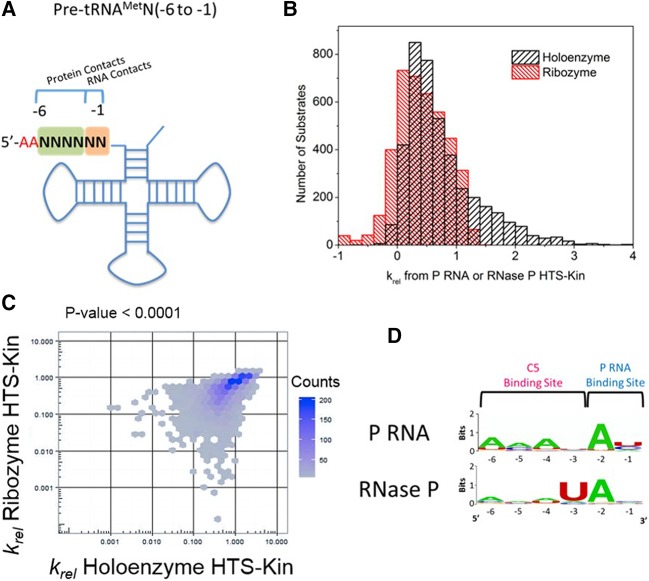
FIGURE 1.
Sequence determinants for holoenzyme and ribozyme reactions are unique and reveal changes in specificity. (A) Secondary structure of pre-tRNAMetN(−6 to −1) with regions of randomization analyzed in this study indicated by N's. As shown, these regions encompass nucleotides of the 5′ leader that are involved in P RNA and C5 protein contact. (B) The affinity distribution of relative rate constants from holoenzyme (black, published in Niland et al. 2016b) and ribozyme (red) with pre-tRNAMetN(−6 to −1) show that while some substrates are processed with rate constants significantly above the genomically encoded reference in holoenzyme reactions, the same substrate population is significantly slower in the ribozyme reaction with no protein subunit. (C) Comparison of the relative rate constants measured for each substrate variant in HTS-Kin reactions with RNase P holoenzyme (x-axis) and P RNA ribozyme (y-axis). (D) Sequence preference in the 5′ leader of pre-tRNAMetN(−6 to −1) examined by creating sequence probability logos of the fastest reacting substrates in holoenzyme or ribozyme reactions. The position in the 5′ leader is indicated on the bottom of the logo; nucleotide preference at each position is indicated by the identity and size of the letter of the nucleobase.