Abstract
Microcephalic primordial dwarfism (MPD) is a group of autosomal recessive inherited single-gene disorders with intrauterine and postnatal global growth failure. Seckel syndrome is the most common form of the MPD. Ten genes are known with Seckel syndrome. Using genome-wide SNP genotyping and homozygosity mapping we mapped a Seckel syndrome gene to chromosomal region 4q28.1-q28.3 in a Turkish family. Direct sequencing of PLK4 (polo-like kinase 4) revealed a homozygous splicing acceptor site transition (c.31-3 A>G) that results in a premature translation termination (p.[=,Asp11Profs*14]) causing deletion of all known functional domains of the protein. PLK4 is a master regulator of centriole biogenesis and its deficiency has recently been associated with Seckel syndrome. However, the role of PLK4 in genomic stability and the DNA damage response is unclear. Evaluation of the PLK4-Seckel fibroblasts obtained from patient revealed the expected impaired centriole biogenesis, disrupted mitotic morphology, G2/M delay, and extended cell doubling time. Analysis of the PLK4-Seckel cells indicated that PLK4 is also essential for genomic stability and DNA damage response. These findings provide mechanistic insight into the pathogenesis of the severe growth failure associated with PLK4-deficiency.
Introduction
Organismal growth is determined mainly by cell number which is under the control of extracellular and intracellular signals that regulate the homeostasis between cell proliferation and death.1 Recent studies proposed microcephalic primordial dwarfism (MPD) as a model disease to understand the molecular mechanism of growth.2 MPD is a group of rare single-gene disease with severe growth failure encompassing Seckel syndrome (OMIM:210600),3 microcephalic osteodysplastic primordial dwarfism types I or III (OMIM:210710), microcephalic osteodysplastic primordial dwarfism II (OMIM:210720),4, 5 and Meier–Gorlin syndrome (OMIM:224690).6
Seckel syndrome is an autosomal recessive disorder characterized by intellectual disability, severe prenatal and proportionate postnatal growth failure, microcephaly with a ‘bird-headed’ profile with receding forehead, large and beaked nose, micrognathia, large eyes and a narrow face. Commonly associated skeletal anomalies are dislocation of the head of the radius and fifth finger clinodactyly. Additional phenotypic features such as delayed bone age, eleven pairs of ribs, strabismus, microphthalmia, optic nerve hypoplasia, high-arched palate, enamel hypoplasia, and hypodontia or oligodontia have been reported.3, 7, 8, 9
Seckel syndrome is a genetically heterogeneous condition and a total of ten genes including ATR,10 RBBP8 (CtlP),11 CEP152 (ref. 7), CENPJ (CPAP),12 CEP63 (ref. 13), DNA2 (ref. 14), ATRIP,15 Ninein (NIN),16 PLK4 (Polo-like kinase 4)8, 9 and CDK5RAP2 (ref. 17) have hitherto been found to be associated with it. Functional evaluation of these genes to identify the cellular pathogenesis underlying Seckel syndrome showed that these genes have critical roles in fundamental cellular processes including centriole biogenesis and centrosome related functions (CEP152, CENPJ, CEP63, NIN and CDK5RAP2), and maintenance of genomic stability (ATR, RBBP8, ATRIP and DNA2). It is suggested that deficiency of these genes can disrupt the homeostasis toward cell proliferation leading to reduced global body mass as seen in MPD.7, 10, 18
In this study, we present the identification of a homozygous PLK4 splice site variant in a consanguineous Turkish family leading to Seckel syndrome. PLK4 was recently associated with Seckel syndrome and abnormalities of cell cycle progression.8, 9 We analyzed the fibroblasts obtained from a PLK4-Seckel patient and show that PLK4 has a critical role in genomic stability, in addition to its well-characterized function in centriole biogenesis and cell cycle progression.
Materials and methods
Patients
In this study, seven individuals of a Turkish family, of which two were affected with Seckel syndrome were evaluated (Figure 1). Genomic DNA was extracted using standard protocols from peripheral blood of the affected individuals and other available family members. A skin biopsy was obtained from the affected individual VIII:2. The study protocol was approved by the Ethics Committee of Karadeniz Technical University, Faculty of Medicine (approval number: 2013/73). The study was conducted in accordance with the Declaration of Helsinki and written informed consent was obtained from the participants.
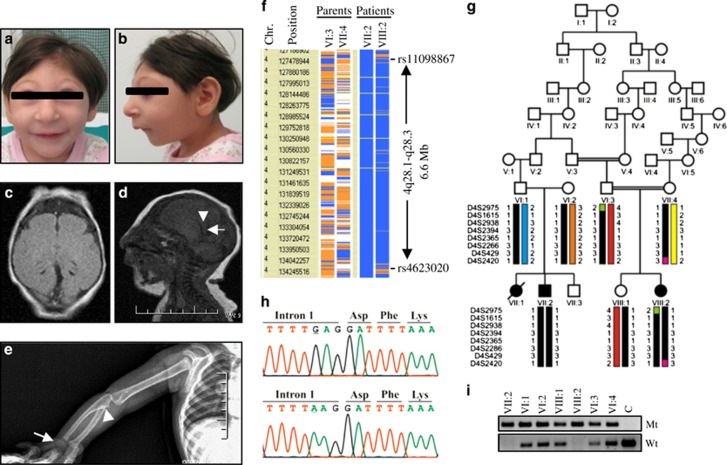
Figure 1.
Phenotype of PLK4-Seckel patient and mutation analysis. (a–b) Frontal and lateral view of the patient VIII:2. (c) T1 weighted axial image of the patient with microcephaly shows pachygyrius. (d) Sagittal T1 weighted sequence shows only the distal part of the body (arrows head) and splenium (arrows) of corpus callosum however the rostrum and genu were absent. (e) View of the right arm showing hypoplasia of the proximal part of the Radius (arrow head), absence of hamate and capitate (arrow). (f) Schematic representation of homozygous segment obtained from genome-wide SNPs screening. Homozygous genotypes seen in affected individual VII-2 are shown in blue, whereas contrasting homozygote genotypes are depicted in white. Heterozygous and noninformative SNPs appear in orange and yellow, respectively. Gray indicates no call. A homozygous 6.6 Mb segment between rs11098867 and rs4623020 SNPs markers on chromosome 4q28.1-q28.3 was seen to be shared by two patients with same haplotype. (g) Family pedigree showing haplotype of critical chromosomal region segregating with disease. Disease-associated region was donated with black bar and colored bars show the wild type haplotypes. (h) Sequence chromatograms showing identified homozygous c.31-3 A>G variation (upper chromatogram) and wild-type sequence (lower chromatogram) in PLK4. (i) Screening of c.31-3 A>G transition in family members with the amplification refractory mutation system. Mt shows mutant allel and Wt shows wild-type allele.
Genotyping and homozygosity mapping
Genome-wide SNP genotyping was carried out by using Illumina Cyto-SNP12 300 K BeadChip platform. To find out the homozygously inherited genomic segments co-segregating with disease in two patients, VIGENOS Plus (Visual Genome Studio Plus) program (Hemosoft, Ankara, Turkey) was used. Homozygosity was confirmed by subsequent microsatellite marker genotyping of all family members.
Mutation analysis
Upon evaluating the functions of the genes located in critical interval, a candidate gene was selected for mutation analyses. The entire exons and exon-intron boundaries of PLK4 (NM_014264.4 and NG_041821.1) were amplified via PCR with primers that were designed by using Primer3 web software (Supplementary Table S1). PCR products were directly sequenced at ABI PRISM 3130 DNA analyzer (Applied Biosystems, Foster City, CA, USA). Screening of the identified sequence variations within the family and ancestrally matched control were carried out via amplification refractory mutation system with allele-specific primers (Supplementary Table S2).
Karyotyping and sister chromatid exchange analysis
Heparinized peripheral blood samples were obtained from patient VIII:2 and a sex and age matched control. Karyotyping and mitomycin-C (MMC)-induced sister chromatid exchange (SCE) analyses were performed as previously described.7
RT-PCR and qPCR analyses
The effects of identified splicing acceptor site variation on splicing of intron 1 were evaluated via RT-PCR. Total RNA was obtained from venous blood samples of patient VIII:2 and her parents and a control by using TRI Reagent (Sigma, St Louis, MO, USA). Synthesis of cDNA was performed with the First-Strand cDNA Synthesis Kit (MBI Fermentas, Vilnius, Lithuania). Subsequently, amplification of cDNA was carried out with PLK4 specific primers designed for sequences corresponding to exons 1 and 3 (Supplementary Table S3). To evaluate the effect of variation on splicing of intron 1, amplicons were extracted from agarose gel and sequenced. Quantitation of wild-type PLK4 transcript in patient’s cDNA was determined with real-time PCR using wild type and mutant transcript specific primers. GAPDH was used as internal control (Supplementary Table S4).
Cell culture and DNA-damaging agents treatment
Primary fibroblast cell culture was established from skin biopsies obtained from patient VIII:2 and an age matched control in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% fetal bovine serum (FBS), (Gibco, Grand Island, NY, USA). Passaging of cells was performed every 3–4 days. To analyze the DNA damage response, exponentially growing PLK4-Seckel and controls cells were treated with 2 mM final concentration of hydroxyurea (HU) (Sigma-Aldrich, St Louis, MO, USA) in DMEM without serum and antibiotics for 4 h. In order to analyze the effect of DNA-damaging agents to basal and activated Chk1 and Chk2 protein level along with p53, PLK4-Seckel cells were treated with 5 and 10 ng/ml concentration of MMC in DMEM supplemented with 5% FBS and antibiotics for 24 h.
Immunofluorescence detection
Fibroblasts were seeded on cover slips with about 50% confluence. Following overnight incubation, fixation and permeabilization were carried out with 4% paraformaldehyde in PBS for 10 min and 0.1% Triton X-100 in PBS for 5 min, respectively. Before and after permeabilization cover slips were rinsed five times with PBS. Blocking was carried out with 3% bovine serum albumin in PBS for 1 h. Appropriate primary and secondary antibodies diluted in blocking solution were used. Cover slips were rinsed five times with PBS after primary and secondary antibody incubations. As primary antibodies; anti-PLK4 (ab137398), anti-pericentrin (ab28144), anti-α-tubulin (ab15246), anti-γ-H2AX (phospho S139) (ab26350), anti-centrin-1 (ab11257) and anti-γ-tubulin (ab27074) (Abcam, Cambridge, UK) were used. Alexa flour 488 goat anti-rabbit IgG (ab96899), and goat anti-mouse IgG (ab150117) and Cy3 conjugated goat anti-rabbit IgG (ab6939) (Abcam) were used as secondary antibodies. Slides were counterstained with DABCO mounting media with DAPI (Life Technologies, Eugene, OR, USA). Samples were analyzed with fluorescence microscope (Nikon eclipse E800, Tokyo, Japan).
Protein isolation and western blotting
Cells were lysed with ice cold RIPA lysis buffer (Santa Cruz, Dallas, TX, USA) supplemented with protease and phosphatase inhibitors (Sigma-Aldrich). Protein concentration was determined by BCA Protein Assay Kit (ThermoFisher Scientific, Rockford, IL, USA) and 25 μg of protein from each samples were loaded on SDS polyacrylamide gel. After SDS-gel electrophoresis, proteins were transferred to PVDF membrane (Bio-Rad, Hercules, CA, USA). As blocking reagent 5% BSA in PBS was used. Primary and secondary antibodies were applied according to manufacturer’s instructions. Membranes were rinsed five times with PBS with 0.05% Tween 20 after primary and secondary antibodies incubations. As primary antibodies; anti-PLK4 (ab137398), anti-α-tubulin (ab15246), anti-Chk1 (ab47574), anti-Chk1-pS345 (ab58567), anti-Chk2 (ab8108), anti-Chk2-pT68 (ab38461), anti-γ-H2AX (phospho S139) (ab26350) (Abcam) and anti-p53 (#9282) (Cell Signaling, Beverly, MA, USA) were used along with following horseradish peroxidase conjugated goat anti-mouse IgG (Bio-Rad, 170-5047) and goat anti-rabbit IgG (Bio-Rad, 170-5045) secondary antibodies (Bio-Rad, Hercules, CA, USA). After the membrane was treated with ECL Plus Western blotting detection kit (Bio-Rad), proteins were visualized and analyzed by ChemiDoc MP Imaging System (Bio-Rad).
Cell cycle and doubling time analysis
PLK4-Seckel and control fibroblasts were seeded into T25 flasks with a density of 5 × 104 cells, and were incubated at 37 °C in humidified 5% CO2 incubator while replenishing the growth medium every 24 h. For cell cycle analysis, after 48 h cells were collected and PI staining was done with Cycletest Plus DNA Reagent Kit (Becton Dickinson, Franklin Lakes, NJ, USA). Cells were analyzed with Accuri C6 flow cytometry (Becton Dickinson, Ann Arbor, MI, USA). For each sample 3 × 104 cells were counted and obtained flow cytometric data were analyzed with software ModFit v4.1. to determine the distribution of cells in G1/G0, S and G2/M phases. Mitotic cell percentage in G2/M phase fraction was determined by using anti-phospho Histone H3-Ser28 (pHH3-Ser28) labeling (for details see Supplementary Method S1). In order to analyze the effect of DNA-damaging agent on cell cycle progression PLK4-Seckel cells were treated with MMC (for details see the Supplementary Method S2). The experiment was repeated six times, averages and standard deviation (SD) were calculated accordingly. To determine the doubling times, PLK4-Seckel and control fibroblast cells were collected in every 24 h for 6 days and counted by flow cytometer. Calculation of doubling times were carried out by using a software freely available (Roth V. 2006 http://www.doubling-time.com/compute.php). Doubling time experiments were repeated three times and averages and SD were calculated accordingly.
Tunel assay
PLK4-Seckel and healthy control fibroblast cells were seeded into T25 flasks and incubated overnight. The cells were treated with hydrogen peroxide (H2O2) at 500 μM final concentration in DMEM for 6 h. Apoptotic cells were analyzed by using APO-BrdU TUNEL Assay Kit (Life Technologies). Tunel positive cells were analyzed by flow cytometer.
Results
Clinical description
We evaluated a seven-generation Turkish family in which three cases (VII:1, VII:2 and VIII:2) were born with Seckel syndrome (Figure 1). According to the information obtained from family members during the interview the family pedigree was constructed. Case VII:1 died within a few months after birth and could not be included in the study. Patient VIII:2 was admitted to the medical genetic department because of microcephaly and growth retardation when she was 45 days old (Figures 1a and b). She was born to consanguineous parents after an uneventful pregnancy at 37 weeks with normal spontaneous vaginal delivery. Her phenotypic features and radiological findings are shown in Table 1 and Figures 1c–e. Her developmental milestones were significantly delayed. She was able to sit without support at 16 months, crawled at 17 months and started walking independently at 31 months. She could say only ‘dad’ and ‘mom’ since 1 year of age but could understand and obey simple commands. Ocular examination showed normal eye size and normal lens, and fundus findings. Audiological, routine laboratory examinations, abdominal ultrasonography and echocardiography were seen to be normal. Karyotype showed 46, XX with 13.3% aneuploidy (data not shown).
Table 1. Phenotypic features of the Seckel patients with PLK4 variations.
| Variations | Patients | Sex | Age | OFC (cm) (SD) | Height (cm) (SD) | Weight (Kg) (SD) | Findings |
|---|---|---|---|---|---|---|---|
| p.Asp11Profs*14 (Present study) | Patient VIII:2 | F | Birth 9 m | 26 (−9.7) 30.8 (−13.6) | 40 (−6.1) 55 (−5.3) | 1.5 (−3.7) 3.4 (−8.3) | Severe intellectual disability, no speech (uses a few words), walked at age 2.7 (31 month) Sloping forehead, prominent metopic suture, upslanting palp. fissures, blepharophimosis, epicantus, large ears, prominent nose, thin upper lip, high arched palate, micrognathia, fifth finger clinodactyly. Skeletal findings: Delayed bone age, radius hypoplasia, epiphysiolysis in distal femur, slight cubbing and ill-defined epiphysis in all the tubular bones. Brain MRI: Microcephaly, simplified gyri |
| Patient VII:2* | M | − | − | − | Severe intellectual disability. Sloping forehead, upslanting palp. fissures, blepharophimosis, epicantus, prominent nose micrognathia | ||
| p.Arg936fs*1 (ref. 8) | F1-P1 | M | Birth 12 y | − 43 (−7.44) | − 115 (−4.64) | − | Profound intellectual disability, no speech, can’t sit unaided, microcornea, cataract (R), contractures, epilepsy |
| F1-P2 | F | Birth 5 y | − 35 (−13.98) | − 78 (−6.9) | − | Profound intellectual disability, can’t sit unaided, no speech. microphthalmia, microcornea, corneal opacity with cataract (R). Skeletal findings: Lumbar scoliosis. Brain MRI: Microcephaly with simplified gyri, large interhemispheric arachnoid cyst, hypoplasia of pons and cerebellum, partial agenesis of corpus callosum (short), enlarged 4th ventricle | |
| F1-P3 | F | Birth 10.5 y | − 33 (−13.46) | − 88 (−8.26) | − | Very severe intellectual disability, no speech (uses a few words), can’t sit unaided, few words. Polydactyly partial duplication right great toe, talipes equinovarus, 2/3 toe syndactyly (R+L), contractures, epilepsy | |
| F1-P4 | F | Birth 1.5 y | − 30 (−15.14) | − 58 (−8.07) | − | Very severe intellectual disability, can’t sit unaided. Microphthalmia, microcornea, vitreous inclusions, contractures. Brain MRI: Microcephaly with markedly simplified gyri, hypoplasia of pons and cerebellum, partial agenesis of corpus callosum (short), venae galeni cyst, enlarged 4th ventricle | |
| F1-P5 | M | Birth 6 m | − 29 (−14.3) | − | − | Very severe intellectual disability, speech with a few words, walked with support at age of 5.5y., died at age 5.5–congenital heart disease (septal defect) | |
| p.Phe433fs*6 (ref. 8) | F2-P6 | F | Birth 20 y | 29 (−8.3) 43 | 46 (−5.5) 130 | 0.18 (−3.9) — | Severe intellectual disability, no speech. Early onset, severe vitreoretinal dystrophy, moderate, mixed deafness, café-au-lait and hypopigmented patches, camptodactyly. Skeletal findings: Severe scoliosis hip dysplasia. Brain MRI: Microcephaly with simplified gyri, short corpus callosum, periventricular heterotopia |
| F3-P7 | M | Birth 5 y | 21 (−4.5) 33.5 (−12.8) | 31 88 (−4.8) | 0.44 (−3.7) — | Severe intellectual disability, can sit, but can’t walk, no speech. IUGR, oligohyraminos, pectus excavatum, brachdactyly, 5th finger clinodactyly and prominent nose, hyperactive, restricted knee extension, cryptorchidism, no ophthalmological assessment. Skeletal findings: Delayed bone age. Brain MRI: Microcephaly with simplified gyri, hypoplasia of pons and cerebellum, interhemispheric arachnoid cyst, agenesis of corpus callosum | |
| p.Phe433Leufs*6 (ref. 9) | IV-4 | F | Birth 40 m | 26 (−5.9) 33 (−10.1) | 40 (−4.2) 68 (−7.2) | − 4.5 (−6.9) | Sloping forehead, prominent metopic suture, prominent eyes, strabismus, optic nerve hypoplasia micrognathia. Skeletal findings: Mild shorting of long bones, no frank dysplastic changes. Brain MRI: Microcephaly, arachnoid cyst |
| IV-5 | M | Birth 19 m | − 31.5 (−11.8) | − 58 (−6.4) | − 3.7−(6.5) | Sloping forehead, prominent metopic suture, prominent eyes, strabismus optic nerve hypoplasia, prominent nose, micrognathia. Skeletal findings: Mild shorting of long bones, no frank dysplastic changes, Brain MRI: Microcephaly arachnoid cyst | |
| IV-2 | F | Birth 18 m | 27 (−5.2) 30.5 (−12.5) | 43 (−2.8) 56 (−7.5) | 1.9 (−2.9) 4 (−7) | Sloping forehead, prominent metopic suture, prominent eyes, strabismus optic nerve hypoplasia, prominent nose, micrognathia |
Homozygosity mapping and mutation analyses
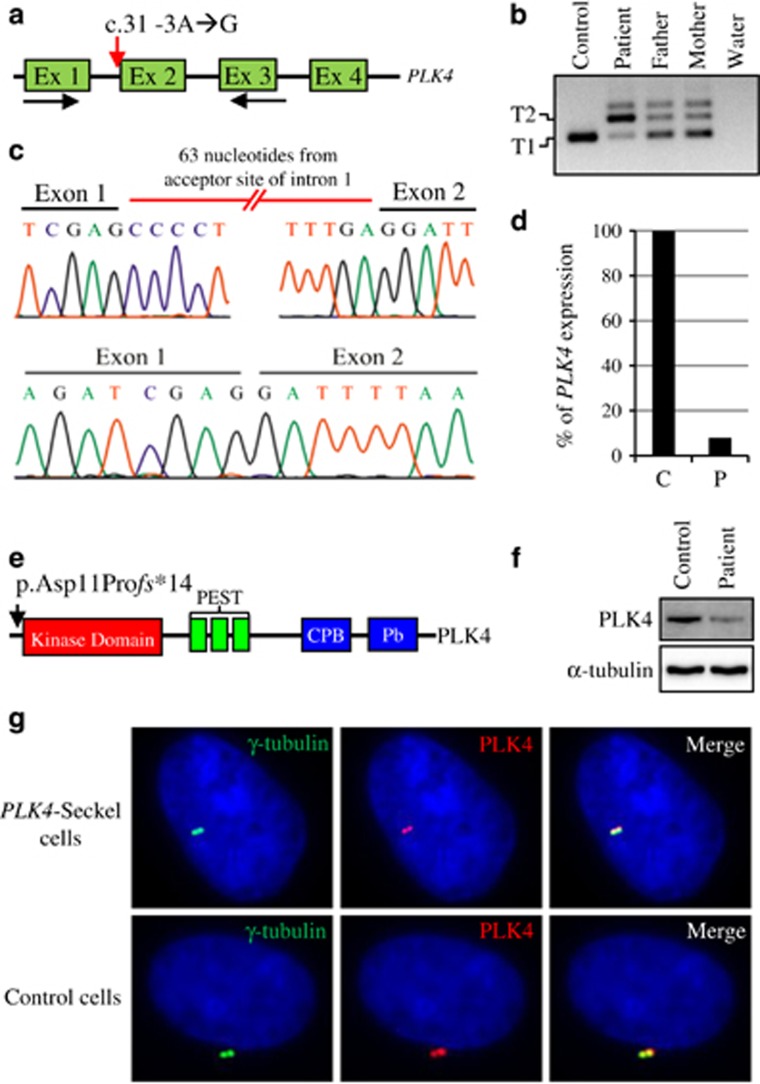
Two affected individuals (VII:2 and VIII:2) and the parents of patient VIII:2 (VI:3 and VII:4) were genotyped using the Illumina Cyto-SNP12 300 K BeadChip platform. Haplotype analyses revealed only one homozygous segment that co-segregates with the disease on chromosome 4q28.1-q28.3 (Figure 1f). Subsequently, microsatellite marker genotyping confirmed the co-segregation of critical region with disease (Figure 1g). PLK4 was selected as the most promising candidate gene from the critical interval because it was previously shown to be master regulator of centriole biogenesis. Moreover, PLK4 was reported to interact with CEP152 which was previously associated with Seckel syndrome.19 Sequence analyses of PLK4 showed a novel homozygous c.31-3 A>G transition on conserved splice acceptor site of first intron of the gene (Figure 1h and Figure 2a). The variation c.31-3 A>G was found to be fully segregated with the disease in the family (Figure 1i) and was not seen in 350 ancestrally matched control and 1000 Genomes Project database. RT-PCR analyses showed that c.31-3 A>G substitution disrupts the splicing of the first intron and leads to transfer of 63 nucleotides from acceptor site of intron 1 to mature mRNA (r.[=, 30_31ins31-63_31-4;31-3_31-1delinsgag]) and result in frameshift and subsequent premature translation termination p.[=, Asp11Profs*14] (Figures 2b, c and e). This novel PLK4 variation was submitted to the Leiden Open Variation Database (individual ID: 00081828). Quantitation of both PLK4 transcripts in patient’s cDNA with Real-time PCR showed that only 8% of the transcripts are wild type and the rest was mutant (Figure 2d). Consistent with transcript level, western blot analysis showed remarkable decrease of PLK4 in PLK4-Seckel cells (Figure 2f). When subcellular localization of PLK4 was analyzed in PLK4-Seckel fibroblasts by immunofluorescence, obvious difference was not detected in terms of PLK4 levels and cellular localization compared to control cells (Figure 2g).
Figure 2.
Expression analysis of PLK4. (a) Amplification of PLK4 from cDNA by using exon 1 and 3 specific primers designated with black arrows. Red arrow indicates the position of splicing acceptor-site variation c.31-3 A>G in partially shown PLK4 gene. (b) Two amplicons (T1 and T2) with different length were obtained from cDNA of affected and carrier individuals. Only a single amplicon (T1) was obtained from cDNA of a healthy control. (c) Sequencing chromatograms of the amplicon T2 showing mutant transcript (upper chromatogram) and T1 showing wild-type transcript (lower chromatogram). (d) Quantitation of wild-type transcript in PLK4-Seckel patient (P; patient, C; control). (e) Schematic representation of the known functional domains of human PLK4 on which black arrow indicates the position of p.Asp11Profs*14 variation. (f) Protein expression analysis of PLK4 in PLK4-Seckel patient’s fibroblasts. (g) Subcellular localization of PLK4 in PLK4-Seckel and control fibroblast cells. Nuclei were counterstained with DAPI (blue).
Disrupted centriole biogenesis and mitotic morphology
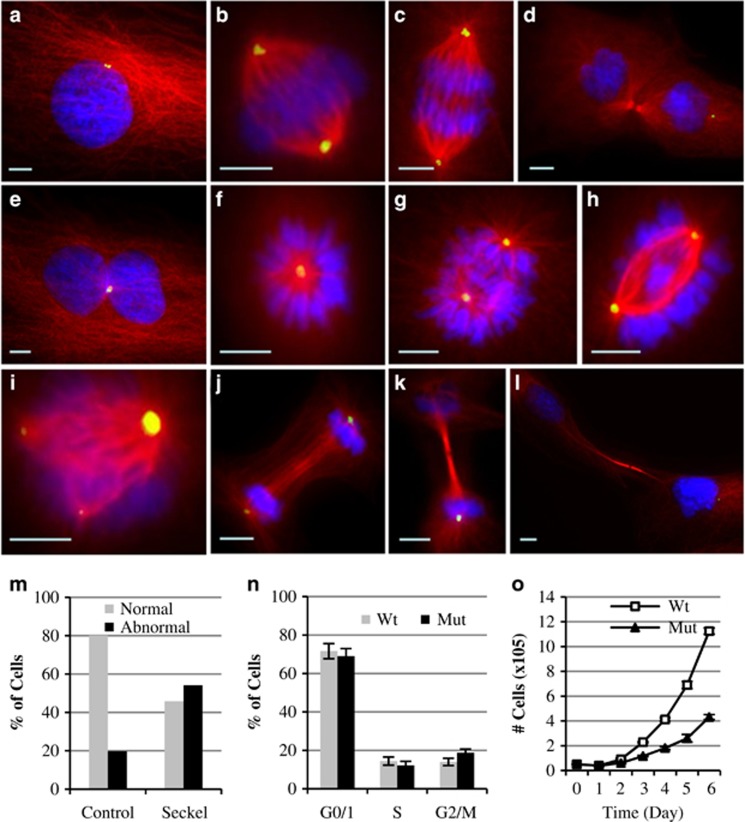
The effect of the described PLK4 variant on centriole biogenesis was evaluated in prophase and prometaphase of asynchronously growing PLK4-Seckel and control fibroblasts by immunofluorescence staining of centrioles using centriole specific proteins γ-tubulin and centrin-1. According to immunofluorescence results while the number of cells with four centrioles was found to be decreased, the number of cells with two or three centriole significantly increased in PLK4-Seckel cells compared to control fibroblasts (Supplementary Figure 1a, b).
In order to determine the functional effect of PLK4 deficiency on mitotic morphology, asynchronously growing PLK4-Seckel fibroblasts were analyzed. Binuclear cells with one centrosome were occasionally observed in interphase of PLK4-Seckel cells (Figure 3e). Frequent abnormalities in metaphase cells included monopolar spindle and improperly aligned chromosomes with irregular central spindle, whereas triple spindles were observed rarely (Figures 3f–i). Cells in telophase showed delayed cleavage furrow with irregular spindle alignment (Figure 3j) and extended telophase bridge (Figures 3k and l). In total, 54% of PLK4-Seckel cells were found to have significant cell division abnormalities compared to normal cells (Figure 3m).
Figure 3.
Mitotic morphology, cell cycle and doubling time analyses of PLK4-Seckel fibroblast cells. Immunostaining of PLK4-Seckel cells with α-tubulin (red) and pericentrin (green) antibodies. Nuclei were counterstained with DAPI (blue). (a–d) Morphologies of interphase, metaphase, anaphase and telophase of control fibroblasts cells, respectively. PLK4-Seckel cells; (e) with two nuclei and one centrosome in interphase, (f) monopolar spindle in metaphase, (g–h) uneven aligned bipolar spindle with improperly aligned chromosomes in metaphases, (i) tripolar mitotic spindle with differently sized centrosomes, (j) delayed cleavage furrow with irregular spindle alignment in telophase, (k–l) extended telophase bridge with midbody ring in telophase. Scale bars, 5 μm. (m) Percentage of cells showing abnormal mitotic morphology in PLK4-Seckel fibroblasts. (n) Increased G2/M phase in PLK4-Seckel cells, P=0.006 (T-test), error bars represent SD. (o) Increased doubling time of PLK4-Seckel fibroblasts compared to control.
Impaired cell cycle and doubling time of PLK4-Seckel fibroblast
Cell cycle analyses of asynchronously growing PLK4-Seckel cells by flow cytometry showed accumulation in G2/M phases (Figure 3n,Supplementary Figure S3). Further evaluation of G2/M fraction by using mitotic marker pHH3 (Ser28) revealed accumulation of PLK4-Seckel cells particularly in G2 phase (Supplementary Figure S2). On the other hand, induction of the PLK4-Seckel fibroblasts with 10 ng/ml final concentration of MMC slightly increased the accumulation of cells in G2/M phases. However, when MMC concentration increased 10-fold PLK4-Seckel cells showed reduced accumulation in G2/M compared to control (Supplementary Figure S3). The doubling time which was 25 h in normal cells, was found to be about 35 h in PLK4-Seckel fibroblasts (Figure 3o).
DNA damage response and apoptosis in PLK4-Seckel fibroblasts
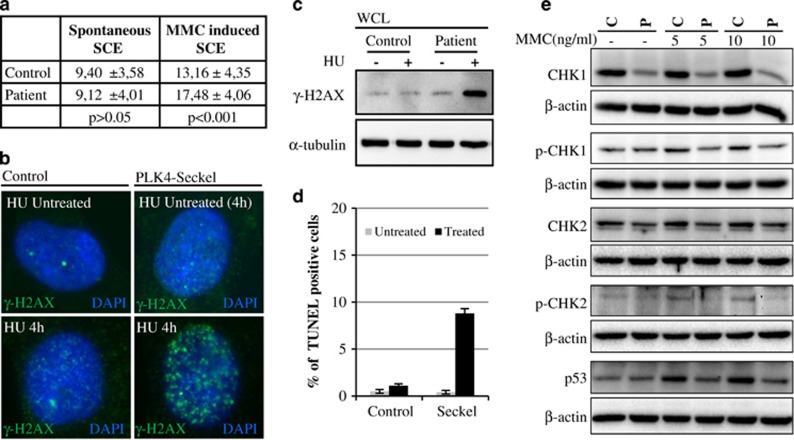
Sister chromatid exchange frequencies were determined in MMC treated PLK4-Seckel lymphocytes and a significant increase of SCE was found in PLK4-Seckel lymphocytes compared to control lymphocytes cells (Figure 4a, Supplementary Figure S4). The response of PLK4-Seckel cells to DNA damage was also analyzed and a remarkable accumulation of phosphorylated H2AX (γ-H2AX) was found after HU treatment compared to control (Figures 4b and c). Moreover, phosphorylated forms of Chk1 and Chk2 along with steady state levels were found to be remarkably, especially Chk1, decreased in PLK4-Seckel cells compared to wild type cells and treatment of cells with MMC did not make an obvious difference (Figure 4e). Phosphorylated forms of Chk1 and Chk2 in control cells increased in response to MMC, whereas in PLK4-Seckel cells they did not change (Figure 4e). As p53 is a well-known target of activated Chk1 and Chk2, MMC-induced p53 stability was analyzed and in contrast to control cells, it was not accumulated (Figure 4e). In addition, an increased rate of apoptosis was seen in H2O2 treated PLK4-Seckel fibroblasts (Figure 4d).
Figure 4.
DNA-damage response and increased sensitivity to oxidative stress in PLK4-Seckel fibroblasts. (a) Mitomycin-C-induced sister chromatid exchanges in PLK4-Seckel lymphocyte. (b) γ-H2AX foci in PLK4-Seckel primary fibroblasts after hydroxyurea treatment. (c) Increased γ-H2AX levels in PLK4-Seckel primary fibroblasts after hydroxyurea treatment. WCL, whole-cell lysate. (d) Enhanced apoptotic cell death in H2O2-induced PLK4-Seckel fibroblasts. Error bars represent SD. (e) Western blot analyses of basal and activated Chk1 and Chk2 along with p53.
Discussion
In this study, genome-wide homozygosity mapping and subsequent mutation analyses revealed a homozygous hypomorphic variant (c.31-3 A>G) in the first intron of the PLK4 in a Turkish family with Seckel syndrome (Figures 1 and 2). This variant disrupts the splice acceptor site of the first intron in about 92% of transcript and leads to a frame-shift and subsequent premature translation termination p.[=,Asp11Profs*14]). The human PLK4 encodes five different splicing variants and all isoforms have an N-terminal kinase domain followed by three PEST domain and a cryptic polo box domain preceding the C-terminal polo-box domain.20 All transcripts include intron one, the splicing of which is disrupted by the present variant. Loss of all functional domains of PLK4 as a result of p.Asp11Profs*14 in protein supports the pathologic nature of the described variant (Figure 2e). Co-segregation of the variant with disease in the family and its absence in the dbSNP databases as well as in Turkish controls provides further evidence that supports its disease causing nature.
Plk4 (Sak), the mouse ortholog of human PLK4, was originally identified in mouse and its expression was shown to be required for cell proliferation.21 PLK4 localizes to the centrioles throughout the cell cycle and was shown to be necessary for centriole duplication and progression of mitosis.22, 23 Homozygous Plk4 knock-out mice die at E7.5 day of development with remarkably increased mitotic and apoptotic cells, whereas heterozygous mutant mice were reported to be phenotypically normal.24 Recently, two PLK4 variants were associated with ten Seckel patients in four families.8, 9 In these patients some ophthalmological anomalies (microcornea, cataract, microphthalmia, retinopathy and optic nerve hypoplasia) that have not been associated with Seckel syndrome previously were reported in more than one patient expanding the phenotype associated with Seckel syndrome (Table 1). The patients we presented here have a characteristic Seckel syndrome phenotype. Ophthalmologic examination of patient VIII:2 was performed when she was 3 years old and her eye size, cornea and optic nerve and retina were found normal (Table 1). Different ocular findings between previously described PLK4-Seckel patients and the patient we presented here can be explained with the type of the variant and the reflected amount of wild-type PLK4 product because of hypomorphic nature of variant.
In animal cells, centrosomes have an important function in organizing the cytoskeleton and the mitotic spindles.25 In dividing cells, each centriole duplicates once every cell cycle to conserve centriole number. Successful centriole duplication is indispensable for animal cell division and its failure promotes abnormal spindle formation and subsequent cell division errors that lead to chromosome missegregation and genomic instability.26 As depletion of PLK4 was shown to cause inhibition of centriole biogenesis the most remarkable characteristic feature of PLK4-depleted cell was described as monopolar spindles with single centriole at the center.23, 27, 28 Therefore, to determine whether the variant that we presented here disrupts centriole biogenesis and mitotic morphology, we analyzed PLK4-Seckel fibroblasts obtained from the patient. In agreement with a previous report, a significant decrease in the number of centrioles was seen in prophase cells of PLK4-Seckel fibroblasts reflecting disrupted centriole biogenesis (Supplementary Figure 1a, b).8 Moreover, improperly aligned chromosomes, uneven spindle assembly and subsequent disrupted mitotic morphology in about 50% of the cells were detected as the characteristic features of the PLK4-Seckel fibroblasts (Figures 3e–m). In accordance with impaired spindle morphology, increased aneuploidy was seen in peripheral blood cells of a PLK4-Seckel patient suggesting impaired chromosome segregation (data not shown). For proper cell cycle progression spatial and temporal organization of mitotic spindles have an indispensable role.29 As expected, PLK4-Seckel fibroblasts displayed an increase in the number of cells in G2/M (Figure 3n). Disrupted centriole biogenesis causes mitotic delay followed by increase in apoptosis and cell doubling time.30 On the other hand, heterozygous Plk4 murine embryonic fibroblasts were also shown to exhibit growth delay compared to wild type counterparts.31 These reports raised the question whether cells obtained from patients proliferate slower than control cells. As expected, doubling time was found to be extended to 35 h in PLK4-Seckel fibroblast compared to 24 h seen in control cells (Figure 3o). Prolonged cell cycle in mammals can cause decrease in progenitor cell proliferation rate.32, 33 Therefore, extended doubling time can be considered as one of the critical mechanisms leading to global developmental failure that causes microcephaly and dwarfism in PLK4-Seckel patients.
In correlation with extended doubling time, accumulation of PLK4-Seckel cells in G2/M phase (Figure 3n, Supplementary Figure S3), particularly in G2 phase (Supplementary Figure S2) showed the G2/M checkpoint activation and raised a question about the possible role of PLK4 in DNA damage response. Despite the well-known role of PLK4 in centriole biogenesis, its contribution to DNA damage response is inconclusive. However, previously described Seckel syndrome genes including ATR, CEP152, ATRIP, CtIP (RBBP8) and DNA2 were shown to contribute to DNA damage response and deficiency of these genes causes developmental failure.7, 10, 11, 14, 34 In order to determine whether PLK4 has a role in DNA repair mechanism, we investigate the MMC-induced SCE frequency and observed a significantly increased SCE showing chromosome instability in PLK4-deficient lymphocytes (Figure 4a and Supplementary Figure S4). Furthermore, considerably strong hydroxyurea-induced H2AX phosphorylation in PLK4-Seckel cells (Figures 4b and c) showed accumulated DNA breaks and subsequent increased activation of DNA damage response.35 In response to DNA damage, downstream signal transducers of DNA damage response such as Chk1, Chk2 and p53 are activated and trigger the cell cycle arrest to provide time for DNA repair.36 However, extended genotoxic stress in response to many types of DNA damage leads to proteasomal degradation of Chk1, although there are some conflicting results regarding to Chk2.37, 38 In consistent with this, inhibition of Chk1 allows cell cycle progression into premature and abnormal mitosis with high levels of DNA damage followed by mitotic catastrophe.39, 40, 41 Reduction of both basal and activated Chk1 and Chk2 in PLK4-Seckel cells (Figure 4e), therefore, emphasize the involvement of PLK4 directly in DNA damage response or regulation of Chk1 and Chk2 basal turnover rates through modulating proteasomal degradation machinery. In accordance with decreased Chk1 and Chk2 levels, decreased accumulation of p53 in response to MMC treatment (Figure 4e) also provide further evidence showing extended DNA damage response in PLK4-Seckel cells. On the other hand, reduced G2/M fraction and increased G1 in PLK4-Seckel cells compared to control after high concentration of MMC (Supplementary Figure S3) suggests mitotic catastrophe which might lead to slippage of mitosis and relative increase of G1 in the subsequent cell cycle following reduction of Chk1 and Chk2 levels.42 In agreement with our result, Chk2 and its downstream target elements Cdc25C and p53 were shown to be substrates of Plk4.20, 43, 44 Moreover, in mammals, in addition to prolonged cell cycle, genomic instability leads to apoptosis.45 However, apoptosis was not reported by Martin et al.8 in PLK4 deficient cells. In line with this report, we have not detected increased apoptosis in untreated PLK4-Seckel fibroblasts as well. Interestingly, in our study, hydrogen peroxide treatment of PLK4-seckel cells increased cell death. Together with the results of enhanced SCE and activated H2AX, this obvious increase in apoptosis upon oxidative stress reflects increased sensitivity to genotoxic stress due to unrepaired DNA lesions in PLK4-Seckel cells (Figure 4d). Taken together, these findings indicate that deficiency of PLK4 causes impaired DNA damage response.
In summary, we describe a novel variant in the PLK4 gene that has recently been associated with Seckel syndrome. Our results show that PLK4 deficiency causes impaired centriole biogenesis, genomic instability and subsequent increased apoptosis correlated with expanded cell doubling time. Together these might lead to insufficient cell proliferation and explain the subsequent global growth failure.
Acknowledgments
We thank all the family members that participated in this study and Dr Ahmet Alver for his kind contribution for statistical analyses. We thank Prof. Dr Han G. Brunner for critical reading and comments. This study was supported by the Karadeniz Technical University Research Fund (grant KTÜ-BAP-10121 and 1061 to E.K.) and Teaching Staff Training Program of Turkish Council of Higher Education in which Gülden Yorgancıoğlu-Budak’s master thesis program is enrolled.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Conlon I, Raff M: Size control in animal development. Cell 1999; 96: 235–244. [DOI] [PubMed] [Google Scholar]
- Klingseisen A, Jackson AP: Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev 2011; 25: 2011–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski F, Goecke T: Studies of microcephalic primordial dwarfism I: approach to a delineation of the Seckel syndrome. Am J Med. Genet. 1982; 12: 7–21. [DOI] [PubMed] [Google Scholar]
- Majewski F, Ranke M, Schinzel A: Studies of microcephalic primordial dwarfism II: the osteodysplastic type II of primordial dwarfism. Am J Med Genet 1982; 12: 23–35. [DOI] [PubMed] [Google Scholar]
- Majewski F, Stoeckenius M, Kemperdick H: Studies of microcephalic primordial dwarfism III: an intrauterine dwarf with platyspondyly and anomalies of pelvis and clavicles—osteodysplastic primordial dwarfism type III. Am J Med Genet 1982; 12: 37–42. [DOI] [PubMed] [Google Scholar]
- Gorlin RJ, Cervenka J, Moller K, Horrobin M, Witkop CJ Jr.: Malformation syndromes. A selected miscellany. Birth Defects Orig Artic Ser 1975; 11: 39–50. [PubMed] [Google Scholar]
- Kalay E, Yigit G, Aslan Y et al: CEP152 is a genome maintenance protein disrupted in Seckel syndrome. Nat. Genet. 2011; 43: 23–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CA, Ahmad I, Klingseisen A et al: Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat. Genet. 2014; 46: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Al Tala S, Almoisheer A, Alkuraya FS: Mutation in PLK4, encoding a master regulator of centriole formation, defines a novel locus for primordial dwarfism. J Med Genet 2014; 51: 814–816. [DOI] [PubMed] [Google Scholar]
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA: A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 2003; 33: 497–501. [DOI] [PubMed] [Google Scholar]
- Qvist P, Huertas P, Jimeno S et al: CtIP mutations cause Seckel and Jawad syndromes. PLoS Genet 2011; 7: e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Dosari MS, Shaheen R, Colak D, Alkuraya FS: Novel CENPJ mutation causes Seckel syndrome. J Med Genet 2010; 47: 411–414. [DOI] [PubMed] [Google Scholar]
- Sir JH, Barr AR, Nicholas AK et al: A primary microcephaly protein complex forms a ring around parental centrioles. Nat Genet 2011; 43: 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen R, Faqeih E, Ansari S et al: Genomic analysis of primordial dwarfism reveals novel disease genes. Genome Res 2014; 24: 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Walker S, Stiff T et al: Identification of the first ATRIP-deficient patient and novel mutations in ATR define a clinical spectrum for ATR-ATRIP Seckel Syndrome. PLoS Genet 2012; 8: e1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber A, Lafranchi SH, Maliga Z et al: Novel microcephalic primordial dwarfism disorder associated with variants in the centrosomal protein ninein. J Clin Endocrinol Metab 2012; 97: E2140–E2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit G, Brown KE, Kayserili H et al: Mutations in CDK5RAP2 cause Seckel syndrome. Mol Genet Genomic Med 2015; 3: 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderton GK, Joenje H, Varon R, Borglum AD, Jeggo PA, O'Driscoll M: Seckel syndrome exhibits cellular features demonstrating defects in the ATR-signalling pathway. Hum Mol Genet 2004; 13: 3127–3138. [DOI] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R et al: Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol 2010; 191: 731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow CJ, Ko MA, Siddiqui NU, Hudson JW, Dennis JW: Sak/Plk4 and mitotic fidelity. Oncogene 2005; 24: 306–312. [DOI] [PubMed] [Google Scholar]
- Fode C, Motro B, Yousefi S, Heffernan M, Dennis JW: Sak, a murine protein-serine/threonine kinase that is related to the Drosophila polo kinase and involved in cell proliferation. Proc Natl Acad Sci USA 1994; 91: 6388–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung GC, Hudson JW, Kozarova A, Davidson A, Dennis JW, Sicheri F: The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat Struct Biol 2002; 9: 719–724. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA: The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 2005; 7: 1140–1146. [DOI] [PubMed] [Google Scholar]
- Hudson JW, Kozarova A, Cheung P et al: Late mitotic failure in mice lacking Sak, a polo-like kinase. Curr Biol 2001; 11: 441–446. [DOI] [PubMed] [Google Scholar]
- Loncarek J, Khodjakov A: Ab ovo or de novo? Mechanisms of centriole duplication. Mol Cell 2009; 27: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW: Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev 2009; 10: 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL: The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol 2009; 184: 225–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne JE, Tack F, Vloemans N et al: Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol Biol Cell 2010; 21: 547–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noatynska A, Gotta M, Meraldi P: Mitotic spindle (DIS)orientation and DISease: cause or consequence? J Cell Biol 2012; 199: 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir JH, Putz M, Daly O et al: Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J Cell Biol 2013; 203: 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko MA, Rosario CO, Hudson JW et al: Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet 2005; 37: 883–888. [DOI] [PubMed] [Google Scholar]
- Pilaz LJ, McMahon JJ, Miller EE et al: Prolonged mitosis of neural progenitors alters cell fate in the developing brain. Neuron 2016; 89: 83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokrani-Benhelli H, Gaillard L, Biasutto P et al: Primary microcephaly, impaired DNA replication, and genomic instability caused by compound heterozygous ATR mutations. Hum Mutat 2013; 34: 374–384. [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J et al: Human CtIP promotes DNA end resection. Nature 2007; 450: 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinner A, Wu W, Staudt C, Iliakis G: Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res 2008; 36: 5678–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S: Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 2004; 73: 39–85. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Otterness DM, Chiang GG et al: Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell 2005; 19: 607–618. [DOI] [PubMed] [Google Scholar]
- Zannini L, Delia D, Buscemi G: CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol 2014; 6: 442–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Xue J, Sowin TJ, Rosenberg SH, Zhang H: A novel mechanism of checkpoint abrogation conferred by Chk1 downregulation. Oncogene 2005; 24: 1403–1411. [DOI] [PubMed] [Google Scholar]
- Poehlmann A, Habold C, Walluscheck D et al: Cutting edge: Chk1 directs senescence and mitotic catastrophe in recovery from G(2) checkpoint arrest. J Cell Mol Med 2011; 15: 1528–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Nagro CJ, Choi J, Xiao Y et al: Chk1 inhibition in p53-deficient cell lines drives rapid chromosome fragmentation followed by caspase-independent cell death. Cell Cycle 2014; 13: 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Gee MM: Targeting the mitotic catastrophe signaling pathway in cancer. Mediators Inflamm 2015; 2015: 146282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrinac S, Ganuelas ML, Bonni S, Nantais J, Hudson JW: Polo-like kinase 4 phosphorylates Chk2. Cell Cycle 2009; 8: 327–329. [DOI] [PubMed] [Google Scholar]
- Bonni S, Ganuelas ML, Petrinac S, Hudson JW: Human Plk4 phosphorylates Cdc25C. Cell Cycle 2008; 7: 545–547. [DOI] [PubMed] [Google Scholar]
- Zhou BB, Bartek J: Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer 2004; 4: 216–225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.