Abstract
Brown midrib (bmr) mutants in sorghum (Sorghum bicolor (L.) Moench) and several other C4 grasses are associated with reduced lignin concentration, altered lignin composition and improved cell wall digestibility, which are desirable properties in biomass development for the emerging lignocellulosic biofuel industry. Studying bmr mutants has considerably expanded our understanding of the molecular basis underlying lignin biosynthesis and perturbation in grasses. In this study, we performed quantitative trait locus (QTL) analysis, identified and cloned a novel cinnamyl alcohol dehydrogenase allele (SbCAD2) that has an 8-bp deletion in its 5′-untranslated region (UTR), conferring the spontaneous brown midrib trait and lignin reduction in the sorghum germplasm line PI 595743. Complementation test and gene expression analysis revealed that this non-coding region alteration is associated with the significantly reduced expression of the SbCAD2 in PI 595743 throughout its growth stages. Moreover, a promoter-GUS fusion study with transgenic Arabidopsis thaliana plants found that SbCAD2 promoter is functionally conserved, driving a specific expression pattern in lignifying vascular tissues. Taken together, our results revealed the genetic basis of bmr occurrence in this spontaneous sorghum mutant and suggested the regulatory region of the SbCAD2 can be a target site for optimizing lignin modification in sorghum and other bioenergy crops.
Introduction
Sorghum is one of the premier biomass feedstocks for biofuel production because of its high biomass yield, outstanding drought tolerance and efficient nutrient usage1. In the renewable bioenergy industry, overcoming the intrinsic recalcitrance of biomass is critical for the development of lignocellulosic biofuel as a cost-effective alternative to fossil fuels2. Biomass recalcitrance is mainly due to the complex configuration of lignin polymer chains and the intertwined network of lignin and polysaccharides in plant cell walls3. In plants, the highly interwoven cell wall matrix rigidified by lignin is crucial for their structural integrity, water conduction and pathogen resistance4, 5. Hence, as a major impediment to plant biomass utilization and an essential component of the secondary cell wall, lignin has become a main research object in plant biology and its biosynthetic pathway has been extensively studied in the past several decades. In general, cell wall lignin is oxidatively polymerized primarily from three types of alcohols (monolignols), which are synthesized in cytosol from phenylalanine through successive deamination, reduction, hydroxylation and methylation steps5. Knowledge from the well-delineated monolignol biosynthetic pathway has led to the successful genetic manipulation of lignin content and composition in bioenergy grasses6, 7. Recently, in order to minimize developmental defects in lignin-modified plants, lignin modification strategy has evolved from simple perturbations of the general monolignol biosynthetic pathways to coordinated orchestrations of the lignin synthesis, deposition and integration into the cell wall network8. Therefore, a profound knowledge and advanced understanding of the underlying regulatory mechanisms of lignin production would benefit the long-term goal of manipulating cell wall structure in biomass feedstocks based on our needs.
Brown midrib (bmr) is a visual trait first documented in maize over eighty years ago and later mutagenesis-induced in sorghum and pearl millet9–11. In grasses, this trait is characterized by the reddish-brown pigment accumulated in leaf midribs and stems during the vegetative stage. In earlier years, this distinct pigmentation was found to be associated with higher ruminant digestion rate and thus became a desired feature in livestock forage development12, 13. Later, the property of reduced lignin content in bmr mutants was appreciated by plant geneticists and utilized for identifying candidate genes for manipulating the lignin production in grasses14. A number of mutagenized bmr mutants of sorghum have been characterized and the corresponding genes involved in specific enzymatic steps of monolignol biosynthetic pathway have been identified in recent years15–18. For instance, bmr6 mutant, originally generated from a diethyl sulfate mutagenesis sorghum population, was found to be associated with reduced cinnamyl alcohol dehydrogenase (CAD) activity19. CAD is a specialized enzyme of the alcohol dehydrogenase family involved in the conversion of the cinnamaldehydes into alcohols, the monomeric precursors of lignin. Downregulation of CAD genes generates atypical lignin polymers with the incorporation of phenolic aldehydes and becomes a promising strategy to improve cell wall digestibility20–22. Later on, a nonsense mutation in a sorghum CAD gene (SbCAD2) was identified to be responsible for the bmr6 phenotype. Specifically, the nonsense mutation truncates the coding region of SbCAD2 prior to the conserved NADPH-binding and C-terminal domains and consequently, abolishes the function of the encoded protein16, 17.
Fiber digestion tests and field evaluation performed on sorghum varieties carrying the mutagenized bmr mutations found that they generally show higher digestibility and inferior agronomic performance relative to their wild-type counterparts, even though the effects of mutations are not uniformly expressed across different genetic backgrounds and are influenced by environmental conditions14, 23. Stacking different bmr mutations in sorghum has an additive effect on lignin content and cellulose-to-ethanol conversion24. Therefore, there are some recent efforts focusing on characterization of novel bmr sorghum lines derived from EMS (ethyl methanesulfonate)-mutagenized populations with the aim to incorporate new sources into the forage and biomass feedstock improvement25, 26. In summary, most sorghum bmr mutants studied so far are from mutagen-induced populations, and no spontaneous bmr phenotypes in sorghum have been characterized in detail at the molecular level.
In this study, we focused on PI 595743, a sorghum germplasm line showing a distinct bmr trait without any observable growth and developmental defects. Phenotypic evaluation confirmed the lignin deficiency in its brown vascular tissues. We next used QTL mapping, DNA sequencing and complementation test to identify that an 8-bp deletion in the 5′ UTR of SbCAD2 is responsible for this novel spontaneous bmr phenotype. This deletion results in the down-regulation of SbCAD2 expression in PI 595743 in a mechanism that is different from that underlying the reduced expression of SbCAD2 in the bmr6 lines. Furthermore, we found that the function of SbCAD2 promoter is conserved in both sorghum and Arabidopsis and this deletion region is implicated in the transcriptional regulation of SbCAD2 expression in lignifying tissue. We expect that this alternative allele of SbCAD2 gene will expand the repertoire of genetic resources for biomass improvement in sorghum, and more importantly, provide new insights into the complex underlying regulatory mechanism of the lignin biosynthetic pathway in grasses.
Results
Phenotypic characterization of the spontaneous bmr mutant
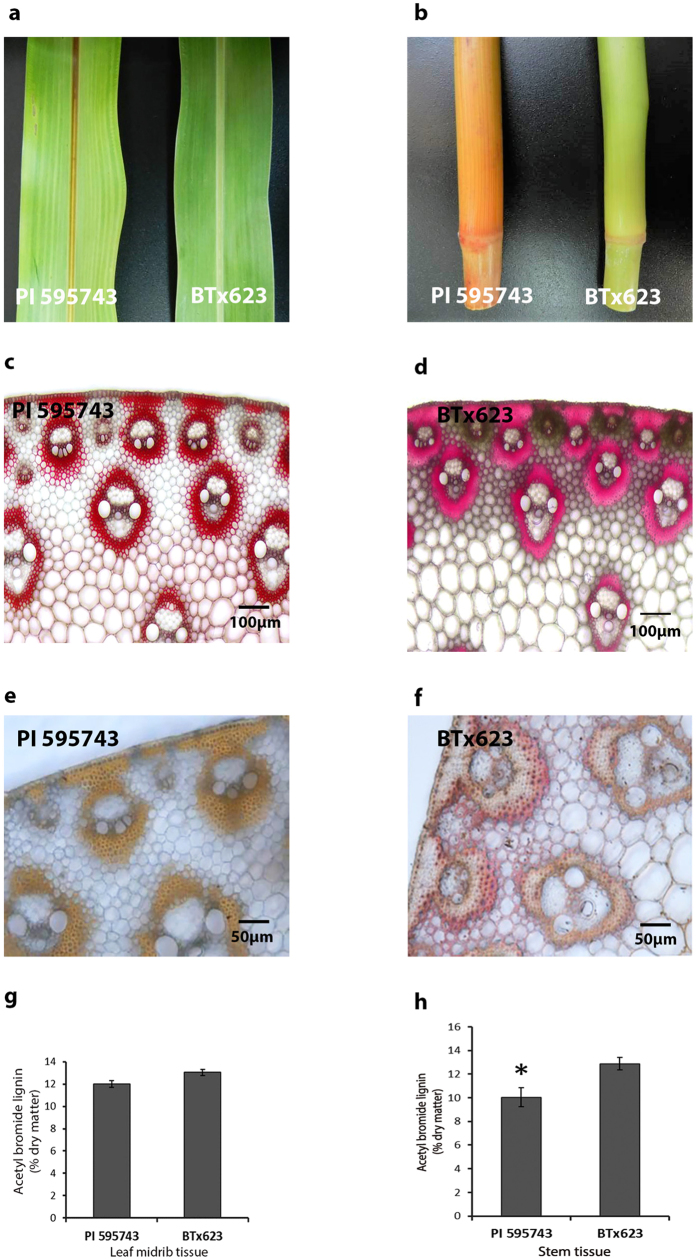
Compared to the non-mutant sorghum plants with pale green midrib, PI 595743 first exhibits the characteristic reddish-brown color in the midrib at the 4, 5-leaf stage. The brown coloration accumulates with the expansion of the leaf blade and gradually fades as plants mature. The base of leaf on the abaxial side has the highest intensity of bmr color. Strong brown pigmentation was also observed throughout the stem of PI 595743 (Fig. 1a,b).
Figure 1.
Phenotypic characterization of PI 595743 mutant. (a,b) Leaf midribs and stems of greenhouse-grown sorghum bmr mutant PI 595743 and non-bmr counterpart BTx623. (c,d) Phloroglucinol staining of cross sections of the second internodes above ground from PI 595743 and BTx623 plants. The microscopic images are representative of multiple experiments. (e,f) Maule staining of the samples used in the phloroglucinol staining. (g,h) Acetyl bromide lignin quantification of leaf midrib tissue and stems collected from PI 595743 and BTx623 lines at the end of vegetative stage. Each bar comprises the mean of three biological and three technical replicates. Error bars indicate the standard error from the mean. Asterisk indicates significant difference at 5% (P < 0.05) level of significance as determined by t-test.
Phloglucinol-HCl staining revealed that the stem cross sections from PI 595743 and its non-bmr counterpart BTx623 displayed red color in lignified tissues, such as the subepidermal region and the scattered vascular bundles (Fig. 1c,d). However, the color contrast was different and PI 595743 had a more intense red coloration, indicating greater incorporation of cinnamaldehyde end-groups (hydroxycinnamyl aldehydes) in the lignin. This result is consistent with that of CAD-deficient mutants in Arabidopsis27. Moreover, when subjected to Maule staining, the pink-red coloration prominently presented in the BTx623 sclerenchyma cells surrounding vascular bundles was missed in the mutant samples (Fig. 1e,f), suggesting that the S unit content was below the detection limit of Maule stain. Together, these results suggested that the total lignin content, especially the composition of lignin, was altered in the vascular tissue of PI 595743. In addition to the histochemical staining, PI 595743 leaf midrib tissue and internode samples were analyzed by acetic bromide, a reagent for determining the total lignin concentration with spectrophotometry-based methods. The means of lignin concentrations of PI 595743 midrib tissue and stem at the end of the vegetative stage were 119.9 and 100.4 mg/g, respectively, representing an average 8% reduction compared to the BTx623 controls (Fig. 1g,h). These results provided quantitative evidence that lignin biosynthesis was indeed impaired in PI 595743 plants.
QTL analysis of bmr trait in the RIL population
The visual brown coloration in bmr mutants has made this trait an ideal candidate for QTL study to reveal the genetic basis of altered lignification. During the process of mapping population development, we found that the F1 plants derived from a cross between the spontaneous mutant PI 595743 and BTx623 showed a non-bmr phenotype, and the F2 progeny revealed a 3:1 Mendelian-like segregation, suggesting that this spontaneous phenotype is controlled by a single recessive gene. This result is consistent with previous findings that brown midrib in grasses is a recessive trait caused by single-gene disruption in the lignin biosynthetic pathway14, 28. Our QTL mapping study integrated with the sequenced BTx623 genome and previously published maps contributed to the rapid development of novel markers within an initially identified QTL interval and the improvement of map resolution. At the end, we constructed a genetic linkage map spanning all ten chromosomes with a marker-saturated chromosome 4 (Fig. S1). The primer sequence information of new SSR markers developed for this study is given in Table S1. After several rounds of iterative QTL analysis, a major QTL explaining 71.2% of the phenotypic variation was determined to be in the interval of markers SASb4546 and SASb4581, with a genetic distance of 2.5 cM from SASb4546 and 1.1 cM from SASb4581 (Fig. 2). This 3.6 cM interval corresponds to 353 kb in terms of physical distance on the sorghum genome sequence (Fig. 3a).
Figure 2.
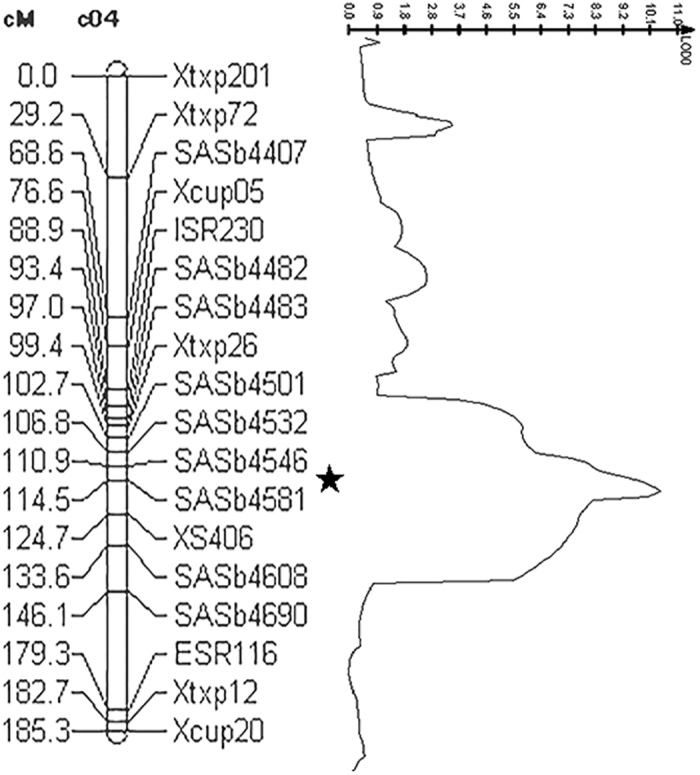
The major QTL underlying the bmr trait in PI 595743 identified in this study. The centiMorgan (cM) distances between marker loci on chromosome 4 are shown along the left and the positions of markers are shown to the right. The LOD (logarithm of the odds) score peak profile using CIM analysis indicates the LOD value for this QTL on chromosome 4 is 10.77.
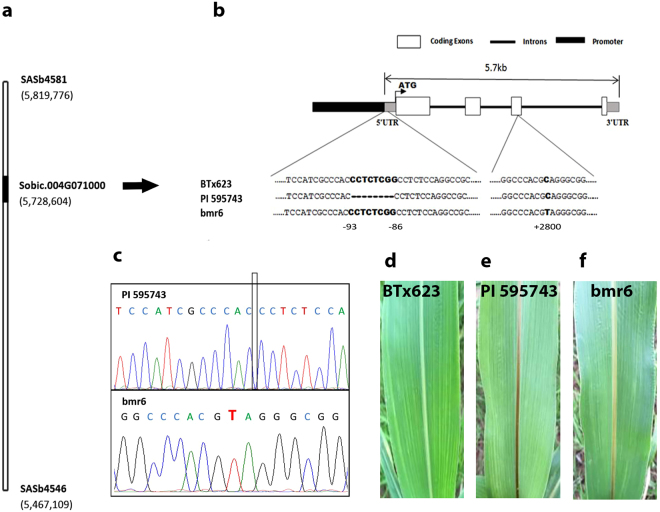
Figure 3.
Map-based cloning of the SbCAD2. (a) Physical map of the target gene flanked by two microsatellite markers with an interval. Target gene Sobic.004G071000 (SbCAD2) is shown in bold. The base pair numbers correspond to the sorghum genome database version 3.1.1 (www.phytozome.net/sorghum). (b) Schematic representation of the SbCAD2 structure and sequence comparison of the SbCAD2 in BTx623, PI 595743 and bmr6. Numbers indicate distances from the start codon. (c) Sequence differences are confirmed by DNA sequencing. Box indicates the 8-bp deletion region in PI 595743. C to T transition in bmr6 is highlighted in red. (d–f) The differences of leaf midrib coloration are observed among BTx623, PI 595743 and bmr6 lines.
Candidate gene selection and validation
Combining the mapping information with sorghum genome sequence database, forty-eight genes have been localized in the region defined by markers SASb4546 and SASb4581. These genes, together with their locations, lengths and functional annotations, are given in Table S2. When we focused on searching for the genes that share functional homology with characterized proteins related to lignin biosynthesis and plant cell wall formation, we found that Sobic.004g071000, the gene encoding a cinnamyl alcohol dehydrogenase (CAD) protein that catalyzes the last step of monolignol biosynthetic pathway, was present in this region. In addition, Sobic.004g071000 (SbCAD2) had been previously identified as the gene underlying the mutagenized bmr6 phenotype in sorghum. Thus, we selected this gene as a promising candidate for further study. Moreover, this selection was supported by the result of our previous phloroglucinol staining which indicates more aldehyde groups in PI 595743 tissues.
A complementation test was performed by crossing PI 595743 with the bmr6 mutant line to determine whether these two mutants are caused by the same gene or two different genes. All twenty-five F1 progeny plants showed the characteristic brown color in their midribs, suggesting these two mutated phenotypes cannot be complemented to each other and they could be either the same mutant alleles, or different mutant alleles of the same gene.
In comparison to the BTx623 reference genome, PI 595743 showed no sequence difference in the exons and introns of SbCAD2. However, an 8-bp nucleotide deletion (CCTCTCGG) was found within its 5′-untranslated region located 93 bp upstream of the start codon (Fig. 3b,c). Furthermore, we designed a specific pair of primers based on this deletion difference and used it to genotype each line in the RIL population. This newly developed maker was mapped to a location that defined the peak of the QTL on chromosome 4 as expected. We also found that this 8-bp deletion co-segregates with the bmr phenotype in the RIL population, which provides further evidence that SbCAD2 is the gene responsible for the expression of bmr trait in the spontaneous mutant PI 595743.
Expression analysis of the SbCAD2
Real-time quantitative PCR (qRT-PCR) was performed to investigate the SbCAD2 expression pattern throughout three representative developmental stages in PI 595743 and the non-bmr control BTx623. Considering the fact that allelic variation in the SbCAD2 between PI 595743 and bmr6 could also affect the expression pattern, along with the observed difference of brown color intensity, we included the bmr6 plants in the gene expression study. Consistent with its role in lignin biosynthesis and cell wall rigidification, transcript levels of SbCAD2 accumulate along with the development of all plants. While compared to the non-bmr line, the accumulation of SbCAD2 transcripts is significantly reduced in both bmr mutants at all three stages. Interestingly, the SbCAD2 expression was further decreased in PI 595743 relative to the bmr6. At the end of the vegetative stage, the SbCAD2 expression was approximately 20-fold lower in PI 595743 relative to the non-bmr counterpart (Fig. 4).
Figure 4.
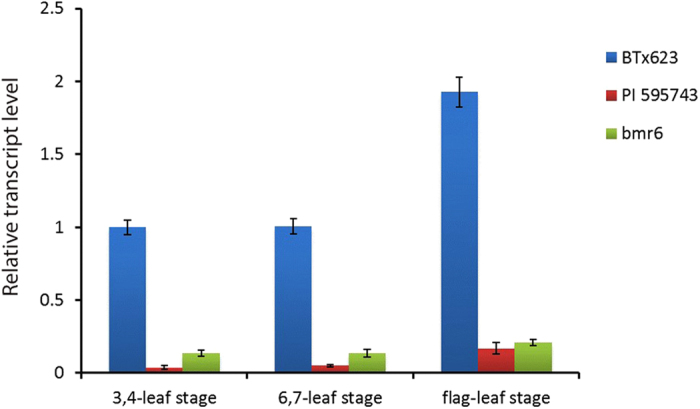
Expression of the SbCAD2 gene at three different developmental stages. Relative expression of SbCAD2 was normalized by geometric means of the housekeeping gene (see Methods). Data are shown as mean ± S.D.
Analysis of the SbCAD2 promoter in transgenic Arabidopsis
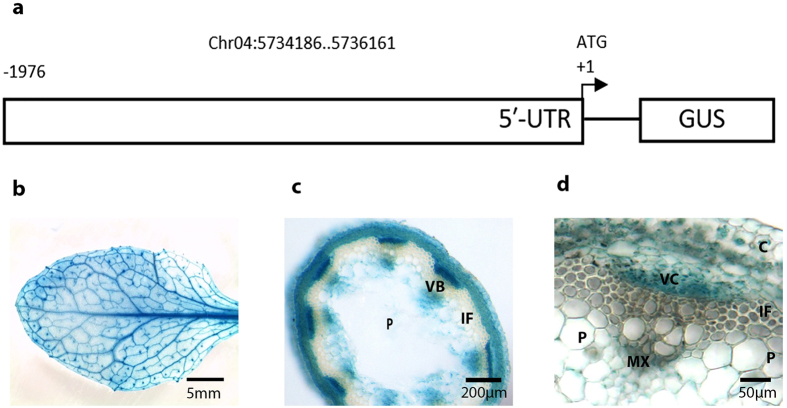
To determine if the SbCAD2 promoter of non-bmr line BTx623 is active in Arabidopsis and its expression pattern is conserved in the lignifying vascular tissues, transgenic Arabidopsis plants containing a pSbCAD2::GUS construct were developed (Fig. 5a) and the expression pattern directed by pSbCAD2 was examined. In fully-expanded leaves, the SbCAD2 promoter directed high specific GUS activity in the midrib and vascular veins (Fig. 5b). Cross sections of the mature inflorescence stems revealed that GUS activity was strongly associated with the developing xylem and the cambial regions adjacent to the metaxylem. No GUS activity was observed in the pith or in the cortex region (Fig. 5c,d). Thus, these data correspond to the function of SbCAD2 and suggest that the mechanism controlling developmental expression of the SbCAD2 promoter in lignifying tissues is conserved among grasses and dicots.
Figure 5.
Histochemical analysis of GUS activity driven by the full-length SbCAD2 in transgenic Arabidopsis plants. (a) Schematic representation of the SbCAD2 promoter-GUS fusion construct. ‘ATG’ indicates the initiation codon site. Numbers indicate the physical position of full promoter sequence. (b–d) SbCAD2 promoter activity is revealed by blue coloration indicating GUS activity in that region. (b) General view of a fully expanded leaf. (c) Cross section of the lower part of inflorescence stem. (d) Enlargement of a vascular bundle, cambial and xylem zone. P, pith; C, cortex; VC, vascular cambium; MX, metaxylem; VB, vascular bundle; IF, interfascicular fibers. All images were obtained from the representatives of 8-week-old independent transgenic lines.
Discussion
Developing brown midrib mutant lines and identifying the genetic basis of lignin deficiency in these mutants are of increased interest due to their potential to reduce biomass recalcitrance. There are more than ten enzymatic steps involved in the monolignol biosynthetic pathway, leading to the synthesis of monolignol precursors for lignin polymerization5, 29. Theoretically speaking, any perturbation in one of these steps could affect the lignin production. To date, only a few steps of the monolignol pathway were found to be directly involved in the exhibition of bmr in sorghum15, 16, 30 and most sorghum bmr mutants developed from mutagenesis are allelic25, 30. Hence, our main objective was to characterize a naturally-occurring bmr mutant in sorghum, with an attempt to isolate novel bmr loci for further study. Interestingly, the causal gene underlying the spontaneous bmr mutant in this study was identified to be SbCAD2, the same gene responsible for the bmr6 phenotype. This high occurrence of allelic bmr mutants could be simply ascribed to the formation of brown color itself. In other words, disruption of a particular step in the monolignol biosynthetic pathway may not be sufficient to induce the generation of a specific bmr phenotype. While the exact cause of brown pigmentation in lignified tissue is elusive, the bmr trait in mutants or transgenic lines with impaired CAD activity has been attributed to the incorporation of cinnamyl aldehydes into lignin in place of cinnamyl alcohols14. This fact is consistent with the phloroglucinol staining result we observed in PI 595743. Another possible explanation of the discrepancy between bmr occurrence and lignin disruption is that modifying some enzymatic steps in the monolignol biosynthetic pathway could result in relatively more severe side-effects on plant growth and development. This hypothesis is supported by several previous studies on lignin-modified mutants. For example, Arabidopsis plants defective in C3H (p-coumarate 3-hydroxylase), the second enzyme in the monolignol pathway, exhibit severe dwarfism and sterility31. Similar deleterious defects were also observed in the diethyl sulfate (DES)-induced bmr lines of sorghum, precluding the subsequent genetic study on those mutants9.
Due to the importance and plasticity of the last step of monolignol pathway, the impacts of CAD disruption on cell wall properties have been intensively studied in several plant species. CAD-deficient mutants32, 33 and transgenic lines with down-regulated CAD activity34, 35 were generally associated with reduced lignin content and/or altered lignin composition change, even though there were some reported inconsistencies partly due to functional redundancy of CAD paralogs or incomplete disruption of CAD expression. In sorghum, the CAD-deficient bmr6 mutant showed reductions in lignin content, altered lignin composition and an improved cell wall digestibility13, 24. Despite the substantial impact on lignin biosynthesis, agronomic evaluation found that the bmr6 mutant was not associated with significant negative impacts on plant fitness in comparison with its near-isogenic counterpart25. In the current work, a spontaneous sorghum bmr mutant, PI 595743, showed a modest decrease in lignin content and dramatic change of lignin composition in certain lignified cell types, in agreement with the aforementioned fact that monolignol biosynthesis is highly plastic as to allow CAD-deficient plants to form lignin polymers directly from cinnamyl aldehydes. The accumulation of aldehydes and reduction of the S lignin subunit in PI 595743 were revealed by phloroglucinol staining and the Maule staining, respectively. These histochemical results provide the first clue that the CAD enzymatic activity is likely affected in PI 595743. Gene expression assay further confirmed the down-regulation of SbCAD2 gene during the development of PI 595743 plants. The significant reduction of CAD activity due to a nonsense mutation was reported in bmr617. Based on the allelic variation, it may prove of interest to examine to what extent the CAD activity is impaired in PI 595743. Furthermore, comparative evaluation of different CAD mutants under the same genetic background, for example, within the near-isogenic lines would be a good starting point to study whether genetic variants in the SbCAD2 gene have different effects on lignin composition, biomass digestibility and the overall agronomic performance.
Previous research on the bmr6 mutant revealed that a nonsense mutation-mediated mRNA decay mechanism was very likely involved in the downregulation of SbCAD2 in its vascular tissues17. In the present work, based on collective evidences from histochemical analysis, QTL mapping, association study and complementation test, an 8-bp deletion in the 5′ UTR of SbCAD2 was linked to the dark-brown pigmentation and the significant reduction of SbCAD2 expression in the spontaneous mutant PI 595743. Thus, an obvious question from these results is: how does this deletion in an untranslated region affect the expression of SbCAD2 and lead to a mutated phenotype? As a fundamental structural and regulatory region of eukaryotic genes, 5′ UTR mainly plays a role in the regulation of mRNA translation, providing a novel layer of coordinated control of gene expression36, 37. Parts of the 5′ UTR may also contain regulatory elements and may be part of the promoter. 5′ UTR-mediated regulation of transcript abundance and translational efficiency has been reported in plant development and stress responses38–40. In the case of PI 595743, the 8-bp deletion within the 5′ UTR could affect its qualitative features, such as length, GC content and potential for secondary structure formation, associated with the translational regulation. There is also a possibility that this deletion is involved in the transcriptional regulation of SbCAD2 by interfering with potential binding sites, an interaction mechanism totally different from the mRNA decay mechanism underlying the reduced expression of SbCAD2 in the bmr6 mutant. Further studies on heterologous expression of a reporter gene fused with this mutated 5′ UTR or promoter deletion analysis are needed to dissect the association between this specific deletion and lignin pathway perturbation.
The active expression of GUS gene in the lignifying tissue of Arabidopsis plants driven by the promoter of the sorghum CAD2 gene reflects the conservation of mechanism that underlies the transcriptional regulation of monolignol biosynthetic pathway in higher plants. The conservative property of lignin biosynthesis in plants has been investigated and discussed previously from different perspectives41–43. Evolutionarily speaking, the signals controlling vascular expression of lignin biosynthetic genes are more likely to be highly conserved in higher plants since lignin biosynthesis is an early adaptation feature for vascular plants to survive in the terrestrial environment41, 44. In addition, the maintenance of conserved motifs in many lignin biosynthetic genes suggested a common catalytic mechanism might be shared during the process of lignification in plants45.
Lignin biosynthesis is a highly coordinated process that involves the expression of multiple monolignol genes and the coordinated regulation of transcription factors44, 46. Knowledge gained from this study could open an avenue for us to decipher the regulatory aspects of the SbCAD2 gene and reveal the occurrence of bmr phenotype and lignin production in grasses at the transcription and post-transcription level.
Methods
Plant materials and mapping population
The plant materials used in this study included the spontaneous sorghum bmr mutant (PI 595743), the elite inbred line (BTx623) and the bmr6 mutant. PI 595743 is a photoperiod insensitive conversion of SC 1201 selected from the World Sorghum Collection. It was developed in Texas, United States and used as a breeding material because of its early maturing, enhanced height and forage potential. In our study, an F6 recombinant inbred line (RIL) population was developed by crossing BTx623 and PI 595743. Specifically, PI 595743 (pollen donor) was crossed with BTx623 (manually emasculated) to obtain heterozygous F1 seeds, and the F1 plants were self-pollinated to produce the F2 segregating population. A set of 188 F6 RILs were obtained from the F2 family by the single-seed descent method.
Lignin detection and quantification
Stem internodes collected from two developmentally-equivalent parental lines were hand-sectioned to a thickness of 200 μm using double-edge razor blades. Phloroglucinol stains the coniferylaldehyde end-groups present in lignin, forming a red-pink color in acidic conditions47. Briefly, sections were stained with phloroglucinol-HCl solution consisting of 1.0% (w/v) phloroglucinol, 3.7% (w/v) HCl, and 10% (v/v) ethanol for 1 min and mounted onto microscope slides for observation. Maule reagent was used to examine the sinapyl alcohol monomers (S lignin) content and localization in vascular tissues as described previously48. Sections were prefixed in 4% glutaraldehyde for 1 h and rinsed with sterile water. After that, sections were immersed in 1% (w/v) potassium permanganate solution for 5 min followed by a water rinse. They were then treated with 10% HCl (w/v) for 5 min, rinsed with water, and mounted on microscope slides in concentrated NH4OH. Stained sections were immediately observed under bright-field lighting on a Nikon 80i microscope and photographed.
For total lignin quantification, stem and midrib materials collected from PI 595743 and BTx623 plants at the end of vegetative stages were dried and pulverized into powder with metal beads in a vibratory solid sample homogenizer. Acetyl bromide (AcBr)-soluble lignin was quantified using the reported procedure49, 50. Briefly, triplicate samples of 10 mg powder from stem and midrib tissues were sequentially washed with 500 μL of water, ethanol, chloroform and acetone for 30 min, respectively. Washed samples were placed in glass 20 mL scintillation vials and heated overnight at 70 °C. Two and a half microliters of 25% AcBr solution was added to each vial and incubated at 50 °C for 2 h with occasional swirling. After cooling to room temperature, each sample was transferred into a 50 mL volumetric flask holding 10 mL of 2 N sodium hydroxide, and 12 mL of acetic acid. More acetic acid was used to bring the solution to the fixed volume. Samples were settled at room temperature overnight and the absorbance of the resultant solution at 280 nm was determined using a spectrophotometer. The lignin percentage of the dry sample was calculated from the regression equation of lignin content in herbaceous samples51.
QTL mapping
Phenotyping was conducted around the time when the brown pigmentation in midrib reaches its maximum intensity, approximately 6 weeks after the planting. The leaf midrib phenotype of each RIL was visually scored and confirmed by the data from three replicates.
For genotyping, genomic DNA was extracted from seedlings of two parental lines and each F6 RIL using a modified CTAB protocol52. Initially, 1019 simple sequence repeat (SSR) markers in sorghum were used to survey their polymorphisms between BTx623 and PI 595743. Among them, 121 were identified as polymorphic and used for genotyping the entire population. PCR products were analyzed by ABI 3730xl DNA Analyzer (Applied Biosystems) and scored using GeneMarker version 4.0 (Soft Genetics LLC).
A linkage map was constructed by MAPMAKER/Exp 3.053 with a logarithm of odds score (LOD) of 3.0 and maximum linkage threshold of 40 cM. The recombination frequency between linked loci was transformed into genetic distance (centimorgan, cM) using the Kosambi’s function. The output files were imported into QTL Cartographer version 2.553 for QTL analysis. Single Marker Analysis, Composite Interval Mapping (CIM) and Multiple Interval Mapping (MIM) analyses were performed by following the lab procedure52.
Candidate gene selection
After the major QTL was localized, the nucleotide sequences of two markers flanking this region were used in BLAST searches against sorghum genome database version 3.1.1 (www.phytozome.net/sorghum) to define the start and end position of the physical interval. All genes within this interval were subject to functional annotation and protein homology detection. A gene was identified as a real candidate only if there was experimental evidence that it is involved with brown pigmentation in vascular tissue, lignin biosynthesis or secondary cell wall formation.
Complementation test and sequencing
Complementation crosses were carried out by emasculating the bmr6 plants at the floral stage followed by hand pollination with PI 595743 pollen. Flowers were bagged to prevent contamination. Seeds were harvested from mature plants and planted in green house for phenotypic evaluation.
Approximately 2 kb of upstream sequence immediately before the coding start codon and the 5.7 kb genomic sequence of SbCAD2 (Sobic.004G071000) in PI 595743 were sequenced as overlapping fragments generated by high-fidelity PCR amplification. Primer information is given in Table S3. The corresponding sequences from BTx623 were retrieved from the sorghum genome database as the reference for comparison.
qRT-PCR analysis
To analyze the expression pattern of SbCAD2 featured in the spontaneous bmr mutant PI 595743, non-bmr BTx623 and bmr6, samples were prepared from each line at the 3,4-leaf stage, the 6, 7-leaf stage and near the end of vegetative stage (emergence of the flag leaf). Fresh leaf tissues from the same location in each plant were harvested, and immediately ground in liquid nitrogen. Total RNA was isolated using TRIzol reagent (Invitrogen) following the manufacturer’s instructions, and 1 μg aliquots were treated with DNase and then reverse transcribed. The transcriptional profile was analyzed by qRT-PCR using the SYBR Green PCR Master Mix (Applied Biosystems) with the Biorad Real-Time PCR System. Primer specificity was verified by cloning and sequencing PCR products obtained from BTx623 cDNA. The melting curve analysis and the evaluation of primer amplification efficiency were performed before running the cDNA samples. Relative expression levels were calculated from the cycle threshold using 2−ΔΔCt method54. A sorghum β-tubulin gene (Sobic.002g350400) was used as the internal control. The data were the average of three biological replicates. Primers used for qRT-PCR are listed in Table S3.
Generation of a SbCAD2 promoter-GUS fusion construct
In order to explore the function of SbCAD2 promoter in lignifying tissue and establish a platform for studying the regulatory mechanism underlying SbCAD2 expression, a construct of the full length SbCAD2 promoter (pSbCAD2) fused to the β-glucuronidase (GUS) reporter gene (uidA) was generated. Specifically, the full length promoter (1976 bp of upstream sequence before the ATG) was PCR amplified from BTx623 genomic DNA by using region-specific primers and the high-fidelity polymerase mix. The PCR product was purified and cloned into the promoterless GUS vector pBI101, producing a fusion construct by following the manual instructions of the In-Fusion HD Cloning Kits (Clontech Laboratories, Inc.). The insertion was PCR amplified and confirmed by sequencing before transformation.
Arabidopsis transformation and GUS activity localization
The full-length promoter-GUS fusion construct was introduced into Agrobacterium tumefaciens strain LBA4404 using the freezing/heat shock method. Transgenic Arabidopsis plants carrying the pSbCAD2::GUS expression cassette were generated by Agrobacterium-mediated transformation using the floral dip method55, 56. In order to confirm the specific expression conferred by the SbCAD2 promoter, a transformation using a CaMV 35 S::GUS construct (pBI 121) was performed in parallel.
Histochemical location of GUS activities in transgenic T1 and T2 plants from different lines was investigated as described in the protocol57. Briefly, the basal region of the floral stems from each equivalently developed plants were collected and incubated for 12–24 h at 37 °C in GUS staining solution: 100 mM sodium phosphate (pH 7), 5 mM EDTA (pH 8), 0.5 mM ferrocyanide, 0.5 mM ferricyanide, 0.5 mg/mL X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronide). A series of ethanol with different concentration (50%, 75%, and 95%) were used to replace the staining solution and remove chloroplast pigments. Crossed sections (80–150 μm) were observed for blue pigment deposition with a Nikon 80i microscope and photographed with an affiliated CCD camera.
Electronic supplementary material
Acknowledgements
We thank Ms. Angela L. Phillips for her excellent technical assistance, Dr. Hengyou Zhang for assistance with vector construction and Drs. Meiqin Liu and Ming Yang for their support on microscopic operation and discussion. This research was supported by the United States Department of Agriculture - National Institute of Food and Agriculture AFRI. Grant Number: 2011–67009–30026 (YH) and additional funding from USDA-ARS, CRIS project, Grant Number: 3072–21000–007–00D (YH). The U.S. Department of Agriculture, Agricultural Research Service, is an equal opportunity/affirmative action employer and all agency services are available without discrimination. Mention of a trademark or proprietary product does not constitute a guarantee or warranty of a product by the U.S. Department of Agriculture and does not imply its approval to the exclusion of other products that may also be suitable.
Author Contributions
H.L. and Y.H. designed the research. H.L. and Y.H. constructed the genetic population and phenotyping. H.L. performed QTL analysis, gene expression and transgenic study. H.L. and Y.H. interpreted the data and wrote the paper. Y.H. supervised the entire study. All authors contributed to the discussion of the results and reviewed the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-10119-1
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rooney WL, Blumenthal J, Bean B, Mullet JE. Designing sorghum as a dedicated bioenergy feedstock. Biofuel Bioprod. Bior. 2007;1:147–157. doi: 10.1002/bbb.15. [DOI] [Google Scholar]
- 2.DeMartini JD, et al. Investigating plant cell wall components that affect biomass recalcitrance in poplar and switchgrass. Energ Environ. Sci. 2013;6:898–909. doi: 10.1039/c3ee23801f. [DOI] [Google Scholar]
- 3.Himmel ME, et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 4.Rogers LA, Campbell MM. The genetic control of lignin deposition during plant growth and development. New Phytol. 2004;164:17–30. doi: 10.1111/j.1469-8137.2004.01143.x. [DOI] [PubMed] [Google Scholar]
- 5.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annual review of plant biology. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 6.Fu CX, et al. Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl. Acad. Sci. USA. 2011;108:3803–3808. doi: 10.1073/pnas.1100310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson LA, et al. Improving saccharification efficiency of alfalfa stems through modification of the terminal stages of monolignol biosynthesis. Bioenerg. Res. 2008;1:180–192. doi: 10.1007/s12155-008-9020-z. [DOI] [Google Scholar]
- 8.Liu CJ, Cai Y, Zhang X, Gou M, Yang H. Tailoring lignin biosynthesis for efficient and sustainable biofuel production. Plant Biotechnol. J. 2014;12:1154–1162. doi: 10.1111/pbi.12250. [DOI] [PubMed] [Google Scholar]
- 9.Porter KS, Axtell JD, Lechtenberg VL, Colenbrander VF. Phenotype, fiber composition, and in-vitro dry-matter disappearance of chemically-induced brown midrib (bmr) mutants of sorghum. Crop Sci. 1978;18:205–208. doi: 10.2135/cropsci1978.0011183X001800020002x. [DOI] [Google Scholar]
- 10.Cherney JH, Axtell JD, Hassen MM, Anliker KS. Forage quality characterization of a chemically-induced brown-midrib mutant in pearl-millet. Crop Sci. 1988;28:783–787. doi: 10.2135/cropsci1988.0011183X002800050012x. [DOI] [Google Scholar]
- 11.Xin ZG, Wang ML, Burow G, Burke J. An induced sorghum mutant population suitable for bioenergy research. Bioenerg. Res. 2009;2:10–16. doi: 10.1007/s12155-008-9029-3. [DOI] [Google Scholar]
- 12.Cherney JH, Cherney DJR, Akin DE, Axtell JD. Potential of brown-midrib, low-lignin mutants for improving forage quality. Adv. Agron. 1991;46:157–198. doi: 10.1016/S0065-2113(08)60580-5. [DOI] [Google Scholar]
- 13.Oliver AL, Grant RJ, Pedersen JF, O’Rear J. Comparison of brown midrib-6 and -18 forage sorghum with conventional sorghum and corn silage in diets of lactating dairy cows. Journal of dairy science. 2004;87:637–644. doi: 10.3168/jds.S0022-0302(04)73206-3. [DOI] [PubMed] [Google Scholar]
- 14.Sattler SE, Funnell-Harris DL, Pedersen JF. Brown midrib mutations and their importance to the utilization of maize, sorghum, and pearl millet lignocellulosic tissues. Plant Science. 2010;178:229–238. doi: 10.1016/j.plantsci.2010.01.001. [DOI] [Google Scholar]
- 15.Bout S, Vermerris W. A candidate-gene approach to clone the sorghum Brown midrib gene encoding caffeic acid O-methyltransferase. Molecular genetics and genomics. 2003;269:205–214. doi: 10.1007/s00438-003-0824-4. [DOI] [PubMed] [Google Scholar]
- 16.Saballos, A., Ejeta, G., Sanchez, E., Kang, C. & Vermerris, W. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics181, 783-795 (2009). [DOI] [PMC free article] [PubMed]
- 17.Sattler, S. E. et al. A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the sorghum brown midrib6 phenotype. Plant Physiology150, 584-595 (2009). [DOI] [PMC free article] [PubMed]
- 18.Saballos, A. et al. Brown midrib2 (Bmr2) encodes the major 4-coumarate:coenzyme A ligase involved in lignin biosynthesis in sorghum (Sorghum bicolor (L.) Moench). Plant J. 70, 818-830 (2012). [DOI] [PubMed]
- 19.Pillonel C, Mulder MM, Boon JJ, Forster B, Binder A. Involvement of cinnamyl-alcohol dehydrogenase in the control of lignin formation in Sorghum bicolor L. Moench. Planta. 1991;185:538–544. doi: 10.1007/BF00202964. [DOI] [PubMed] [Google Scholar]
- 20.Saathoff AJ, Sarath G, Chow EK, Dien BS, Tobias CM. Downregulation of cinnamyl-alcohol dehydrogenase in switchgrass by RNA silencing results in enhanced glucose release after cellulase treatment. Plos One. 2011;6:e16416. doi: 10.1371/journal.pone.0016416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu CX, et al. Downregulation of cinnamyl alcohol dehydrogenase (CAD) Leads to improved saccharification efficiency in switchgrass. Bioenerg. Res. 2011;4:153–164. doi: 10.1007/s12155-010-9109-z. [DOI] [Google Scholar]
- 22.Anderson NA, et al. Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell. 2015;27:2195–2209. doi: 10.1105/tpc.15.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliver AL, Pedersen JF, Grant RJ, Klopfenstein TJ. Comparative effects of the sorghum bmr-6 and bmr-12 genes: I. Forage sorghum yield and quality. Crop Sci. 2005;45:2234–2239. doi: 10.2135/cropsci2004.0644. [DOI] [Google Scholar]
- 24.Dien BS, et al. Improved sugar conversion and ethanol yield for forage sorghum (Sorghum bicolor L. Moench) lines with reduced lignin contents. Bioenerg. Res. 2009;2:153–164. doi: 10.1007/s12155-009-9041-2. [DOI] [Google Scholar]
- 25.Sattler SE, et al. Characterization of novel sorghum brown midrib mutants from an EMS-mutagenized population. G3. 2014;4:2115–2124. doi: 10.1534/g3.114.014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scully ED, et al. Characterization of novel brown midrib 6 mutations affecting lignin biosynthesis in sorghum. J. Integr. Plant Biol. 2016;58:136–149. doi: 10.1111/jipb.12375. [DOI] [PubMed] [Google Scholar]
- 27.Sibout, R. et al. CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell17, 2059–2076 (2005). [DOI] [PMC free article] [PubMed]
- 28.Bittinger TS, Cantrell RP, Axtell JD. Allelism tests of the brown-midrib mutants of sorghum. J. Hered. 1981;72:147–148. doi: 10.1093/oxfordjournals.jhered.a109455. [DOI] [Google Scholar]
- 29.Vanholme R, et al. Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science. 2013;341:1103–1106. doi: 10.1126/science.1241602. [DOI] [PubMed] [Google Scholar]
- 30.Sattler SE, et al. Identification and characterization of four missense mutations in Brown midrib 12 (Bmr12), the caffeic O-methyltranferase (COMT) of sorghum. Bioenerg. Res. 2012;5:855–865. doi: 10.1007/s12155-012-9197-z. [DOI] [Google Scholar]
- 31.Franke R, et al. The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant Journal. 2002;30:33–45. doi: 10.1046/j.1365-313X.2002.01266.x. [DOI] [PubMed] [Google Scholar]
- 32.Sibout R, et al. Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiology. 2003;132:848–860. doi: 10.1104/pp.103.021048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halpin C, et al. Brown-midrib maize (bm1) - a mutation affecting the cinnamyl alcohol dehydrogenase gene. Plant Journal. 1998;14:545–553. doi: 10.1046/j.1365-313X.1998.00153.x. [DOI] [PubMed] [Google Scholar]
- 34.Trabucco GM, et al. Functional characterization of cinnamyl alcohol dehydrogenase and caffeic acid O-methyltransferase in Brachypodium distachyon. Bmc Biotechnol. 2013;13:61. doi: 10.1186/1472-6750-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fornale S, et al. Altered lignin biosynthesis improves cellulosic bioethanol production in transgenic maize plants down-regulated for cinnamyl alcohol dehydrogenase. Mol. Plant. 2012;5:817–830. doi: 10.1093/mp/ssr097. [DOI] [PubMed] [Google Scholar]
- 36.Bailey-Serres, J. Selective translation of cytoplasmic mRNAs in plants. Trends in Plant Science4, 142–148 (1999). [DOI] [PubMed]
- 37.Barrett LW, Fletcher S, Wilton SD. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol. Life Sci. 2012;69:3613–3634. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samadder, P., Sivamani, E., Lu, J. L., Li, X. G. & Qu, R. D. Transcriptional and post-transcriptional enhancement of gene expression by the 5′UTR intron of rice rubi3 gene in transgenic rice cells. Molecular Genetics and Genomics279, 429–439 (2008). [DOI] [PubMed]
- 39.Matsuura H, et al. A computational and experimental approach reveals that the 5′-proximal region of the 5′-UTR has a Cis-regulatory signature responsible for heat stress-regulated mRNA translation in Arabidopsis. Plant Cell Physiol. 2013;54:474–483. doi: 10.1093/pcp/pcs189. [DOI] [PubMed] [Google Scholar]
- 40.Tian JX, Du QZ, Li BL, Zhang DQ. Single-nucleotide polymorphisms in the 5′ UTR of UDP-glucose dehydrogenase (PtUGDH) associate with wood properties in Populus tomentosa. Tree Genet. Genomes. 2014;10:339–354. doi: 10.1007/s11295-013-0689-6. [DOI] [Google Scholar]
- 41.Weng JK, Chapple C. The origin and evolution of lignin biosynthesis. New Phytol. 2010;187:273–285. doi: 10.1111/j.1469-8137.2010.03327.x. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar P, Bosneaga E, Auer M. Plant cell walls throughout evolution: towards a molecular understanding of their design principles. J. Exp. Bot. 2009;60:3615–3635. doi: 10.1093/jxb/erp245. [DOI] [PubMed] [Google Scholar]
- 43.Li X, Wu HX, Southerton SG. Comparative genomics reveals conservative evolution of the xylem transcriptome in vascular plants. BMC Evol. Biol. 2010;10:190. doi: 10.1186/1471-2148-10-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, Q. & Dixon, R. A. Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends in Plant Science16, 227–233 (2011). [DOI] [PubMed]
- 45.Zhong, R. & Ye, Z. H. Transcriptional regulation of lignin biosynthesis. Plant Signaling & Behavior4, 1028–1034 (2009). [DOI] [PMC free article] [PubMed]
- 46.Grima-Pettenati J, Soler M, Camargo ELO, Wang H. Transcriptional regulation of the lignin biosynthetic pathway revisited: New players and insights. Adv. Bot. Res. 2012;61:173–218. doi: 10.1016/B978-0-12-416023-1.00006-9. [DOI] [Google Scholar]
- 47.Clifford MN. Specificity of acidic phloroglucinol reagents. J. Chromatogr. 1974;94:321–324. doi: 10.1016/S0021-9673(01)92389-1. [DOI] [Google Scholar]
- 48.Franke R, et al. Modified lignin in tobacco and poplar plants over-expressing the Arabidopsis gene encoding ferulate 5-hydroxylase. Plant J. 2000;22:223–234. doi: 10.1046/j.1365-313x.2000.00727.x. [DOI] [PubMed] [Google Scholar]
- 49.Hatfield RD, Grabber J, Ralph J, Brei K. Using the acetyl bromide assay to determine lignin concentrations in herbaceous plants: Some cautionary notes. J. Agr.Food Chem. 1999;47:628–632. doi: 10.1021/jf9808776. [DOI] [PubMed] [Google Scholar]
- 50.Hatfield R, Fukushima RS. Can lignin be accurately measured? Crop Sci. 2005;45:832–839. doi: 10.2135/cropsci2004.0238. [DOI] [Google Scholar]
- 51.Morrison IM. A semi-micro method for the determination of lignin and its use in predicting the digestibility of forage crops. J Sci Food Agric. 1972;23:791. [PubMed] [Google Scholar]
- 52.Punnuri S, Huang YH, Steets J, Wu YQ. Developing new markers and QTL mapping for greenbug resistance in sorghum [Sorghum bicolor (L.) Moench] Euphytica. 2013;191:191–203. doi: 10.1007/s10681-012-0755-4. [DOI] [Google Scholar]
- 53.Lander ES, et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 54.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 55.Zhang XR, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 56.Horsch RB, et al. A simple and general-method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- 57.Jefferson RA, Kavanagh TA, Bevan MW. Gus fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher-plants. Embo. J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.