–1.24) 0.589 |
0 |
| Taqman | 4 | 1.15 (0.83–1.61) 0.403 |
81.1 | 0.59 (0.28–1.28) 0.184 | 34.6 | 1.18 (0.83–1.66) 0.356 | 80.2 | 0.69 (0.33–1.45) 0.325 |
48.4 | 1.19 (0.85–1.66) 0.317 |
78.5 |
| MALDI-TOF-MS | 3 | 1.21 (1.05–1.39) 0.009 |
0 | 1.39 (0.92–2.09) 0.116 |
0 | 1.17 (1.00–1.36) 0.055 |
0 | 1.64 (1.12–2.41) 0.012 |
0 | 1.10 (0.93–1.30) 0.264 |
0 |
| Type of cancer | |||||||||||
| Estrogen-dependent | 3 | 1.18 (1.02–1.37) 0.022 |
42.2 | 1.36 (0.86–2.14) 0.192 | 0 | 1.15 (0.98–1.35) 0.091 |
4.7 | 1.66 (1.08–2.55) 0.022 |
0 | 1.09 (0.92–1.30) 0.303 |
0 |
| Gastrointestinal | 5 | 1.09 (0.90–1.33) 0.369 |
64.3 | 0.69 (0.35–1.34) 0.270 | 0 | 1.11 (0.90–1.36) 0.326 |
63.5 | 0.77 (0.41–1.48) 0.437 |
5.3 | 1.11 (0.91–1.36) 0.295 |
61.7 |
| Age and sex matched | 11 | 1.12 (0.98–1.28) 0.099 |
54.8 | 1.11 (0.79–1.56) 0.540 |
0 | 1.11 (1.01–1.22) 0.027 |
49.4 | 1.31 (0.95–1.81) 0.098 |
0 | 1.10 (1.00–1.21) 0.060 |
43.7 |
| Controls in HWE | 10 | 1.09 (0.95–1.26) 0.237 |
51.8 | 0.84 (0.51–1.38) 0.493 |
0 | 1.10 (0.95–1.28) 0.200 |
50.4 | 0.94 (0.58–1.53) 0.815 |
0 | 1.09 (0.99–1.21) 0.092 |
47.6 |
* Numbers of comparisons. RFLP: Restriction Fragment Length Polymorphism. MOLDI-TOF-MS: Matrix-Assisted Laser Desorption/ Ionization Time of Flight Mass Spectrometry. HWE: Hardy-Weinberg Equilibrium.
Figure 3. Forests for HOTAIR rs4759314 polymorphism and cancer.
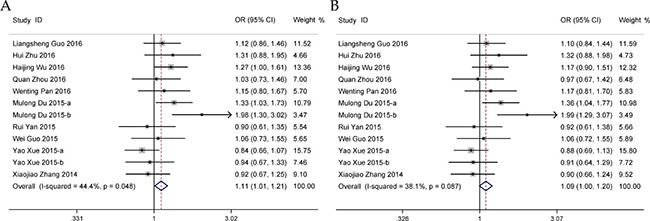
(A) dominant model (GA + GG vs. AA); (B) heterozygous model (GA vs. AA).
Meta-analysis for HOTAIR rs7958904 G>C polymorphism
A total of six studies consisting of 4,387 cases and 4,812 controls were included in the quantitative analysis. No significant heterogeneity was detected across studies in all five genetic models and the fixed-effects model was therefore selected to summarize the ORs. Overall, a decreased risk of cancer was observed for Chinese (Table 3, Figure 4). When stratified by the type of cancers, a similarly decreased risk was identified in gastrointestinal cancer group (Table 3). Moreover, according to the results of subsequent subgroup analyses, the decreased risk does not obviously influenced by genotyping method and HWE status (Table 3). Sensitivity analysis revealed that the pooled ORs were not conspicuously changed by any single study (Supplementary Figure 5). No publication bias was detected in all genetic models (C vs. G: P = 0.362; CC vs. CG+GG: P = 0.305; CG+CC vs. GG: P = 0.416; CC vs. GG: P = 0.298; CG vs. GG: P = 0.523; Supplementary Figure 6).
Table 3. Summary ORs and 95% CIs of HOTAIR rs7958904 polymorphism and cancer risk.
| Locus | N* | Allele (C vs. G) | Recessive (CC vs. CG+GG) | Dominant (CG+CC vs. GG) | Homozygote (CC vs. GG) | Heterozygote (CG vs. GG) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | ||
| Total | 6 | 0.82 (0.77–0.87) < 0.01 |
0 | 0.79 (0.67–0.93) < 0.01 |
0 | 0.81 (0.75–0.88)< 0.01 |
0 | 0.64 (0. 54–0.75) < 0.01 |
0 | 0.85 (0.78–0.93) < 0.01 |
0 |
| Method | |||||||||||
| Taqman | 3 | 0.85 (0.78–0.93) < 0.01 |
0 | 0.85 (0.69–1.06) 0.143 |
0 | 0.84 (0.76–0.94) < 0.01 |
0 | 0.72 (0.58–0.89) < 0.01 |
0 | 0.87 (0.78–0.98) 0.017 |
0 |
| MOLDI-TOF-MS | 3 | 0.77 (0.70–0.86) < 0.01 |
0 | 0.71 (0.54–0.91) < 0.01 |
0 | 0.78 (0.68–0.88) < 0.01 |
0 | 0.55 (0.42–0.70) < 0.01 |
0 | 0.83 (0.73–0.95) < 0.01 |
0 |
| Type of cancer | |||||||||||
| Estrogen-dependent | 1 | 0.77 (0.67–0.89) < 0.01 |
NA | 0.67 (0.47–0.98) 0.037 |
NA | 0.78 (0.65–0.93)< 0.01 |
NA | 0.53 (0.37–0.76) < 0.01 |
NA | 0.84 (0.70–1.01) 0.063 |
NA |
| Gastrointestinal | 3 | 0.85 (0.78–0.93) < 0.01 |
0 | 0.85 (0.69–1.06) 0.143 |
0 | 0.84 (0.76–0.94) < 0.01 |
0 | 0.72 (0.58–0.89) < 0.01 |
0 | 0.87 (0.78–0.98) 0.017 |
0 |
| Controls in HWE | 5 | 0.83 (0.77–0.89) < 0.01 |
0 | 0.79 (0.66–0.94) < 0.01 |
0 | 0.82 (0.75–0.90) < 0.01 |
0 | 0.65 (0.55–0.77) < 0.01 |
0 | 0.86 (0.78–0.94) < 0.01 |
0 |
* Numbers of comparisons. MOLDI-TOF-MS: Matrix-Assisted Laser Desorption/ Ionization Time of Flight Mass Spectrometry. HWE: Hardy-Weinberg Equilibrium. NA: not available.
Figure 4. Forests for HOTAIR rs7958904 polymorphism and cancer.
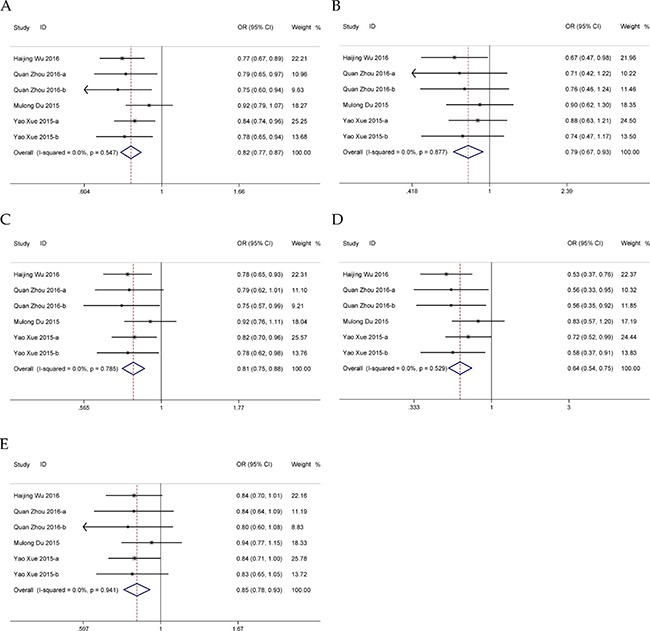
(A) allele model (C vs. G); (B) recessive model (CC vs. CG + GG); (C) dominant model (CG + CC vs. GG); (D) homozygous model (CC vs. GG); (E) heterozygous model (CG vs. GG).
Meta-analysis for HOTAIR rs874945 G>A polymorphism
Five eligible case-control studies with 3,800 cases and 4,160 controls were included in the meta-analysis. Overall, we found a significant association between rs874945 G>A polymorphism and cancer risk in allelic, dominant and homozygous models (Table 4, Figure 5). Stratification accounting for genotyping method revealed increased cancer risk existed in MALDI-TOF-MS genotyping method group (Table 4). Further subgroup analyses of cancer type and HWE status were conducted and no significant result was observed (Table 4). Sensitivity analysis indicated that the pooled ORs remained robust only in recessive and heterozygous models (Supplementary Figure 7). No publication bias was detected in all genetic models (A vs. G: P = 0.495; AA vs. AG+GG: P = 0.362; AG+AA vs. GG: P = 0.715; AA vs. GG: P = 0.503; AG vs. GG: P = 0.962; Supplementary Figure 8).
Table 4. Summary ORs and 95% CIs of HOTAIR rs874945 polymorphism and cancer risk.
| Locus | N* | Allele (A vs. G) | Recessive (AA vs. AG+GG) | Dominant (AG+AA vs. GG) | Homozygote (AA vs. GG) | Heterozygote (AG vs. GG) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | OR (95%CI) P | I2(%) | ||||||||||
| Total | 5 | 1.11 (1.02–1.20) 0.013 |
0 | 1.14 (0.91–1.42) 0.262 |
0 | 1.10 (1.01–1.21) 0.039 |
0 | 1.26 (1.02–1.56) 0.035 |
0 | 1.08 (0.98–1.19) 0.120 |
0 | ||||||||
| Method | |||||||||||||||||||
| Taqman | 2 | 1.09 (0.98–1.22) 0.132 |
0 | 1.07 (0.76–1.50) 0.717 |
0 | 1.10 (0.97–1.25) 0.148 |
0 | 1.17 (0.84–1.64) 0.353 |
0 | 1.09 (0.96–1.25) 0.199 |
0 | ||||||||
| MOLDI-TOF-MS | 3 | 1.12 (1.00–1.26) 0.046 |
0 | 1.19 (0.89–1.60) 0.245 |
0 | 1.11 (0.97–1.27) 0.139 |
0 | 1.33 (1.00–1.76) 0.050 |
0 | 1.07 (0.93–1.23) 0.365 |
0 | ||||||||
| Type of cancer | |||||||||||||||||||
| Estrogen-dependent | 1 | 1.07 (0.91–1.25) 0.419 |
NA | 1.14 (0.74–1.75) 0.551 |
NA | 1.06 (0.88–1.27) 0.568 |
NA | 1.20 (0.79–1.82) 0.383 |
NA | 1.03 (0.85–1.26) 0.749 |
NA | ||||||||
| Gastrointestinal | 2 | 1.09 (0.98–1.22) 0.132 |
0 | 1.07 (0.76–1.50) 0.717 |
0 | 1.10 (0.97–1.25) 0.148 |
0 | 1.17 (0.84–1.64) 0.353 |
0 | 1.09 (0.96–1.25) 0.199 |
0 | ||||||||
| Controls in HWE | 2 | 1.09 (0.98–1.22) 0.132 |
0 | 1.07 (0.76–1.50) 0.717 |
0 | 1.10 (0.97–1.25) 0.148 |
0 | 1.17 (0.84–1.64) 0.353 |
0 | 1.09 (0.96–1.25) 0.199 |
0 | ||||||||
*Numbers of comparisons. MOLDI-TOF-MS: Matrix-Assisted Laser Desorption/ Ionization Time of Flight Mass Spectrometry. HWE: Hardy-Weinberg Equilibrium. NA: not available.
Figure 5. Forests for HOTAIR rs874945 polymorphism and cancer.
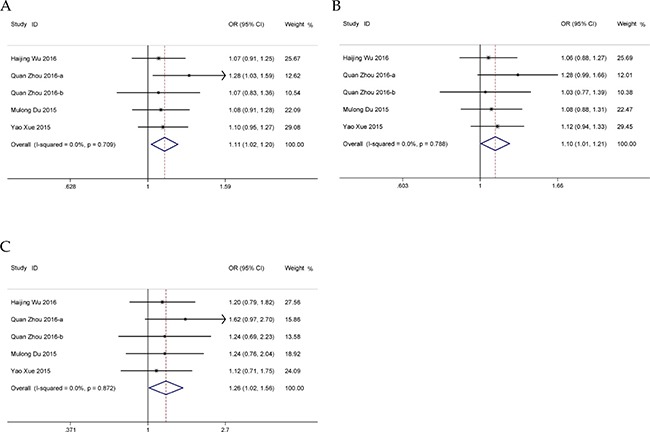
(A) allele model (A vs. G); (B) dominant model (AG + AA vs. GG); (C) homozygous model (AA vs. GG).
Meta-analysis for HOTAIR rs1899663 G>T polymorphism
Five studies with 3,112 cases and 3,817 controls were included to estimate the association between rs1899663 G>T polymorphism and cancer risk. The results of this meta-analysis were shown in Table 5, and no significant association was identified in all five genetic models.
Table 5. Summary ORs and 95% CIs of HOTAIR rs1899663 polymorphism and cancer risk.
| Locus | N* | Allele (T vs. G) | Recessive (TT vs. TG+GG) | Dominant (TG+TT vs. GG) | Homozygote (TT vs. GG) | Heterozygote (TG vs. GG) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) P | I2(%) | OR (95%CI) P | I2 (%) | OR (95%CI) P | I2 (%) | OR (95%CI) P | I2 (%) | OR (95%CI) P | I2 (%) | ||
| Total | 5 | 0.93 (0.84–1.02) 0.102 |
0 | 0.78 (0.54–1.11) 0.163 |
0 | 0.93 (0.84–1.04) 0.193 |
0 | 0.73 (0.51–1.03) 0.071 |
0 | 0.95 (0.85–1.06) 0.350 | 0 |
| Source of controls | |||||||||||
| Population | 3 | 0.93 (0.83–1.04) 0.208 |
0 | 0.79 (0.52–1.20) 0.265 |
0 | 0.94 (0.82–1.07) 0.334 |
0 | 0.74 (0.49–1.11) 0.147 |
0 | 0.96 (0.83–1.10) 0.512 | 0 |
| Hospital | 1 | 1.05 (0.84–1.31) 0.654 |
NA | 0.82 (0.33–2.02) 0.658 |
NA | 1.08 (0.84–1.39) 0.548 |
NA | 0.88 (0.36–2.15) 0.778 |
NA | 1.09 (0.85–1.41) 0.494 | NA |
| Method | |||||||||||
| RFLP | 4 | 0.90 (0.81–1.00) 0.044 |
0 | 0.77 (0.52–1.13) 0.184 |
0 | 0.90 (0.80–1.02) 0.086 |
0 | 0.70 (0.48–1.02) 0.066 |
0 | 0.92 (0.82–1.04) 0.176 | 0 |
| MOLDI-TOF-MS | 1 | 1.05 (0.84–1.31) 0.654 |
NA | 0.82 (0.33–2.02) 0.658 |
NA | 1.08 (0.84–1.39) 0.548 |
NA | 0.88 (0.36–2.15) 0.778 |
NA | 1.09 (0.85–1.41) 0.494 | NA |
| Type of cancer | |||||||||||
| Estrogen-dependent | 2 | 0.96 (0.82–1.13) 0.620 |
23.1 | 0.78 (0.45–1.37) 0.395 |
0 | 0.98 (0.82–1.17) 0.822 |
18.0 | 0.75 (0.43–1.29) 0.296 |
0 | 1.00 (0.83–1.21) 0.991 | 0 |
| Gastrointestinal | 2 | 0.95 (0.83–1.09) 0.443 |
0 | 0.80 (0.47–1.35) 0.400 |
0 | 0.96 (0.82–1.12) 0.576 |
0 | 0.77 (0.46–1.29) 0.325 |
0 | 0.97 (0.83–1.14) 0.729 | 0 |
* Numbers of comparisons. MOLDI-TOF-MS: Matrix-Assisted Laser Desorption/ Ionization Time of Flight Mass Spectrometry. RFLP: Restriction Fragment Length Polymorphism. NA: not available.
DISCUSSION
Cancer is a polygenic and multifactorial disease which is thought to be caused by complex genetic factors and gene-environment interactions. The lncRNA polymorphisms have been demonstrated to be involved in carcinogenesis and progression of different types of cancers. Recently, the association between lncRNA HOTAIR polymorphisms and cancer risk has been developed gradually. However, the results remain contradictory and inconclusive, especially in different geographical location and ethnicity groups. In 2016, three similar meta-analyses involving eight articles explored the association of HOTAIR polymorphisms with several kinds of cancers among worldwide population [34–36]. However, none of them drew a conclusion for Chinese population. Since then, six new articles with nine individual studies for Chinese have been published [23, 24, 26–29]. Therefore, it is necessary to summarize all eligible individual studies and conduct a comprehensive meta-analysis to determine the correlations of HOTAIR polymorphisms with cancer susceptibility in Chinese population.
To the best of our knowledge, this is the first meta-analysis which attempt to explore the association between HOTAIR polymorphisms and cancer risk in population of Chinese ethnicity. A total of twenty-one eligible studies comprising 12,278 cases and 14,532 controls were included into current meta-analysis and it provided the most comprehensive assessment of the correlations of five common polymorphisms in the HOTAIR gene with cancer risk for Chinese to date.
Overall, our results demonstrated that rs920778 polymorphism of HOTAIR was significantly associated with increased cancer risk among Chinese population. Compared with those previous meta-analyses, some advantages of our analysis should be highlighted. First, although our combined results were consistent with most individual studies and meta-analyses, we definitely expended the sample size and boosted the statistical power dramatically by adding another seven individual studies with 7,119 subjects on the basis of the updated data. Second, we conducted a more comprehensive subgroup analysis. Noteworthy, in the analysis based on cancer type, we observed an increased risk for this polymorphism both in estrogen-dependent cancer group and gastrointestinal cancer group. Third, we conducted all the analyses for five genetic models including allelic, recessive, dominant, homozygous and heterozygous models to draw a comprehensive assessment. Fourth, when we excluded the studies involving Caucasian population, the heterogeneity of the analyses was significantly decreased. That helped to enhance the reliability of our conclusion.
As for rs4759314 polymorphism, even though no association was found in previous meta-analyses [34–36], the evidence from our meta-analysis supported a significant association with increased cancer risk in dominant and heterozygous models for Chinese. The discrepancy between our findings and previous meta-analyses might be due to the inclusion of another four recent studies involving 5,423 subjects in our analysis. Noteworthy, further subgroup analyses by cancer type revealed that significantly increased risks were only found in the estrogen-dependent cancer group, and it suggested the increased cancer risk might be tumor type-specific. Conversely, rs7958904 polymorphism exhibited as a potential protective factor for cancer risk on the basis of our results that this polymorphism was associated with the decreased cancer susceptibility in Chinese population. And the associations were further confirmed by stratified analyses according to genotyping method, cancer type and HWE status.
In addition, our pooled analyses also showed a significantly increased cancer risk for rs874945 polymorphism in allelic, dominant and homozygous models. However, potentially due to the relatively small sample size with only 5 studies involved, the elevated cancer risk for rs874945 did not exhibit tumor type specificity. So these results should be interpreted with caution and further studies are needed to clarify the accurate association. Lastly, as for rs1899663 polymorphism, our results could not provide any evidence of such an association with cancer risk under any genetic model.
There are a few potential limitations existed in the present meta-analysis. First, all published articles retrieved were written in English in our study, which may cause potential language bias. Second, the estimations about interactions between gene-gene, gene-environment, and multiple polymorphic loci in the same gene were not performed. Third, most of the studies included in our meta-analysis were concerning estrogen-dependent and gastrointestinal cancers. Thus this limited the general application of the results to other types of cancers, such as lung cancer, liver cancer and so on. Fourth, sensitivity analyses for rs4759314 and rs874945 polymorphisms revealed that omission of each study made some significant changes on the findings, which could be explained by the limited number of studies involved. Finally, as for rs920778, most of the studies included reported positive results and the publication bias was detected except in homozygous model. Thus, the comprehensive analyses should be interpreted with caution.
In summary, the current meta-analysis provides evidence that four functional polymorphisms of HOTAIR involving rs920778, rs7958904, rs4759314, and rs874945 might contribute to genetic susceptibility to cancer risk in Chinese population, whereas rs1899663 may have no impact. Accordingly, large scale and well-designed studies are warranted to confirm the association of above polymorphisms in HOTAIR and cancer risk in the future.
MATERIALS AND METHODS
Literature search strategy
Eligible studies regarding the association between HOTAIR polymorphisms and cancer risk in Chinese population were systematically searched from Pubmed, Embase, Wanfang and CNKI databases up to October 31, 2016. The terms used for search were as follows: “HOX transcript antisense RNA OR HOTAIR” and “polymorphism OR variant OR SNP OR genotype OR allele” and “cancer OR carcinoma OR tumor OR neoplasm”. In addition, citation lists of all relevant articles were manually searched for additional eligible publications.
Inclusion and exclusion criteria
Studies were all selected according to the following criteria: (1) case-control design study; (2) evaluating the association between HOTAIR polymorphisms and cancer risk in Chinese population; (3) available genotype distribution data for calculating the odds ratios (ORs) with 95% confidence intervals (CIs). Reviews, letters, conference abstracts, duplicate studies and studies without sufficient genotype information were excluded. In addition, we finally selected the study with larger sample size from duplicate publications.
Data extraction
The following data were extracted by two independent investigators: first author, publication year, source of controls, sample size, genotype frequency, genotyping methods, age and sex matched status, type of cancers, P value for Hardy-Weinberg equilibrium (HWE) in the control group. Disputes were resolved through group discussion.
Statistical analysis
The crude ORs with 95% CIs were calculated to determine the relationship between the HOTAIR polymorphisms and cancer susceptibility in Chinese population. For the rs4759314 polymorphism, the pooled ORs were estimated by allelic (G vs. A), recessive (GG vs. GA+AA), dominant (GA+GG vs. AA), homozygous (GG vs. AA) and heterozygous (GA vs. AA) models. As for rs920778, rs7958904, rs874945, and rs1899663 polymorphisms, similar five genetic models were assessed. Subgroup analyses based on source of controls, type of cancers, genotyping methods, HWE status of controls and case-control matched status were subsequently performed. HWE was examined by chi-square test in the controls. The I2 statistic was used to evaluate the heterogeneity between studies. If I2 > 50%, significant heterogeneity was found and the random-effects model should be applied. Otherwise, the fixed-effects model should be used. Sensitivity analysis was conducted by sequentially omitting each single study to evaluate the stability of our results. Publication bias was assessed by both visual inspection of funnel plot and Egger's test. The STATA software 12.0 (StataCorp LP, College Station, TX) was used for all the statistical analyses. P < 0.05 was considered to be statistically significant.
SUPPLEMENTARY MATERIALS FIGURES AND TABLES
ACKNOWLEDGMENTS AND FUNDING
This work was supported by the National Natural Science Foundation of China (Grant No. 81302127, 81400950) and the Joint Specialized Research Fund for the Doctoral Program of Higher Education in China (Grant No. 20132104120018).
Footnotes
CONFLICTS OF INTEREST
No conflicts of interest were disclosed.
REFERENCES
- 1.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Chen J, Zhang J, Wang Z, Shao T, Jiang C, Xu J, Li X. Construction and analysis of lncRNA-lncRNA synergistic networks to reveal clinically relevant lncRNAs in cancer. Oncotarget. 2015;6:25003–25016. doi: 10.18632/oncotarget.4660. https://doi.org/10.18632/oncotarget.4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, Weinstein JN, Liang H. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 2015;75:3728–3737. doi: 10.1158/0008-5472.CAN-15-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, Chu LW, Zhang Z, Zhao H, Zheng SL, Isaacs WB, Xu J. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis. 2011;32:1655–1659. doi: 10.1093/carcin/bgr187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu L, Zhu G, Zhang C, Deng Q, Katsaros D, Mayne ST, Risch HA, Mu L, Canuto EM, Gregori G, Benedetto C, Yu H. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res Treat. 2012;136:875–883. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 11.Emadi-Andani E, Nikpour P, Emadi-Baygi M, Bidmeshkipour A. Association of HOTAIR expression in gastric carcinoma with invasion and distant metastasis. Adv Biomed Res. 2014;3:135. doi: 10.4103/2277-9175.133278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, Zhou L, Wu LM, Lai MC, Xie HY, Zhang F, Zheng SS. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 14.Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu ZL, Zhou GZ, Cao G, Jin L, Xie HW, Wang CM, Lv J, De W, et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Dang Q, Xie H, Yang Z, He D, Liang L, Song W, Yeh S, Chang C. Infiltrating mast cells enhance prostate cancer invasion via altering LncRNA-HOTAIR/PRC2-androgen receptor (AR)-MMP9 signals and increased stem/progenitor cell population. Oncotarget. 2015;6:14179–14190. doi: 10.18632/oncotarget.3651. https://doi.org/10.18632/oncotarget.3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, An J, Wu M, Zheng Q, Gui X, Li T, Pu H, Lu D. LncRNA HOTAIR promotes human liver cancer stem cell malignant growth through downregulation of SETD2. Oncotarget. 2015;6:27847–27864. doi: 10.18632/oncotarget.4443. https://doi.org/10.18632/oncotarget.4443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai B, Song XQ, Cai JP, Zhang S. HOTAIR: a cancer-related long non-coding RNA. Neoplasma. 2014;61:379–391. doi: 10.4149/neo_2014_075. [DOI] [PubMed] [Google Scholar]
- 18.Du M, Wang W, Jin H, Wang Q, Ge Y, Lu J, Ma G, Chu H, Tong N, Zhu H, Wang M, Qiang F, Zhang Z. The association analysis of lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese population. Oncotarget. 2015;6:31255–31262. doi: 10.18632/oncotarget.5158. https://doi.org/10.18632/oncotarget.5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Zhou L, Fu G, Sun F, Shi J, Wei J, Lu C, Zhou C, Yuan Q, Yang M. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis. 2014;35:2062–2067. doi: 10.1093/carcin/bgu103. [DOI] [PubMed] [Google Scholar]
- 20.Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen H, Zhou L, Yuan Q, Zhou C, Yang M. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol Carcinog. 2016;55:90–96. doi: 10.1002/mc.22261. [DOI] [PubMed] [Google Scholar]
- 21.Xue Y, Gu D, Ma G, Zhu L, Hua Q, Chu H, Tong N, Chen J, Zhang Z, Wang M. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 2015;30:303–310. doi: 10.1093/mutage/geu076. [DOI] [PubMed] [Google Scholar]
- 22.Guo W, Dong Z, Bai Y, Guo Y, Shen S, Kuang G, Xu J. Associations between polymorphisms of HOTAIR and risk of gastric cardia adenocarcinoma in a population of north China. Tumour Biol. 2015;36:2845–2854. doi: 10.1007/s13277-014-2912-y. [DOI] [PubMed] [Google Scholar]
- 23.Guo L, Lu X, Zheng L, Liu X, Hu M. Association of Long Non-Coding RNA HOTAIR Polymorphisms with Cervical Cancer Risk in a Chinese Population. PloS One. 2016;11:e0160039. doi: 10.1371/journal.pone.0160039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu H, Shang X, Shi Y, Yang Z, Zhao J, Yang M, Li Y, Xu S. Genetic variants of lncRNA HOTAIR and risk of epithelial ovarian cancer among Chinese women. Oncotarget. 2016;7:41047–41052. doi: 10.18632/oncotarget.8535. https://doi.org/10.18632/oncotarget.8535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan R, Cao J, Song C, Chen Y, Wu Z, Wang K, Dai L. Polymorphisms in lncRNA HOTAIR and susceptibility to breast cancer in a Chinese population. Cancer Epidemiol. 2015;39:978–985. doi: 10.1016/j.canep.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 26.Qiu H, Wang X, Guo R, Liu Q, Wang Y, Yuan Z, Li J, Shi H. HOTAIR rs920778 polymorphism is associated with ovarian cancer susceptibility and poor prognosis in a Chinese population. Future Oncol. 2016;13:347–355. doi: 10.2217/fon-2016-0290. https://doi.org/10.2217/fon-2016-0290 [DOI] [PubMed] [Google Scholar]
- 27.Qiu H, Liu Q, Li J, Wang X, Wang Y, Yuan Z, Li J, Pei DS. Analysis of the association of HOTAIR single nucleotide polymorphism (rs920778) and risk of cervical cancer. Apmis. 2016;124:567–573. doi: 10.1111/apm.12550. [DOI] [PubMed] [Google Scholar]
- 28.Zhu H, Lv Z, An C, Shi M, Pan W, Zhou L, Yang W, Yang M. Onco-lncRNA HOTAIR and its functional genetic variants in papillary thyroid carcinoma. Sci Rep. 2016;6:31969. doi: 10.1038/srep31969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Q, Chen F, Fei Z, Zhao J, Liang Y, Pan W, Liu X, Zheng D. Genetic variants of lncRNA HOTAIR contribute to the risk of osteosarcoma. Oncotarget. 2016;7:19928–19934. doi: 10.18632/oncotarget.7957. https://doi.org/10.18632/oncotarget.7957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayram S, Sumbul AT, Dadas E. A functional HOTAIR rs12826786 C>T polymorphism is associated with breast cancer susceptibility and poor clinicopathological characteristics in a Turkish population: a hospital-based case-control study. Tumour Biol. 2016;37:5577–5584. doi: 10.1007/s13277-015-4430-y. [DOI] [PubMed] [Google Scholar]
- 31.Gong WJ, Yin JY, Li XP, Fang C, Xiao D, Zhang W, Zhou HH, Li X, Liu ZQ. Association of well-characterized lung cancer lncRNA polymorphisms with lung cancer susceptibility and platinum-based chemotherapy response. Tumour Biol. 2016;37:8349–8358. doi: 10.1007/s13277-015-4497-5. [DOI] [PubMed] [Google Scholar]
- 32.Bayram S, Ulger Y, Sumbul AT, Kaya BY, Rencuzogullari A, Genc A, Sevgiler Y, Bozkurt O, Rencuzogullari E. A functional HOTAIR rs920778 polymorphism does not contributes to gastric cancer in a Turkish population: a case-control study. Familial cancer. 2015;14:561–567. doi: 10.1007/s10689-015-9813-0. [DOI] [PubMed] [Google Scholar]
- 33.Bayram S, Sumbul AT, Batmaci CY, Genc A. Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumour Biol. 2015;36:3863–3870. doi: 10.1007/s13277-014-3028-0. [DOI] [PubMed] [Google Scholar]
- 34.Qi Q, Wang J, Huang B, Chen A, Li G, Li X, Wang J. Association of HOTAIR polymorphisms rs4759314 and rs920778 with cancer susceptibility on the basis of ethnicity and cancer type. Oncotarget. 2016;7:38775–38784. doi: 10.18632/oncotarget.9608. https://doi.org/10.18632/oncotarget.9608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian T, Li C, Xiao J, Shen Y, Lu Y, Jiang L, Zhuang X, Chu M. Quantitative Assessment of the Polymorphisms in the HOTAIR lncRNA and Cancer Risk: A Meta-Analysis of 8 Case-Control Studies. PloS One. 2016;11:e0152296. doi: 10.1371/journal.pone.0152296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Liu X, You LH, Zhou RZ. Significant association between long non-coding RNA HOTAIR polymorphisms and cancer susceptibility: a meta-analysis. Onco Targets Ther. 2016;9:3335–3343. doi: 10.2147/OTT.S107190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


