Abstract
Arrhythmogenic cardiomyopathy (ACM) and hypertrophic cardiomyopathy (HCM) are genetically and phenotypically distinct disorders of the myocardium. Here we describe for the first time co-inheritance of mutations in genes associated with ACM or HCM in two families with recurrence of both cardiomyopathies. Among the double heterozygotes for mutations in desmoplakin (DSP) and myosin binding protein C (MYBPC3) genes identified in Family A, two were diagnosed with ACM and two with HCM. In Family B, one patient was identified to carry mutations in α-T-catenin (CTTNA3) and β-myosin (MYH7) genes, but he does not fulfill the current diagnostic criteria neither for ACM nor for HCM. Interestingly, the double heterozygotes showed a variable clinical expression of both cardiomyopathies and they do not exhibit a more severe phenotype than family members carrying only one of the two mutations.
Introduction
Arrhythmogenic Cardiomyopathy (ACM) is characterized by segmental loss of cardiomyocytes, myocardial degeneration, and replacement by fibrofatty scar tissue, predominantly in the right ventricle (RV).1, 2 It shows a prevalence ranging from 1:2000 to 1:5000 in the general population. Hypertrophic Cardiomyopathy (HCM) is defined by the presence of increased left ventricular (LV) wall thickness that is not solely explained by abnormal loading conditions.3, 4 Its prevalence is of 1:500 in the general population. Both ACM and HCM are associated with an increased risk of heart failure, heart transplantation, malignant cardiac arrhythmias and stroke. These autosomal dominant cardiomyopathies represent the most common causes of sudden cardiac death in the young and athletes.1, 5 HCM is frequently described as a disease of the sarcomere,6 whereas, most ACM mutations affect proteins of the intercalated disc, in particular of the desmosome.7
Here we report two Italian families showing co-inheritance of mutations in genes associated with ACM and HCM. To the best of our knowledge, patients carrying mutated ACM and HCM genes have not been described yet.
Materials and methods
The study was approved by the institutional review board and all subjects gave written informed consent after counselling in accordance with the ethical standards of the Declaration of Helsinki. Clinical evaluation was carried out as previously described.8, 9 The diagnosis of definite ACM or HCM was made according to the 2010 ITF criteria10 or to the 2011 ACCF/AHA11 and 2014 ESC3 HCM guidelines, respectively. The DNA analyses were carried out by denaturing high-performance liquid chromatography and direct sequencing. In the ACM probands IV-4 of the Family A and III-3 of the Family B, PKP2, DSP, DSC2, DSG2, JUP and CTNNA3 genes were screened, whereas HCM patients III-4 of the Family A and II-1 of the Family B, were screened for mutations in MYBPC3, MYH7, TNNI3 and TNNT2 genes. All the available family members were subsequently analyzed for the identified mutations. In addition, a member of Family A affected with HCM (III-6) was screened for 133 genes with Blueprint Genetics Heart panel (version 1.1, updated May 6, 2014; http://cardiology.blueprintgenetics.com/panels/cardiomyopathy-panel/). The targeted sequencing was performed using a MiSeq Desktop Sequencer (Illumina). All the variants with a MAF≥1% in locus-specific databases dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/), Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/), the 1000 Genomes Project (http://www.1000genomes.org) and Exome Variant Server (EVS; http://evs.gs.washington.edu/EVS/) were excluded. Variants with a coverage <15 ×, synonymous single-nucleotide variants and TTN variants were filtered out. Moreover, all the variants identified in genes not directly associated nor with ACM neither with HCM were considered variants of unknown significance and excluded from further analysis. Taking into account that the allele frequency of ACM in the general population spans from 0.01 and 0.025% and the allele frequency of HCM in the general population is about 0.1%, a genetic variant has been considered a mutation with a possible clinical significance if its minor allele frequency is ≤0.01% for ACM and ≤0.1% for HCM. In silico prediction tools such as Condel (http://bg.upf.edu/fannsdb/query/condel), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (http://sift.jcvi.org/) and Mutation Taster (http://www.mutationtaster.org/index.html) were used. In order to classify the identified variants, we used the Pathogenicity Calculator software (http://calculator.clinicalgenome.org/site/cg-calculator), developed to enable a more accurate application of the American College of Medical Genetics (ACMG) consensus guidelines.12 The criteria for classifying a variant as pathogenic, likely pathogenic, uncertain significance, benign or likely benign are based on different weighted levels of evidences and on the rules for combining these evidences, provided in the ACMG guidelines.12 The identified mutations associated with cardiomyopathies were submitted to ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/).
Results and discussion
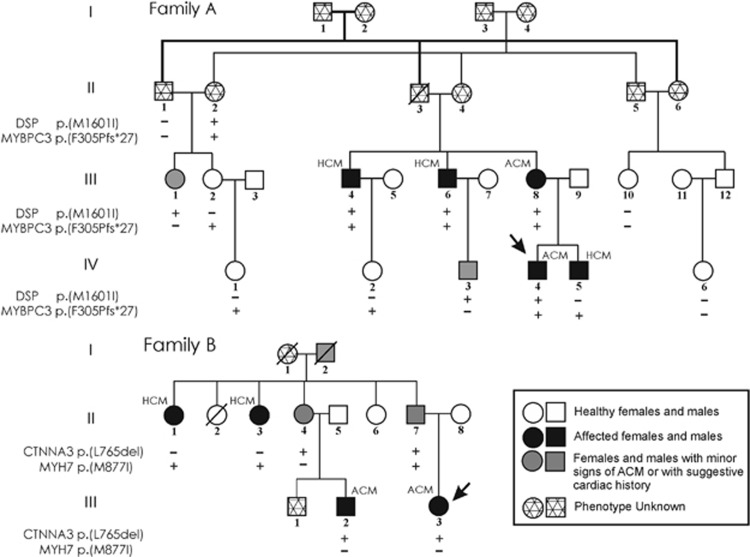
The proband IV-4 of Family A (Figure 1) was diagnosed to be affected by ACM at the age of 19 (Table 1; Supplementary Figure S1). He resulted to carry the variant NM_004415.2: c.4803G>A p.(Met1601Ile) in DSP gene (ClinVar SCV000564076)(data previously reported13). This missense variant is absent in all consulted databases of genetic variations and involves a highly conserved residue in the central rod domain. The prediction analysis with four different in-silico tools provided conflicting results, only Mutation Tester predicted this variant as damaging. The prediction data should be used carefully in sequence variant interpretation and particularly in cases with opposite results. For this reason, the prediction was considered as a minor evidence. The Pathogenicity Calculator software classified this variant as a likely pathogenic mutation. To provide additional evidences in support of the pathogenicity of this missense mutation, functional studies will be needed. Subject III-6 of the same family (Figure 1; Table 1) was a competitive athlete up to 40 years. At first visit two D-echocardiogram revealed minor kinetic abnormalities of RV and an asymmetric septal hypertrophy with a maximal left ventricular wall thickness (LVWT) of 15 mm, initially interpreted as a possible secondary effect of intense physical activity. After two years of follow-up, he complained of palpitations. His electrocardiogram showed ST segment elevation in the precordial lead V1 suggesting Brugada Syndrome, but a provocative flecainide test was negative. At the same time a HCM phenotype persisted, despite stopping training, with a maximal LVWT of 17 mm. As he was 60 years old, he had permanent atrial fibrillation (AF). Instrumental findings of patient III-6 are reported in Supplementary Figure S2. The genetic screening of 133 cardiomyopathy genes identified a total of 890 variants. After the filtering for quality and frequency and discarding TTN and synonymous single-nucleotide variants, the mutation in DSP gene, previously detected in the proband, and an additional variant NM_000256.3: c.913_914delTT p.(F305Pfs*27) in MYBPC3 gene (ClinVar SCV000564398) were reported. The Pathogenicity Calculator software classified this MYBPC3 variant as a pathogenic mutation. The genetic analysis extended to the family members identified three individuals negative for both mutations, four carrying the MYBPC3 mutation, two carrying the DSP mutation and four harboring both mutations (Figure 1). Among the double heterozygotes, subject III-8 was diagnosed with ACM (Table 1) at the age of 48 years. Subject III-4 was a competitive athlete until he was 32 years old. A non-obstructive HCM was diagnosed with an asymmetric maximal LVWT of 18 mm (Supplementary Figure S3). Cardiac magnetic resonance (CMR) was not feasible due to claustrophobia. Subject II-2 is 80 years old and declined a clinical evaluation.
Figure 1.
Pedigree of Family A and B showing co-inheritance of ACM and HCM mutations. Arrows indicate the ACM probands.
Table 1. Clinical data of ACM and HCM mutation carriers in Families A and B.
| ID | Mutated Gene | Gender | Age first visit | Age last FU | ECG | Late potentials | max LVWT, (mm) | LA (mm) | EF (%) | RV | CMR data | Symptoms | Arrhytmias | Events | Comorbidities | Diagnosis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family A | ||||||||||||||||
| III-4 | DSP MYBPC3 | M | 48 | 52 | LVH | negative | 18 | 49 | 54 | Mild dilation | NP | Chest pain and dyspnea | Sporadic polymorphic PVCs. | None | Gastritis, athlete since 32 yrs | HCM |
| III-6 | DSP MYBPC3 | M | 44 | 60 | Neg inferior T waves (first), LAD and septal pseudonecrosis (last) | positive | 18 | 52 | 66 | Minor kinetic abnormalities | NP | Palpitations | Rare PVCs, Permanent AF | None | None, athlete since 40 yrs | HCM |
| III-8 | DSP MYBPC3 | F | 48 | 65 | normal | positive | 12 | 41 | 68 | Mild dilation, regional akinesia | NP | None | Frequent PVCs, persistent AF (Pharm CV) | None | Hypertension hypercholesterolemic | ACM |
| IV-4 Proband | DSP MYBPC3 | M | 19 | 36 | Neg T waves V1-V3 | positive | 10 | 33 | 60 | Mild dilation, regional akinesia | Structural and dynamic alterations of RV, spot of LGE | Chest pain | Frequent PVCs | None | None | ACM |
| IV-2 | MYBPC3 | F | 15 | 18 | LAD | negative | 11 | 27 | 56 | Normal | Septal crypts, normal LV and RV dimensions, thickness and function | None | Rare PVCs | None | None | − |
| IV-3 | DSP | M | 16 | 29 | normal | negative | 9 | 32 | 67 | Mild dilation,regionalhypokinesia | Mild RV dilation with normal function, mitral valve prolapse | None | None | None | None | Borderline ACM |
| IV-5 | MYBPC3 | M | 30 | 33 | normal | negative | 13 | 34 | 61 | normal | LV hypertrophy (14 mm) | None | None | None | None | HCM |
| III-1 | DSP | F | 25 | 35 | normal | positive | 7 | 31 | 61 | normal | NP | None | None | None | None | Borderline ACM |
| III-2 | MYBPC3 | F | 28 | 40 | normal | positive | 9 | 29 | 58 | normal | NP | None | None | None | None | − |
| IV-1 | MYBPC3 | F | 18 | 23 | normal | negative | 8 | 30 | 57 | normal | NP | None | None | None | None | − |
| Family B | ||||||||||||||||
| II-7 | CTNNA3 MYH7 | M | 50 | 57 | normal | negative | 12 | 45 | 62 | Mild dilation | NP | None | None | None | None | Borderline ACM |
| III-3 Proband | CTNNA3 | F | 15 | 32 | normal | positive | 7 | 34 | 64 | Mild dilation, regional akinesia | Mild RV dilatation and kineticalterations of RV apex | None | NSVT | None | None | ACM |
| II-1 | MYH7 | F | 69 | 77 | LVH + Repolarisation abnorm | NP | 20 | 54 | 56 | Normal | HCM (max LVWT=18 mm) with apical aneurism, septal and apical LGE | Dyspnea (NYHAclassII), palpitations | Paroxysmal AF, frequent PVCs and NSVT | Syncope in the past | Hypothyroidism, latent TB | HCM |
| II-3 | MYH7 | F | 64 | 65 | normal | NP | 19 | 42 | 64 | Normal | NP | Dyspnea | None | None | None | HCM |
| II-4 | CTNNA3 | F | 64 | 66 | normal | negative | 8 | 31 | 65 | normal | NP | None | None | None | None | Borderline ACM |
| III-2 | CTNNA3 | M | 36 | 44 | normal | positive | 9 | 38 | 60 | Mild dilation, regional akinesia | NP | None | None | None | None | ACM |
Abbreviations: AF, atrial fibrillation; CMR, cardiac magnetic resonance; ECG, electrocardiogram; EF, ejection fraction; FU, follow-up; LA, left atrium; LAD, left anterior descending; LGE, late gadolinium enhancement; LVH, Left ventricular hypertrophy; LVWT, Left Ventricular Wall Thickness; NP, not performed; NSVT, non-sustained ventricular tachycardia; NYHA class, New York Heart Association classification; Pharm CV, Pharmacological cardioversion; PVCs, premature ventricular contractions; RV, right ventricle; TB, tuberculosis.
In summary, among the four clinically evaluated double heterozygotes for mutations in DSP and MYBPC3 genes, two were diagnosed with ACM and two with HCM (Table 1). The MYBPC3 variant was previously described by our group as a founder mutation in about 20% of Italian HCM patients.8 The onset of the disease in patients carrying this mutation is more likely between ages 30 and 40. The penetrance in males is higher than 50% after the third decade of life and reaches 85% in subjects ≥60 years, whereas in females is <50% during all the lifetime.8 According to these data, it is interesting to note that the two double heterozygote subjects diagnosed with HCM are men older than 40 years, even if the RV was mildly dilated in both cases, whereas the subjects diagnosed with ACM, are a 66 year old woman and a man younger than 40 years of age. The female subject showed also mild LV hypertrophy (in the presence of hypertension) with paroxysmal AF whereas the male proband showed ACM phenotype without LV hypertrophy. A follow-up program for this subject is mandatory considering that he is a carrier of the founder mutation with a significant high risk to develop HCM.
Moreover, among the four subjects carrying only the MYBPC3 mutation, the male IV-5 was diagnosed with HCM at the age of 30, whereas III-2, IV-1 and IV-2 were all asymptomatic females, with age ranging from 18 to 40 years (Table 1). Finally, the two single-DSP mutation carriers were considered borderline for ACM because they showed only 1 major (family history) and 1 minor criterion. Late potentials were present at 40 and 80 filter settings in one of them (III-1) and minor structural alterations were detected in the other one (IV-3) (Table 1).
In Family B, the proband III-2 (Figure 1) was diagnosed with ACM at the age of 15 years and she resulted to carry the in frame deletion NM_001127384.2: c.2296_2298delTTG p.(Leu765del) in CTNNA3 gene (ClinVar SCV000564396)(data previously reported9). This variant is absent in 250 ethnically matched healthy controls and in dbSNP, 1000Genomes, EVS and ExAC databases. It is localized in an important domain of αT-catenin, and the affected residue is strongly conserved among species. Moreover, the mutant protein p.(L765del) showed a much stronger dimerization potential and formed aggresomes in HEK293T cells.9 The Pathogenicity Calculator software classified this variant as a pathogenic mutation. Only later, individuals II-1 and II-3 were referred to our Cardiological Centre as members of the Family B. In subject II-1 a non-obstructive HCM was diagnosed at the age of 64 years. She had frequent episodes of AF and non-sustained ventricular tachycardia. CMR showed widespread septal and apical late gadolinium enhancement. The genetic screening showed the presence of the variant NM_000257.3: c.2631G>T p.(Met877Ile) in MYH7 gene (ClinVar SCV000564076). The MYH7 variant, previously identified in another Italian HCM patient,14 is absent in dbSNP, in 1000Genomes, in EVS and ExAC and involves a highly conserved residue in subfragment 2 domain of the neck region. The prediction analysis with four different in-silico tools provided conflicting results, only Mutation Tester predicted this variant as damaging. The Pathogenicity Calculator software classified this MYH7 variant as a pathogenic mutation.
The genetic analysis in four family members revealed another carrier of MYH7 mutation, two individuals with the CTTNA3 mutation and one subject carrying both mutations (Figure 1). The female II-3 carrying the MYH7 mutation showed non-obstructive HCM at 69 years old. Among the CTNNA3 mutation carriers, patient III-2 was diagnosed with ACM when he was 36 years old, whereas his mother showed only minor echocardiographic abnormalities not fulfilling diagnostic criteria for ACM. Interestingly subject II-7 (57 years old) carrying both mutations does not fulfill the current diagnostic criteria neither for ACM nor for HCM. His echocardiogram demonstrates a mild right ventricular dilatation (right ventricular outflow tract (RVOT) diameter in parasternal long-axis (PLAX)=30 mm) and a LVWT of 12 mm with an abnormal filling pattern (Table 1).
In the affected members of both families we cannot exclude the presence of additional variants in cardiomyopathy genes or in other novel genes, which could contribute to disease expression, in particular in affected subjects of Family B not screened using a next generation sequencing approach. Of note, few papers reported the identification of non-synonymous rare variants of unknown clinical significance in desmosomal genes in HCM patients. Lopes et al. found a large number of desmosomal candidate variants in HCM cases, most of which occurred in patients who had also variants in genes encoding for sarcomere proteins, making difficult to establish their pathogenic role.15 Botillo et al. identified at least a desmosomal variant in 14% of their HCM patients, negative for mutations in sarcomeric genes.16 The impact of these desmosomal variants on the HCM phenotype is still unknown.
The results of the present study show that the clinical expression of both cardiomyopathies in these rare patients seems to be different from individual to individual, probably due in part to the age and gender related incomplete penetrance of the co-inherited mutations. Moreover, these double heterozygotes do not seem to show a more severe form of cardiomyopathy than family members carrying only one of the two mutations. This could be explained by the fact that these mutations involve proteins belonging to different cell structures, avoiding a possible synergic effect as supposed in ACM or HCM multiple mutations affecting the desmosome or the sarcomere, respectively.17, 18, 19, 20 The possibility to perform a comprehensive clinical and genetic study is of pivotal importance for clarifying the effect of co-occurring mutations on the cardiac phenotype allowing better diagnosis, prognosis and a well-defined management of patients. However, due to the dramatically increased number of the variants identified with next generation sequencing approaches and the frequent identification of missense variants in ACM genes in the general population (16%),21 it is of primary importance that the final interpretation of the genetic results is carried out in close collaboration among the experienced professionals of a recognized center. An adequate pre- and post-genetic test counselling for the patients and their relatives is also essential; at the same time, the genetic heterogeneity, incomplete penetrance and variable expressivity related to the inherited cardiomyopathies heighten the complexity of the genetic counselling. This underlines the pressing need for a better understanding of the genotype-phenotype correlation and the modifier factors involved in modulating the clinical phenotype in patients affected with inherited cardiac diseases.
Acknowledgments
We would like to thank the patients and their families for participation in this study. This study was funded by the TRANSAC Strategic Research Grant CPDA133979/13, University of Padua, Italy; Target Projects 331/12, RP 2014-00000394, Regional Health System, Venice, Italy; The University of Padua Research Project (PRAT) CPDA133979.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Thiene G, Nava A, Corrado D, Rossi L, Pennelli N: Right ventricular cardiomyopathy and sudden death in young people. N Engl J Med 1988; 318: 129–133. [DOI] [PubMed] [Google Scholar]
- Basso C, Bauce B, Corrado D, Thiene G: Pathophysiology of arrhythmogenic cardiomyopathy. Nat Rev Cardiol 2011; 9: 223–233. [DOI] [PubMed] [Google Scholar]
- Elliott PM, Anastasakis A, Borger MA et al: ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC). Eur Heart J 2014; 35: 2733–2779. [DOI] [PubMed] [Google Scholar]
- Maron BJ, Ommen SR, Semsarian C, Spirito P, Olivotto I, Maron MS: Hypertrophic cardiomyopathy: present and future, with translation into contemporary cardiovascular medicine. J Am Coll Cardiol 2014; 64: 83–99. [DOI] [PubMed] [Google Scholar]
- Corrado D, Basso C, Schiavon M, Thiene G: Screening for hypertrophic cardiomyopathy in young athletes. N Engl J Med 1998; 339: 364–369. [DOI] [PubMed] [Google Scholar]
- Ho CY, Charron P, Richard P, Girolami F, Van Spaendonck-Zwarts KY, Pinto Y: Genetic advances in sarcomeric cardiomyopathies: state of the art. Cardiovasc Res 2015; 105: 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calore M, Lorenzon A, De Bortoli M, Poloni G, Rampazzo A: Arrhythmogenic cardiomyopathy: a disease of intercalated discs. Cell Tissue Res 2015; 360: 491–500. [DOI] [PubMed] [Google Scholar]
- Calore C, De Bortoli M, Romualdi C et al: A founder MYBPC3 mutation results in HCM with a high risk of sudden death after the fourth decade of life. J Med Genet 2015; 52: 338–347. [DOI] [PubMed] [Google Scholar]
- van Hengel J, Calore M, Bauce B et al: Mutations in the area composita protein αT-catenin are associated with arrhythmogenic right ventricular cardiomyopathy. Eur Heart J 2013; 34: 201–210. [DOI] [PubMed] [Google Scholar]
- Marcus FI, McKenna WJ, Sherrill D et al: Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the task force criteria. Circulation 2010; 121: 1533–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersh BJ, Maron BJ, Bonow RO et al: ACCF/AHA guideline for the diagnosis and treatment of hypertrophic cardiomyopathy: executive summary: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Circulation 2011; 124: 2761–2796. [DOI] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S et al: Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauce B, Rampazzo A, Basso C et al: Clinical phenotype and diagnosis of arrhythmogenic right ventricular cardiomyopathy in pediatric patients carrying desmosomal gene mutations. Heart Rhythm 2011; 8: 1686–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melacini P, Corbetti F, Calore C et al: Cardiovascular magnetic resonance signs of ischemia in hypertrophic cardiomyopathy. Int J Cardiol 2008; 128: 364–373. [DOI] [PubMed] [Google Scholar]
- Lopes LR, Zekavati A, Syrris P et al: Genetic complexity in hypertrophic cardiomyopathy revealed by high-throughput sequencing. J Med Genet 2013; 50: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottillo I, D'Angelantonio D, Caputo V et al: Molecular analysis of sarcomeric and non-sarcomeric genes in patients with hypertrophic cardiomyopathy. Gene 2016; 577: 227–235. [DOI] [PubMed] [Google Scholar]
- Bauce B, Nava A, Beffagna G et al: Multiple mutations in desmosomal proteins encoding genes in arrhythmogenic right ventricular cardiomyopathy/dysplasia. Heart Rhythm 2010; 7: 22–29. [DOI] [PubMed] [Google Scholar]
- Rigato I, Bauce B, Rampazzo A et al: Compound and digenic heterozygosity predicts lifetime arrhythmic outcome and sudden cardiac death in desmosomal gene-related arrhythmogenic right ventricular cardiomyopathy. Circ Cardiovasc Genet 2013; 6: 533–542. [DOI] [PubMed] [Google Scholar]
- Maron BJ, Maron MS, Semsarian C: Double or compound sarcomere mutations in hypertrophic cardiomyopathy: a potential link to sudden death in the absence of conventional risk factors. Heart Rhythm 2012; 9: 57–63. [DOI] [PubMed] [Google Scholar]
- Biagini E, Olivotto I, Iascone M et al: Significance of sarcomere gene mutations analysis in the end-stage phase of hypertrophic cardiomyopathy. Am J Cardiol 2014; 114: 769–776. [DOI] [PubMed] [Google Scholar]
- Kapplinger JD, Landstrom AP, Salisbury BA et al: Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia associated mutations from background genetic noise. J Am Coll Cardiol 2011; 57: 2317–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.