Abstract
CRISPR–Cas is a bacterial and archaeal adaptive immune system that uses short, invader-derived sequences termed spacers to target invasive nucleic acids. Upon recognition of previously encountered invaders, the system can stimulate secondary spacer acquisitions, a process known as primed adaptation. Previous studies of primed adaptation have been complicated by intrinsically high interference efficiency of most systems against bona fide targets. As such, most primed adaptation to date has been studied within the context of imperfect sequence complementarity between spacers and targets. Here, we take advantage of a native type I-C CRISPR–Cas system in Legionella pneumophila that displays robust primed adaptation even within the context of a perfectly matched target. Using next-generation sequencing to survey acquired spacers, we observe strand bias and positional preference that are consistent with a 3′–5′ translocation of the adaptation machinery. We show that spacer acquisition happens in a wide range of frequencies across the plasmid, including a remarkable hotspot that predominates irrespective of the priming strand. We systematically characterize protospacer sequence constraints in both adaptation and interference and reveal extensive flexibilities regarding the protospacer adjacent motif in both processes. Lastly, in a strain with a genetically truncated CRISPR array, we observe increased interference efficiency, which, when coupled with forced maintenance of a targeted plasmid, provides a useful experimental system to study spacer loss. Based on these observations, we propose that the Legionella pneumophila type I-C system represents a powerful model to study primed adaptation and the interplay between CRISPR interference and adaptation.
Keywords: CRISPR–Cas, type I-C, primed adaptation, spacer loss, Legionella
INTRODUCTION
Bacteria and archaea constantly interact with mobile genetic elements including bacteriophages, plasmids, transposons, and other conjugative elements (for review, see Burrus and Waldor 2004; Frost et al. 2005). With their genomes greatly shaped by these mobile elements, these microbes can benefit from acquisition of foreign DNA, but also suffer detrimental effects from “selfish” elements such as lytic bacteriophages. To combat deleterious horizontal gene transfer, bacteria and archaea harbor multiple resistance mechanisms (for a broad review, see Labrie et al. 2010), including “adaptive immunity” provided by CRISPR–Cas (clustered regularly interspaced short palindromic repeats and CRISPR-associated genes) systems (Barrangou et al. 2007; Brouns et al. 2008; Marraffini and Sontheimer 2008). Several different types of CRISPR–Cas systems have been identified that each use distinct protein compositions to function as adaptive immunity against invasive nucleic acids (Makarova et al. 2011, 2015). A CRISPR array consists of distinct short spacers separated by repeat sequences (Ishino et al. 1987; Mojica et al. 2000; Jansen et al. 2002) and is transcribed as a noncoding RNA that undergoes further processing by Cas proteins to form individual repeat-spacer units (crRNAs) (Brouns et al. 2008). These crRNAs are loaded as guide sequences into Cas–crRNA interference complexes that bind to targeted nucleic acids (termed protospacers) (Brouns et al. 2008; Jore et al. 2011; Wiedenheft et al. 2011). In type I systems (the focus of this study), these interference complexes mediate target cleavage by recruiting the Cas3 nuclease, a process known as interference (Sinkunas et al. 2011, 2013; Mulepati and Bailey 2013; Hochstrasser et al. 2014; Huo et al. 2014). In both type I and type II CRISPR–Cas systems, an appropriate protospacer adjacent motif (PAM) is required to distinguish between alien protospacers and self-spacer sequences (Deveau et al. 2008; Horvath et al. 2008; Mojica et al. 2009). One key feature of CRISPR–Cas immunity is its ability to adapt to new threats through the acquisition of new spacers derived during encounters with foreign DNA (Barrangou et al. 2007; Erdmann and Garrett 2012; Yosef et al. 2012; Diez-Villasenor et al. 2013; for review, see Jackson et al. 2017), such as from nonproductive bacteriophage infection (Hynes et al. 2014). Spacer acquisition can be either “naïve” (where the invader has not been previously cataloged in the array) (Yosef et al. 2012; Levy et al. 2015; Wei et al. 2015) or “primed” (where, upon recognition of invaders previously targeted by CRISPR–Cas, secondary spacers are acquired that can enhance protection) (Datsenko et al. 2012; Swarts et al. 2012; Fineran et al. 2014). Compared with naïve adaptation, primed adaptation is much more efficient (Datsenko et al. 2012; Swarts et al. 2012; Staals et al. 2016) and reliant on recruitment of the interference machinery to a preexisting target (Blosser et al. 2015; Redding et al. 2015; Vorontsova et al. 2015).
Our understanding of primed spacer acquisition is based upon the studies of type I-E (Datsenko et al. 2012; Swarts et al. 2012; Savitskaya et al. 2013; Fineran et al. 2014; Xue et al. 2015), type I-F (Richter et al. 2014; Westra et al. 2015; Staals et al. 2016), and type I-B systems (Li et al. 2014a, b; 2017) in the presence of targeted DNA such as plasmids and bacteriophages. During CRISPR adaptation, the conserved proteins Cas1 and Cas2 form a protein complex that plays a key role in prespacer capture and insertion into the CRISPR array (Richter et al. 2012; Nunez et al. 2014, 2015; Wang et al. 2015). Regarding the generation of prespacers from invasive DNA, characterizations of acquired spacers under priming conditions have revealed nonconserved patterns in different type I CRISPR–Cas systems. Primed spacer acquisition in the E. coli type I-E system showed a clear preference (>90%) from the untargeted strand and either an extensively long positional gradient on large bacteriophages or a lack of positional gradient on small-sized priming plasmids (Datsenko et al. 2012; Savitskaya et al. 2013; Fineran et al. 2014; Strotskaya et al. 2017). In the Haloarcula hispanica type I-B system, ∼70% of new spacers were derived from the untargeted strand and a moderate preference was seen for the priming-proximal region (Li et al. 2014b, 2017). In the Pectobacterium atrosepticum type I-F system, ∼65% of spacers were acquired from the targeted strand with a clear gradient centered at the priming site (Richter et al. 2014; Staals et al. 2016). A similar gradient in spacer acquisition efficiency was also observed in the Pseudomonas aeruginosa and E. coli type I-F systems (Vorontsova et al. 2015). A “sliding” model has been proposed to explain these patterns: A complex is recruited to the targeted sequence and subsequently slides away from the priming site in a 3′–5′ direction, preferentially on one strand, and stops at an appropriate PAM site where Cas1 and Cas2 assist in spacer extraction (Heler et al. 2014). The proposed translocation directionality of this “sliding complex” is consistent with the 3′–5′ helicase and endonuclease activity of Cas3 in vitro (Westra et al. 2012b; Mulepati and Bailey 2013; Sinkunas et al. 2013; Huo et al. 2014), and the concept that Cas3 is the basis of primed acquisition directionality is further supported by single-molecule imaging of Cas3 translocation (Redding et al. 2015). Besides the sliding model that describes the overall patterns of primed acquisition, another model regarding the molecular basis of spacer extraction suggests that double-stranded Cas3 degradation products are preferentially used as donors for Cas1–Cas2 (Swarts et al. 2012; Kunne et al. 2016; Severinov et al. 2016). These two models are not necessarily mutually exclusive: It is possible that, if Cas3 nuclease activity is regulated (by some as-yet-unclear signal), the processing of targeted DNA could contribute to spacer acquisition through either Cas3-directed sliding (the sliding model) or Cas3-mediated degradation followed by Cas1–Cas2 recycling for protospacer extraction (the alternative model).
Despite a growing body of work on primed adaptation, a number of factors limit direct comparison between most of the previous studies. Specifically, due to the high interference efficiency against bona fide targets, most studies have used mismatched priming sequences with either a noncanonical PAM or mutations in the seed sequence of a protospacer (Datsenko et al. 2012; Savitskaya et al. 2013; Fineran et al. 2014; Li et al. 2014b; Richter et al. 2014). Such target mismatches not only affect interference (Wiedenheft et al. 2011; Xue et al. 2015), but influence the efficiency of primed adaptation (Fineran et al. 2014; Li et al. 2014a; Kunne et al. 2016; Xue et al. 2016), which could potentially pose an impact on how spacers are acquired during priming (Redding et al. 2015; Vorontsova et al. 2015). To prime with a bona fide target, either inducible expression or anti-CRISPR regulated systems to control interference have been used (Vorontsova et al. 2015; Semenova et al. 2016; Staals et al. 2016). Notably, one of these studies also used a native system (without overexpression of cas genes or an anti-CRISPR) to show that, with both naïve and primed CRISPR adaptation, the first newly acquired spacer (interference-proficient) stimulates acquisition from bona fide targeting (Staals et al. 2016). The focus of the current study is on the type I-C CRISPR-Cas system in L. pneumophila (Rao et al. 2016), a relatively permissive system that allows for efficient priming by bona fide, perfect-match targets. Along with this inherent experimental strength, type I-C systems remain relatively understudied, despite representing the second most abundant type of CRISPR-Cas systems in prokaryotes (Makarova et al. 2011, 2015).
RESULTS
Priming of the permissive L. pneumophila type I-C CRISPR–Cas system induces robust spacer acquisition
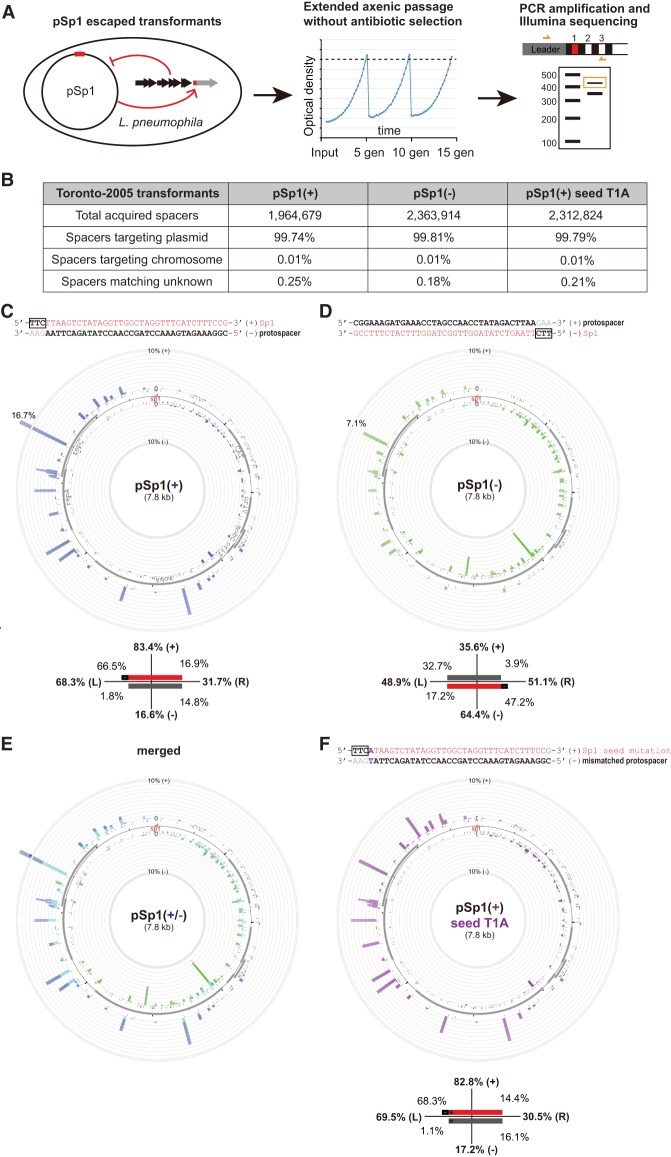
In our previous work, we experimentally showed that a perfectly targeted plasmid can temporarily coexist, without detectable mutations (in either plasmid or CRISPR-Cas locus), with the type I-C CRISPR-Cas system in L. pneumophila str. Toronto-2005 (Rao et al. 2016). We previously observed that these transformants display plasmid loss during nonselective axenic passage that corresponds with an enrichment for spacer acquisition events (Rao et al. 2016). Here we exploited this experimental adaptation system to study spacer acquisition in the type I-C system in depth (Fig. 1A). A targeted plasmid that includes the protospacer sequence for CRISPR spacer 1 (Sp1) and a canonical TTC PAM (Mojica et al. 2009; Leenay et al. 2016) was used to prime spacer acquisition (we refer to the protospacer as the targeted sequence complementary to the crRNA spacer and PAM as the 5′–3′ sequence complementary to the downstream from the protospacer [Westra et al. 2012a; Leenay et al. 2016]). We placed the spacer sequence on either the plus strand (pSp1[+]) or the minus strand (pSp1[−]) (with the corresponding priming protospacer on the opposite strand). These targeted plasmids showed a ∼1% relative transformation efficiency compared with untargeted control plasmids, and the resulting transformants were passaged without antibiotic selection for 15 generations to enrich for spacer acquisition events that we subsequently cataloged by PCR amplification, gel extraction, and deep sequencing (Fig. 1A). Around 2 million new spacers were extracted from Illumina raw reads in each priming experiment and mapped to potential sources, including the priming plasmid or the bacterial chromosome. The vast majority (>99.7%) of spacers were derived from the plasmid, with the remaining few from the chromosome or unknown sources (possibly due to chimeric sequences or sequencing errors) (Fig. 1B). Collectively, these numerous spacer sequences covered all available TTC canonical PAM sites on the plasmid (Supplemental Table S1), suggesting a sufficient sequencing depth to represent the CRISPR-adapted population.
FIGURE 1.
Primed spacer acquisition by L. pneumophila type I-C CRISPR-Cas occurs in a strand-biased manner. (A) Schematic workflow to characterize primed spacer acquisition. Bacterial transformants of targeted plasmids were passaged for 15 generations without antibiotic selection to enrich for spacer acquisition. CRISPR loci were PCR amplified and adapted arrays were further isolated through gel size selection. Amplicons were subjected to Illumina sequencing, and acquired spacers (complementary to the original protospacers) were extracted from raw reads and mapped to either the plasmid or the bacterial chromosome. (B) The vast majority of acquired spacers during priming were derived from the plasmid instead of the chromosome. (C,D) Circos plots of acquired spacers mapped to the pSp1 priming plasmid where the priming protospacer (complementary to the spacer 1 sequence from the type I-C system) is either on the minus strand (C) or on the plus strand (D). In the strand-specific mappings, bars protruding inside and outside of each plasmid circle represent spacers matching the minus and plus strand of the plasmid, respectively, and the height of bars indicates the number of spacers mapped to indicated positions. Note that a secondary scale was used for plasmid loci acquired at a frequency of over 10% of all spacers. The frequency of the major spacer acquisition hotspot is indicated. To numerically represent the overall spacer acquisition patterns, the plasmid is divided into four geographic fractions relative to the priming site (denoted by the colored rectangle): the 5′ half (left) and the 3′ half (right) on the (+) strand, and the 3′ half (left) and the 5′ half (right) on the (−) strand. In these schematics, the protospacer strand is represented by a dark gray bar, the spacer strand is the red bar, and the corresponding PAM is the adjacent black rectangle. (E) A merged view of the two mappings was created where overlapped coverages were shown in cyan. (F) Priming by an imperfect target with a seed mismatch showed similar overall patterns of spacer acquisition as priming using a bona fide target. Each Circos plot in the figure represents the average of two independent biological replicates.
Primed spacer acquisition occurs in a strand-biased fashion but is influenced by local hotspots
Through Sanger sequencing of 23 acquired spacers, we previously observed a moderate preference (74%) of spacers derived from the untargeted strand (Rao et al. 2016). With a much higher sequencing depth, we comprehensively reexamined the patterns of spacer acquisition from the plasmid. When the priming protospacer is on the minus strand of the plasmid (pSp1[+]), a majority (83%) of spacers are mapped to the plus strand, and an obvious enrichment of acquisitions is seen from the 5′ region proximal to the priming site on both strands (Fig. 1C). When the priming protospacer sequence is flipped to the plus strand (pSp1[−]), the preferred strand of acquisition is also switched, with 64% of new spacers derived from the minus strand, and as before, more spacers are mapped to the 5′ region of the priming site relative to the 3′ region (Fig. 1D). A clear correlation between the directionality of the priming sequence and the strand preference of acquired spacers is shown from the merged view of pSp1(+) and pSp1(−) mappings (Fig. 1E). These observations are consistent with a strand-specific 3′–5′ translocation of Cas3 starting from the priming site.
Besides the strand bias and positional gradient, we observed a wide range of acquisition frequencies across the plasmid. Among all 238 TTC PAM sites, 30 positions each accounted for >1% of all acquisitions in at least one priming experiment, and 62 were acquired at <0.05% frequencies in both priming settings (Supplemental Table S1). Strikingly, we identified one locus in the coding strand of repC that consistently ranked as one of the most frequently acquired spacers regardless of the primed strand (Fig. 1C,D).
Lastly, we examined if a mismatched protospacer primes the type I-C system differently from a perfect match. While a single T1A mutation in the seed sequence increased the plasmid transformation efficiency from ∼1% to ∼9% relative to the untargeted plasmid, the overall patterns (strand bias and positional gradient) of acquired spacers were largely unchanged (Fig. 1C,F). These data are consistent with models in which primed spacer acquisition using perfect (interference-driven) or imperfect matches involves shared molecular mechanisms (Semenova et al. 2016; Staals et al. 2016).
Sequence specificity contributes to the acquisition hotspot
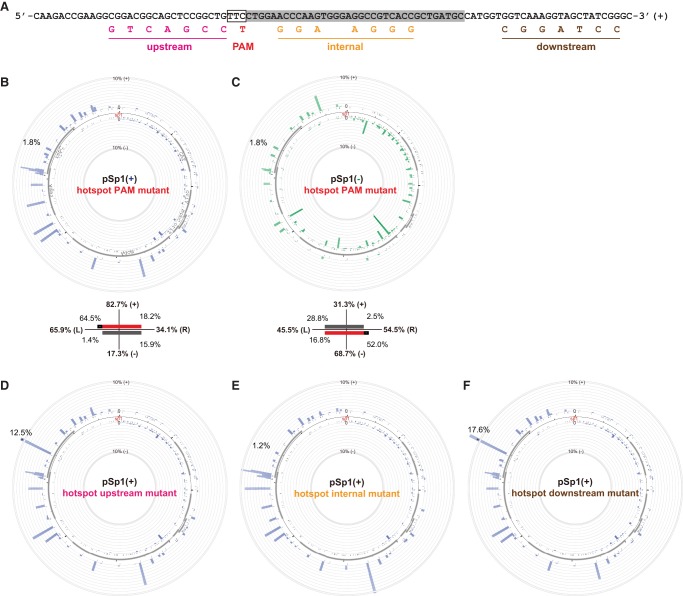
To identify the factors contributing to the high acquisition efficiency of the observed primed acquisition hotspot, we first examined this hotspot region for the presence of any outstanding feature: PAM density, GC content, origin of replication, or predicted small RNA transcription. In the absence of an obvious signal from any of these features, we hypothesized that some other sequence specificity of the hotspot region underlies the high acquisition efficiency. Thus, we generated a set of plasmids carrying mutations upstream, downstream, or within the hotspot while maintaining the repC open reading frame to avoid any side effects due to amino acid changes (Fig. 2A).
FIGURE 2.
The spacer acquisition hotspot is reliant on its internal sequence. (A) Mutations were introduced, with the repC open reading frame maintained, at the upstream, PAM, internal, or downstream sequences of the major spacer acquisition hotspot to examine factors contributing to the high acquisition frequency. (B,C) The mutation at the PAM dramatically reduced the acquisition frequency at the hotspot, while the overall patterns of spacer acquisition remained largely unaffected. Note that the mutation does not eliminate the available PAM, but shifted the PAM 1 nt away. (D–F) Mutations within, but not flanking, the hotspot also largely decreased the acquisition frequency at the hotspot. Each Circos plot in the figure represents the average of two independent biological replicates.
We first tested the PAM mutant in pSp1(+) where the TTC PAM of the hotspot is changed to TTT (where coincidentally another TTC motif is made with +1 nt shift). The acquisition efficiency of the mutant sequence decreased by ninefold (16.7%–1.8%) (Fig. 2B). We also observed a large reduction of acquisition efficiency at the hotspot by introducing the same mutation in pSp1(−) (Fig. 2C). Importantly, by comparing these hotspot PAM mutants with the wild-type plasmids, we did not observe a major difference in the overall strand bias due to the elimination of the hotspot, i.e., the imperfect mirroring of strand bias (∼80% plus in pSp1[+] and ∼60%–70% minus in pSp1[−]) is retained.
We next examined the other regions of the hotspot by introducing different sets of mutations in pSp1(+). The acquisition efficiency of the hotspot was dramatically eliminated by internal mutations, slightly reduced by changes upstream, and not reduced at all by the downstream perturbations (Fig. 2D–F). Elimination of the hotspot increases acquisition frequencies of other plasmid loci (Fig. 2B,E), suggesting that its loss modifies the availability of the adaptation machinery to other loci. The major impacts of the PAM mutation and the internal substitutions suggest that some sequence specificity within the hotspot contributes to the acquisition preference at this locus.
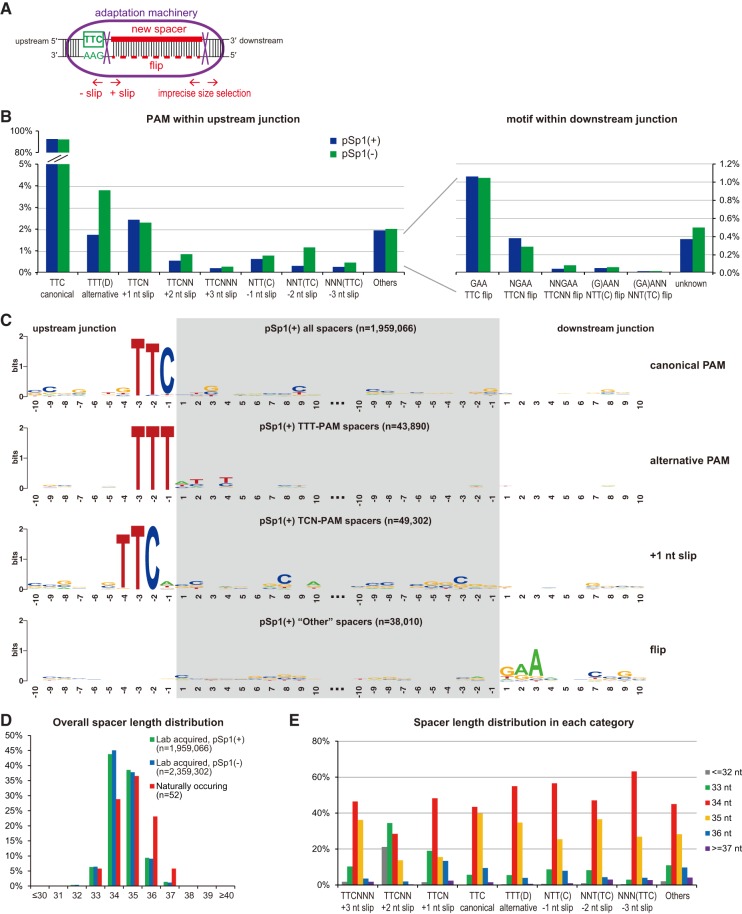
Analysis of acquired spacers reveals an alternative PAM and extensive acquisition inaccuracies
Of all acquired spacers from the plasmid, those having a TTC PAM account for 92.5% and 90.0% in the pSp1(+) and pSp1(−) priming experiment, respectively (Fig. 3B). We examined the trinucleotide sequence upstream of all acquired spacers from pSp1(+) for the abundance of other PAMs (Supplemental Fig. S1A). The ∼2 million acquired spacers from pSp1(+) are next to 2978 different PAM loci. While most (90%) of these PAM loci were acquired rarely (with <0.01% frequencies), some PAM sequences other than TTC were overrepresented, suggesting one or more alternative PAMs. Based on the frequency rankings of the trinucleotide PAM sequences, the second most frequent PAM is TTT, followed by four TCN motifs (Supplemental Fig. S1A). As these less frequent PAMs share a 2 nt identity with TTC, which could derive from slipping events where the real PAM is still a TTC located nearby (Shmakov et al. 2014; Staals et al. 2016; Li et al. 2017), we first suspected that the TTT and TCN PAMs might be due to −1 nt slips (upstream) and +1 nt slips (downstream), respectively (Fig. 3A). Thus, we separately reanalyzed acquired loci with each of these PAMs with respect to their flanking sequence. Spacers with a TCN PAM showed a major T signal further upstream, consistent with +1 nt slips from TTC (Fig. 3C). However, spacers with a TTT PAM did not show an outstanding signal next to the trinucleotide, suggesting that TTT is an alternative PAM other than TTC (Fig. 3C). Consistent with this interpretation, acquired spacers with a TTT PAM showed independent localizations relative to those with a TTC PAM (Supplemental Fig. S1B).
FIGURE 3.
PAM preference and acquisition inaccuracies in primed adaptation. (A) Schematic representation of spacer selection by the adaptation machinery. In most cases, the machinery selects spacer sequences immediately downstream from the PAM, with an inexact molecular ruler at the PAM-distal end. Less frequently, the machinery shifts a few nucleotides downstream (+ slip) or upstream (− slip) at the PAM-proximal end, causing slipping events. Flipping events were also observed where the double-stranded DNA substrates were incorporated in an opposite orientation into the CRISPR array. (B) Based on the PAM localization within the upstream or downstream junction, spacer acquisition events were categorized into different types, with their frequencies shown for both pSp1(+) and pSp1(−) priming. Note that for the alternative TTT PAM, spacers with the first nucleotide being C were excluded as these were classified as potential −1 nt slipping events. (C) Sequence Logo of the upstream and downstream 20 nt junctions of indicated categories of spacer acquisitions from pSp1(+). (D) Length distribution of all acquired spacers primed by pSp1(+) and pSp1(−), compared with the native spacers from type I-C CRISPR loci in L. pneumophila ST222 strains. (E) Length distribution of acquired spacers from each slipping category.
As we observed +1 nt slips, we wondered if there were other types of acquisition errors. We systematically examined + slips where the acquisition machinery extracts protospacers further downstream at the PAM and − slips where cleavage happens further upstream (Fig. 3A). Indeed, besides +1 nt slips that occur at a ∼2% frequency, other types of slips do happen—though at 0.1%–1% frequencies—with a decreasing trend as the slipping goes further (Fig. 3B). Apart from the aforementioned classes of spacer acquisition where the upstream sequence of the plasmid contains either a TTC or a TTT PAM, ∼2% of spacers remained unexplained. When we examined the target sequence upstream and downstream from these spacers, we observed a clear GAA signal directly downstream (Fig. 3C). This downstream GAA signal is consistent with a phenomenon known as “flipping”—where the prespacer substrate next to a TTC PAM is integrated in an opposite direction into the CRISPR array so that the reverse complementary strand is used as a spacer (Shmakov et al. 2014; Staals et al. 2016; Li et al. 2017). Indeed, we identified ∼1% spacers derived from potential flips with an original TTC PAM, 0.3%–0.4% spacers from a combination of flips and +1 nt slips, and even rarer still—combinations of flips and other types of slips (Fig. 3B). When combined, the canonical TTC PAM, the alternative TTT PAM, and slipping and flipping events explain >99.5% of all acquired spacers from the priming plasmid (Fig. 3B).
The native spacers in L. pneumophila type I-C CRISPR arrays range from 33 to 37 nt, with 35 nt being the most frequent length. We next asked if acquired spacers had a similar length distribution. Compared with native spacers, laboratory acquired spacers showed a slight shift toward shorter lengths, with 34 nt being the most frequent (Fig. 3D). Compared with type I-E and type I-F systems that acquire spacers mostly (∼90%) with a uniform length (Datsenko et al. 2012; Savitskaya et al. 2013; Fineran et al. 2014; Richter et al. 2014; Staals et al. 2016), the type I-C system acquired spacers with a broader range of lengths—which could indicate a less precise molecular ruler in the adaptation machinery. Spacer-length precision did not appear influenced by either slipping or flipping events, as spacers with a canonical TTC PAM showed a similar distribution of length (Fig. 3E). Interestingly, in +1 nt slips and +2 nt slips, we observed a distinct distribution of spacer length, with an increasing preference for shorter (≤33 nt) ones (Fig. 3E). This is in contrast with the observations in the P. atrosepticum type I-F system where − slips instead of + slips correlated with aberrant spacer lengths (Staals et al. 2016), indicating another molecular distinction between these two adaptation machineries. Taken together, we identified a broad set of potential acquisition inaccuracies in the L. pneumophila type I-C system. A representative example of these inaccuracies can also be found at the major spacer acquisition hotspot (Supplemental Fig. S1C).
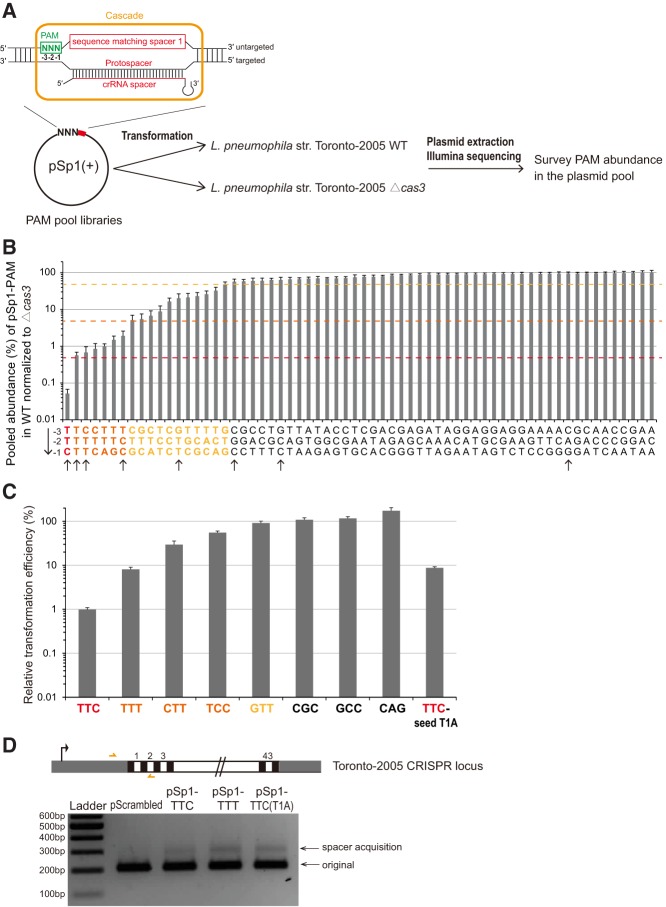
Systematic quantification of interference efficiencies confirms a hierarchy of preferred PAMs
PAM recognition in spacer acquisition is attributed to the adaptation machinery—and this process is likely independent from the Cascade interference complex that by itself recognizes the PAM and binds to target. To examine the possible coevolution of PAM recognition by these two machineries (Kunne et al. 2016), we asked if the Cascade interference complex also recognizes alternative PAMs other than the canonical TTC motif. Indeed, using an in vivo positive screen, Leenay et al. (2016) recently identified TTC, CTC, TCC, and TTT, with decreasing preferences, as functional PAMs for interference in the Bacillus halodurans type I-C system (Leenay et al. 2016). Here, we performed a plasmid-removal based screen to examine functional PAMs for interference in L. pneumophila (Fig. 4A). We transformed a plasmid library containing a full spacer 1 match with a randomized trinucleotide PAM into either L. pneumophila str. Toronto-2005 wild-type or Δcas3. By analyzing the PAM abundance in the survived plasmid pools using high-throughput sequencing, we identified PAMs that were depleted to different degrees by the wild-type type I-C system. Among the 64 PAM sequences, TTC achieved the highest protection efficiency of >99.9%, six others (TTT, CTT, CTC, TTA, TTG, and TCC) within the range of 95%–99.5%, and 11 more above 50% (Fig. 4B). It is noteworthy that TTT is the second most interference-efficient PAM, consistent with our observation that TTT is also the second most frequent PAM used in spacer acquisition. Many of the less protective PAMs share a 2 nt identity with TTC, suggesting that a 1 nt perturbation of the PAM would still allow some functionality. We confirmed the observed hierarchy of PAM activities using a CFU-based plasmid transformation efficiency assay of eight selected PAMs (Fig. 4C). Inspection of the transformants of TTT PAM plasmids showed spacer acquisition events similar to the transformants of the bona fide target and mismatched protospacer plasmids, suggesting that the alternative PAM primes in a similar manner (Fig. 4D). Together, our assay suggests that the L. pneumophila type I-C system possesses a broader range of active PAMs in interference than in primed adaptation. Such flexibility in PAM selection during interference was also observed in the E. coli type I-E system (Fineran et al. 2014; Xue et al. 2015).
FIGURE 4.
PAM preference for L. pneumophila type I-C CRISPR-Cas interference. (A) Schematic workflow to characterize functional PAMs for CRISPR interference. A pool of plasmids containing the protospacer sequence for spacer 1 and a random trinucleotide PAM was generated and transformed into either L. pneumophila str. Toronto-2005 wildtype or Δcas3, and the abundance of each PAM sequence in the pool was quantified through Illumina sequencing. (B) Pooled abundances were derived by normalizing the ratio of each PAM in the wildtype transformant pool to that in the Δcas3 pool. These relative abundances categorized PAM sequences into different preferences for CRISPR interference. (C) Individual plasmid transformation efficiency assay confirmed, with a lower sensitivity, the observations in the Illumina-based pooled assay. Plasmids containing either spacer 1 and an indicated PAM or a scrambled control sequence were electroporated into L. pneumophila str. Toronto-2005 wildtype. The relative transformation efficiency is calculated by normalizing transformation efficiency of the spacer 1 plasmids to that of the control plasmid. Error bars represent the SEM of three biological replicates. (D) Spacer acquisition was also observed in transformants of targeted plasmids with the alternative TTT PAM.
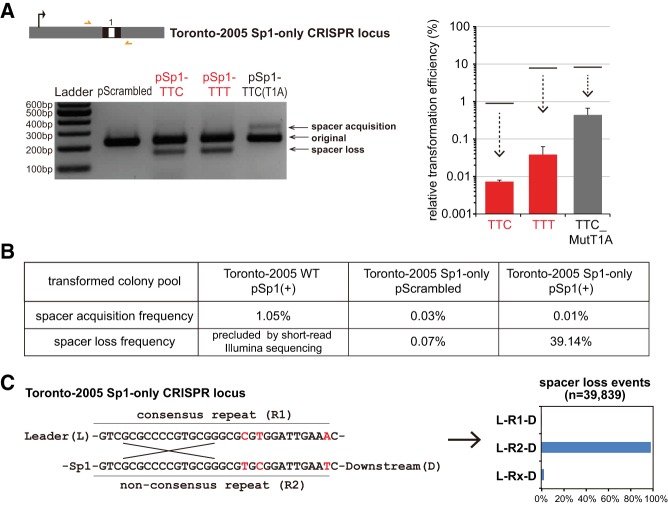
Truncation of the type I-C array leads to an increase in interference that experimentally highlights spacer loss events
As L. pneumophila type I-C CRISPR-Cas is relatively permissive for interference, we wondered how spacer acquisition efficiency would change if the system could be modified to more efficiently cleave targets. While studying a minimized type I-C array that contains only a single spacer, we made an unexpected observation that allowed us to further explore the system within the context of increased interference. We generated a CRISPR array-minimized strain in which all 43 spacers except the spacer 1 were deleted in L. pneumophila str. Toronto-2005. Remarkably, the sole spacer in this strain showed a ∼100-fold increased protection efficiency against its matching protospacer as compared to the parental strain that contains a full-length (43 spacers) array (Fig. 5A). When examining the CRISPR loci in the less frequent transformants, no spacer acquisition was observed, in contrast to what we observed for the more permissive, full-length array strain. Instead, the transformants that we recovered under these experimental conditions were enriched for spacer loss events (Fig. 5A). To test if the modified CRISPR array remained adaptable, we next transformed the spacer 1 only strain with a mismatched target plasmid (carrying a T1A seed mutation). Use of this mismatched target led to both decreased interference efficiency and robust spacer acquisition, demonstrating the adaptability of the minimized CRISPR array under certain conditions (Fig. 5A).
FIGURE 5.
Truncating the I-C CRISPR array increases interference efficiency and provides an experimental system to study loss. (A) An array-minimized (Sp1-only) CRISPR-Cas system is highly efficient (>99.9% by relative transformation efficiency) at protecting against targeted plasmids. The plasmid transformation efficiency assay was performed to measure interference efficiencies of the modified CRISPR-Cas system (compared with those of the original system, shown by the upper lines for reference—see Fig. 4C). Error bars represent the SEM of three biological replicates. The resulting transformants were examined by PCR amplification for any spacer acquisition or loss in the CRISPR loci and analyzed by agarose gel. (B) Illumina quantification of spacer acquisition and spacer loss frequencies in plasmid transformants of L. pneumophila str. Toronto-2005 wildtype or the Sp1-only strain. Spacer loss frequency was not determined for wildtype transformants because only the leader-end of the CRISPR array was amplified for sequencing, an experimental limitation necessitated by the short read-length of current Illumina technologies. (C) Most spacer loss events retained the downstream nonconsensus repeat (R2), shown in the bar graph, consistent with a mechanism of homologous recombination between the two flanking repeats (R1 and R2). The few Rx repeats contain mismatches to both R1 and R2 and may derive from sequencing errors.
To confirm these observations, we next performed Illumina deep sequencing on the PCR product flanking this one spacer. Taking advantage of the short length of the array in a single-spacer strain (which is amenable to Illumina short-read sequencing), we observed 39% spacer loss frequency in the CRISPR loci and a ∼100-fold lower spacer acquisition frequency relative to the wild-type strain transformants (Fig. 5B). As the minimized array was designed to contain two similar but distinct repeat sequences flanking spacer 1, we next examined the spacer loss events to determine the nature of the single repeat sequence left behind after such events. We found that most loci retained the downstream repeat, likely due to a 3′ positioning of polymorphisms whose retention would be favored by the majority of homologous recombination events (Fig. 5C). Lastly, we took advantage of this experimental system to quantify spacer dynamics in the single-spacer strain transformed with an untargeted plasmid. We observed a low, but detectable (<0.1%) spacer loss frequency when the selection for CRISPR mutants was relaxed through the use of an untargeted plasmid (Fig. 5B). Taken together, these observations suggest that spacer loss events naturally occur in CRISPR loci at a low frequency within the population—and that using antibiotic selection to maintain a plasmid that is undergoing highly efficient CRISPR-Cas targeting can select for this rare subgroup. As predicted by this model, we also observed similar enrichment for spacer loss in a wild-type (full-length) type I-C system in L. pneumophila str. Toronto-2000 when we experimentally forced the maintenance of a plasmid targeted by a newly acquired spacer that drives high interference (Supplemental Fig. S2). These observations are consistent with a previous study showing that preexisting CRISPR mutants exist at >10−4 frequency in Staphylococcus epidermidis and thus providing an avenue by which beneficial targeted plasmids can be transferred (Jiang et al. 2013).
DISCUSSION
A clear interplay between CRISPR interference and adaptation has been established (Xue et al. 2015; Semenova et al. 2016; Staals et al. 2016). For CRISPR-Cas systems that execute the interference very efficiently (interference-strict), a slow or delayed target degradation (by target mismatches or other means) is often necessary to maintain a high enough copy number of the target plasmid in order to achieve an efficient primed adaptation (Kunne et al. 2016; Semenova et al. 2016; Severinov et al. 2016). Here we confirmed that when the cleaving efficiency of a system allows the temporary coexistence of target and a functional CRISPR-Cas, robust spacer acquisition predominates. Using the interference-permissive L. pneumophila type I-C system, we also showed that a bona fide target primes similarly to a mismatched protospacer, further supporting the concept that mismatched priming and “interference-driven” adaptation share similar pathways and differ mainly in efficiencies in some experimental systems (Semenova et al. 2016; Staals et al. 2016).
Our analyses of spacer acquisition patterns (Fig. 6) are consistent with the sliding model in which during the beginning stages of primed acquisition, one or more proteins (including, at least, Cas3) translocates 3′–5′ preferentially on the untargeted strand for prespacer selection (Heler et al. 2014). Remarkable insight into the likely nature and activity of the downstream stages of acquisition comes from recent work in type I-F systems, in which Cas2 and Cas3 are fused and the resulting “Cas2–3” has long provided a tantalizing link between interference and adaptation. In these studies, Cas1 and Cas2–3 were shown to form a complex (Fagerlund et al. 2017; Rollins et al. 2017) that integrates new spacers into a CRISPR array in vitro (Fagerlund et al. 2017). This in vitro activity depended on Cas1 integrase activity but was independent of Cas3 nuclease or helicase activity (Fagerlund et al. 2017)—indeed Cas1 itself was shown to inhibit Cas2–3 nuclease activity within the complex (Rollins et al. 2017). Together, these results are consistent with a model in which Cas3 3′–5′ helicase activity underlies the directionality of primed acquisition, with other members of the complex acting at the subsequent site along the plasmid where new spacers are derived.
FIGURE 6.
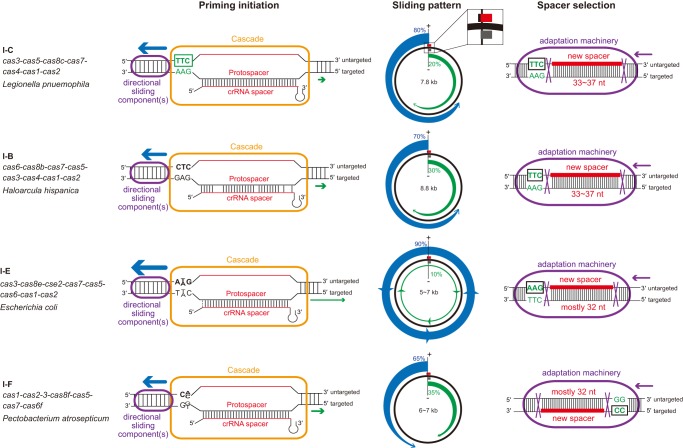
Schematic summary of primed spacer acquisition in type I CRISPR-Cas. Primed spacer sampling is separated into three steps: (i) priming initiation where the Cascade–crRNA complex binds to the targeted DNA and recruits one or more sliding complex component(s); (ii) a surveying of the plasmid out from that site, consistent with 3′–5′ movement of the sliding component(s) coupled with variable strand specificity; (iii) spacer selection from another plasmid locus by the adaptation complex upon recognition of an appropriate PAM sequence. Comparisons of each step in the type I-C system versus type I-B (Li et al. 2014b, 2017), type I-E (Datsenko et al. 2012; Savitskaya et al. 2013; Fineran et al. 2014), and type I-F systems (Richter et al. 2014; Staals et al. 2016), show similarities and distinctions in molecular mechanisms. An inset shows the strand location of each protospacer (gray bar), spacer (red), and PAM (black bar) in each sliding pattern schematic.
Several similarities and differences exist between L. pneumophila type I-C acquisition and what has been described for other systems in other bacteria (Fig. 6). We observe similar spacer acquisition patterns between the L. pneumophila type I-C system and the H. hispanica type I-B system (Li et al. 2014b): Both systems show a moderate (70%–80%) overall bias toward the untargeted strand and a positional preference for the 5′ region on both strands relative to the priming site. In contrast, the P. atrosepticum type I-F system prefers spacers on the targeted strand (Richter et al. 2014; Staals et al. 2016). Type I-F's opposite strand preference with respect to spacer selection could possibly be due to an opposite spatial organization of the adaptation complex relative to the PAM recognition. The E. coli type I-E system shows robust spacer acquisitions from the untargeted strand, like type I-C and type I-B, but the spacer acquisition efficiency gradient is relatively longer (Datsenko et al. 2012; Savitskaya et al. 2013; Fineran et al. 2014; Strotskaya et al. 2017). To explain the discrepancies of positional preference (sampling distance) in different systems, variable processivity of Cas3 in different systems has been proposed (Redding et al. 2015). This model would suggest that the type I-C and type I-B systems should have an intermediate level of Cas3 processivity (between a highly processive type I-E system and a less processive type I-F system). The different levels of strand bias in these systems, on the other hand, may be attributed to different degrees of “PAM-independent processing” in which Cas3 is recruited with the help of Cas1–Cas2 and travels bidirectionally (Redding et al. 2015).
Using next-generation sequencing, we have achieved a high survey depth, thus enabling a comprehensive examination of L. pneumophila type I-C spacer acquisition patterns. With respect to PAM preference, in addition to the canonical TTC PAM (∼90% of all acquired spacers), we identified an alternative TTT PAM (2%–4%) and extensive slipping and flipping events (6%–8%). With respect to spacer size selection, we observed a flexible choice more similar to the type I-B system (Li et al. 2017) than the slightly more stringent type I-E (Savitskaya et al. 2013; Fineran et al. 2014) and type I-F (Richter et al. 2014; Staals et al. 2016) systems. A recent study on the length variation of acquired spacers in the H. hispanica type I-B system showed that the nucleotide specificity at both ends of the PAM-protospacer sequence could influence the molecular ruler in the adaptation machinery (Li et al. 2017). The observed similarities between type I-C and type I-B acquisition patterns is also consistent with their closer Cas1-based phylogenetic relationship relative to the other two systems (Supplemental Fig. S3). Further insights into the molecular basis of spacer acquisition stringency may be derived from a detailed structure-based comparison of Cas1 and Cas2 from each system.
It is known that different CRISPR-Cas systems, as well as different spacers within one array, often show a wide range of interference efficiencies (Marraffini and Sontheimer 2008; Bikard et al. 2012; Cady et al. 2012; Li et al. 2014a; Xue et al. 2015; Qiu et al. 2016; Rao et al. 2016). Both technical and biological factors could contribute to this variation. On the one hand, transformation methods, plasmid copy number, and bacterial culture conditions could all affect how efficiently invasive DNA is cleaved (Majsec et al. 2016; Rao et al. 2016; Severinov et al. 2016). On the other hand, innate factors could also influence interference efficiency—such as expression levels of Cas proteins, transcription and processing efficiencies of individual spacers, and binding affinities between Cascade and crRNA (Xue et al. 2015; Patterson et al. 2016; Rao et al. 2016; Hoyland-Kroghsbo et al. 2017). We observed a dramatic increase in interference efficiency for the same spacer (Sp1) within a strain in which the CRISPR array was minimized from 43 to one spacer. This could be due to a higher abundance of Sp1 crRNA, a relatively increased availability of Cas proteins for Sp1 (due to lack of competition with other spacers for loading), or a combination of both. Future experiments to examine the crRNA abundance and to over-express each Cas functional group (to determine limiting factors) will be necessary to test these hypotheses. Notably, these results also demonstrate the experimental strength of mini-array strains with respect to the study of spacer loss, given their accessibility to short read-length technologies.
We observed a great range of spacer acquisition frequencies at different locations of the same element. While this variation could be affected by PAM specificity, strand specificity, and strand-specific distance from the priming site, our examination of the major spacer acquisition hotspot points toward other factors that directly contribute to prespacer capture by the adaptation machinery. By introducing different mutations in the hotspot neighborhood, we found that the internal sequence, but not the flanking nucleotides, contributes to the high acquisition frequency of this hotspot sequence. Based on these data, we speculate that some DNA motif or ssDNA secondary structure within the hotspot sequence (likely at the PAM-proximal end) could attract the adaptation machinery, with further systematic mutation experiments required to identify the exact contributor. As has been done in other systems (Yosef et al. 2013; Staals et al. 2016), subsequent analyses of our type I-C data could be extended to examine similar classes of acquired spacers (e.g., most frequently acquired, least frequently acquired) for common features that influence their acquisition frequencies.
Going forward, several features make the L. pneumophila type I-C system a good model system to study CRISPR-Cas functionality. First, type I-C systems represent one of the most common types of CRISPR-Cas systems, yet nevertheless remain relatively understudied (Makarova et al. 2011, 2015). Second, our earlier comparative genomics data suggest that the system is naturally adaptable (Rao et al. 2016). Third, the relatively permissive interference of the system facilitates the laboratory study of primed spacer acquisition and interference using the same set of perfectly matched target sequences. Lastly, based on our initial characterizations, the system displays several features that distinguish it from type I-E and type I-F systems, the two systems most exhaustively studied to date.
MATERIALS AND METHODS
Bacterial strains and plasmids
Legionella pneumophila strain Toronto-2005 is a clinical isolate of Sequence Type 222 from Toronto, Canada, with a circularized genome available (GenBank CP012019) (Rao et al. 2016). An RpsLK43R streptomycin resistant derivate of the clinical isolate is used as wildtype in this study. From this RpsLK43R strain, a Δcas3 deletion mutant and an array-minimized (Sp1-only) strain were generated by allelic exchange as previously described (Ensminger et al. 2012; Rao et al. 2016). Specifically, in the Sp1-only strain, only the first repeat, Sp1, and the last repeat of the original array were retained. The priming plasmids were generated by cloning the insert (see Supplemental Table S2) into the ApaI/PstI-cut pMMB207 backbone (Rao et al. 2013, 2016) (see Supplemental File for the full pSp1[+] sequence). Our previous study using Illumina sequencing showed that this plasmid has an average copy number of 7.6 in L. pneumophila str. Philadelphila-1 (Rao et al. 2013). Site-directed mutagenesis (QuickChange II) was used to mutate the spacer acquisition hotspot in the original plasmid. Bacterial electroporation and axenic passage were performed as previously described (Rao et al. 2016). After axenic passage for 15 generations, the CRISPR adaptation ratio in the bacterial population increased from ∼1% to ∼24%, as quantified by Illumina sequencing (data not shown). Each priming experiment was performed in two biological replicates, and these replicates were largely consistent in spacer mappings (data not shown). Unless specified, data shown are averages of two replicates.
PCR of CRISPR loci and preparation of Illumina libraries
Roughly 1 OD unit (∼1 × 109) bacterial cells from either colony pool (containing at least 50 independent colonies) or axenic passage were used for genomic DNA extraction using the NucleoSpin Tissue kit (Machery-Nagel). CRISPR loci were amplified using the Kapa HiFi Polymerase (Kapa Biosystems) and primers listed in Supplemental Table S2. Raw PCR products of 20 amplification cycles were used for library preparation. In addition, to enrich for adapted CRISPR arrays, 30-cycle PCR products were concentrated by ethanol precipitation and separated in 6% acrylamide gel by running at 60V for 3 h. An ∼70 bp higher band than the original array (∼350 bp) was extracted and DNA purified from the extraction was subjected to another 10-cycle PCR to increase the yield. These further size-selection steps to enrich for adapted arrays did not introduce significant bias relative to the raw PCR products (data not shown). Purified PCR amplicons were normalized by PicoGreen to 1 ng and processed using the Nextera XT kit (Illumina). Multiplexed libraries were subjected to Illumina NextSeq sequencing at 2 × 150 bp read-length (CAGEF, University of Toronto).
Illumina reads processing and data analyses
Paired-end raw reads were first attempted to merge by FLASH (Magoc and Salzberg 2011) using “-m 50 -M 100 -x 0.02” settings. The unassembled single-end reads were quality trimmed by Trimmomatic (Bolger et al. 2014) using “SLIDINGWIN DOW:3:20 MINLEN:50” settings. These preprocessed reads were combined and processed using a Perl script (available upon request) to annotate the presence of leader sequence (L), CRISPR repeats (R), existing spacers (S), new spacers (X), and downstream sequence (D) in each read. The new spacers were extracted and aligned using BLASTN to either the priming plasmid or L. pneumophila str. Toronto-2005 genome. BLASTN results were then summarized into coverages of each nucleotide in the plasmid and subjected to Circos visualization (Krzywinski et al. 2009). To examine the PAM preference, slipping, and flipping of acquired spacers, flanking sequences of acquired spacers were extracted from the plasmid and subjected to Sequence Logo (Crooks et al. 2004) visualization. To avoid potential redundancy, flipping cases were only examined from spacers without a TTC or TTT PAM in the upstream junction. To quantify spacer acquisition and spacer loss frequencies, the following formulas were used, in which each item denotes the count of reads with the indicated annotation:
Preparation and analyses of PAM plasmids pool
Oligos (see Supplemental Table S2) with a randomized trinucleotide upstream of Sp1 sequence were annealed, digested, and ligated into the ApaI/PstI-cut pMMB207 vector. A total of ∼3000 E. coli colonies were obtained after transformation and combined into a pool. Plasmids were extracted from the E. coli pool using the PureYield Plasmid Midiprep Kit (Promega), and a control plasmid with a scrambled insert was spiked into the plasmid pool at ∼1% ratio. Roughly 1 µg of the pooled plasmids was electroporated into 4 OD units of L. pneumophila str. Toronto-2005 wildtype or Δcas3 overnight culture. Three biological replicates of electroporation were performed. With 5 µg/mL chloramphenicol selection, over 3000 colonies were obtained from each electroporation. Plasmids were then extracted from these L. pneumophila transformants using the EZ-10 Spin Column Miniprep Kit (Biobasic). Without any PCR amplification, these plasmid pools were subjected to the Nextera XT library preparation and Illumina NextSeq sequencing. After quality filtering, reads containing the Sp1 sequence (or the scrambled sequence) were extracted and PAM sequences were identified from these reads. PAM frequencies in L. pneumophila transformants were normalized to both the scrambled control and the E. coli plasmid pool.
DATA DEPOSITION
Raw Illumina NextSeq 500 data used to catalog spacer acquisition have been deposited in the NCBI Sequence Read Archive under the BioProject PRJNA360289.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Project Grant from the Canadian Institutes of Health Research (PHT-148819), the University of Toronto, Connaught Fund (NR-2015-16), and an infrastructure grant from the Canada Foundation for Innovation and the Ontario Research Fund (30364) to A.W.E. The authors thank members of the Centre for the Analysis of Genome Evolution and Function (CAGEF) at the University of Toronto for performing Illumina sequencing. We thank members of the Ensminger laboratory for helpful discussions and for careful reading of the manuscript and the reviewers for their careful and constructive suggestions.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.062083.117.
REFERENCES
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712. [DOI] [PubMed] [Google Scholar]
- Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. 2012. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12: 177–186. [DOI] [PubMed] [Google Scholar]
- Blosser TR, Loeff L, Westra ER, Vlot M, Kunne T, Sobota M, Dekker C, Brouns SJ, Joo C. 2015. Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex. Mol Cell 58: 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrus V, Waldor MK. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol 155: 376–386. [DOI] [PubMed] [Google Scholar]
- Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O'Toole GA. 2012. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol 194: 5728–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. 2012. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 3: 945. [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190: 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez-Villasenor C, Guzman NM, Almendros C, Garcia-Martinez J, Mojica FJ. 2013. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol 10: 792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensminger AW, Yassin Y, Miron A, Isberg RR. 2012. Experimental evolution of Legionella pneumophila in mouse macrophages leads to strains with altered determinants of environmental survival. PLoS Pathog 8: e1002731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann S, Garrett RA. 2012. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol 85: 1044–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerlund RD, Wilkinson ME, Klykov O, Barendregt A, Pearce FG, Kieper SN, Maxwell HWR, Capolupo A, Heck AJR, Krause KL, et al. 2017. Spacer capture and integration by a type I-F Cas1-Cas2-3 CRISPR adaptation complex. Proc Natl Acad Sci 114: E5122–E5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Gerritzen MJ, Suarez-Diez M, Kunne T, Boekhorst J, van Hijum SA, Staals RH, Brouns SJ. 2014. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci 111: E1629–E1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3: 722–732. [DOI] [PubMed] [Google Scholar]
- Heler R, Marraffini LA, Bikard D. 2014. Adapting to new threats: the generation of memory by CRISPR-Cas immune systems. Mol Microbiol 93: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser ML, Taylor DW, Bhat P, Guegler CK, Sternberg SH, Nogales E, Doudna JA. 2014. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci 111: 6618–6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol 190: 1401–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyland-Kroghsbo NM, Paczkowski J, Mukherjee S, Broniewski J, Westra E, Bondy-Denomy J, Bassler BL. 2017. Quorum sensing controls the Pseudomonas aeruginosa CRISPR-Cas adaptive immune system. Proc Natl Acad Sci 114: 131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Nam KH, Ding F, Lee H, Wu L, Xiao Y, Farchione MD Jr, Zhou S, Rajashankar K, Kurinov I, et al. 2014. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol 21: 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes AP, Villion M, Moineau S. 2014. Adaptation in bacterial CRISPR-Cas immunity can be driven by defective phages. Nat Commun 5: 4399. [DOI] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. 1987. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169: 5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SA, McKenzie RE, Fagerlund RD, Kieper SN, Fineran PC, Brouns SJ. 2017. CRISPR-Cas: adapting to change. Science 356: eaal5056. [DOI] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43: 1565–1575. [DOI] [PubMed] [Google Scholar]
- Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. 2013. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet 9: e1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, et al. 2011. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 18: 529–536. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunne T, Kieper SN, Bannenberg JW, Vogel AI, Miellet WR, Klein M, Depken M, Suarez-Diez M, Brouns SJ. 2016. Cas3-derived target DNA degradation fragments fuel primed CRISPR adaptation. Mol Cell 63: 852–864. [DOI] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8: 317–327. [DOI] [PubMed] [Google Scholar]
- Leenay RT, Maksimchuk KR, Slotkowski RA, Agrawal RN, Gomaa AA, Briner AE, Barrangou R, Beisel CL. 2016. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol Cell 62: 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R. 2015. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520: 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang R, Xiang H. 2014a. Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res 42: 7226–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Wang R, Zhao D, Xiang H. 2014b. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res 42: 2483–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gong L, Zhao D, Zhou J, Xiang H. 2017. The spacer size of I-B CRISPR is modulated by the terminal sequence of the protospacer. Nucleic Acids Res 45: 4642–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27: 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majsec K, Bolt EL, Ivancic-Bace I. 2016. Cas3 is a limiting factor for CRISPR-Cas immunity in Escherichia coli cells lacking H-NS. BMC Microbiol 16: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. 2011. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322: 1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Soria E, Juez G. 2000. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol 36: 244–246. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155: 733–740. [DOI] [PubMed] [Google Scholar]
- Mulepati S, Bailey S. 2013. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem 288: 22184–22192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. 2014. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol 21: 528–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez JK, Harrington LB, Kranzusch PJ, Engelman AN, Doudna JA. 2015. Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature 527: 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AG, Jackson SA, Taylor C, Evans GB, Salmond GP, Przybilski R, Staals RH, Fineran PC. 2016. Quorum sensing controls adaptive immunity through the regulation of multiple CRISPR-Cas systems. Mol Cell 64: 1102–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Wang S, Chen Z, Guo Y, Song Y. 2016. An active type I-E CRISPR-Cas system identified in Streptomyces avermitilis. PLoS One 11: e0149533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao C, Benhabib H, Ensminger AW. 2013. Phylogenetic reconstruction of the Legionella pneumophila Philadelphia-1 laboratory strains through comparative genomics. PLoS One 8: e64129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao C, Guyard C, Pelaz C, Wasserscheid J, Bondy-Denomy J, Dewar K, Ensminger AW. 2016. Active and adaptive Legionella CRISPR-Cas reveals a recurrent challenge to the pathogen. Cell Microbiol 18: 1319–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding S, Sternberg SH, Marshall M, Gibb B, Bhat P, Guegler CK, Wiedenheft B, Doudna JA, Greene EC. 2015. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell 163: 854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Gristwood T, Clulow JS, Fineran PC. 2012. In vivo protein interactions and complex formation in the Pectobacterium atrosepticum subtype I-F CRISPR/Cas System. PLoS One 7: e49549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Dy RL, McKenzie RE, Watson BN, Taylor C, Chang JT, McNeil MB, Staals RH, Fineran PC. 2014. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res 42: 8516–8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins MF, Chowdhury S, Carter J, Golden SM, Wilkinson RA, Bondy-Denomy J, Lander GC, Wiedenheft B. 2017. Cas1 and the Csy complex are opposing regulators of Cas2/3 nuclease activity. Proc Natl Acad Sci 114: E5113–E5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitskaya E, Semenova E, Dedkov V, Metlitskaya A, Severinov K. 2013. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol 10: 716–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenova E, Savitskaya E, Musharova O, Strotskaya A, Vorontsova D, Datsenko KA, Logacheva MD, Severinov K. 2016. Highly efficient primed spacer acquisition from targets destroyed by the Escherichia coli type I-E CRISPR-Cas interfering complex. Proc Natl Acad Sci 113: 7626–7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severinov K, Ispolatov I, Semenova E. 2016. The influence of copy-number of targeted extrachromosomal genetic elements on the outcome of CRISPR-Cas defense. Front Mol Biosci 3: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Savitskaya E, Semenova E, Logacheva MD, Datsenko KA, Severinov K. 2014. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Res 42: 5907–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. 2011. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 30: 1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Waghmare SP, Dickman MJ, Barrangou R, Horvath P, Siksnys V. 2013. In vitro reconstitution of Cascade-mediated CRISPR immunity in Streptococcus thermophilus. EMBO J 32: 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RH, Jackson SA, Biswas A, Brouns SJ, Brown CM, Fineran PC. 2016. Interference-driven spacer acquisition is dominant over naive and primed adaptation in a native CRISPR-Cas system. Nat Commun 7: 12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotskaya A, Savitskaya E, Metlitskaya A, Morozova N, Datsenko KA, Semenova E, Severinov K. 2017. The action of Escherichia coli CRISPR-Cas system on lytic bacteriophages with different lifestyles and development strategies. Nucleic Acids Res 45: 1946–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MW, Brouns SJ. 2012. CRISPR interference directs strand specific spacer acquisition. PLoS One 7: e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorontsova D, Datsenko KA, Medvedeva S, Bondy-Denomy J, Savitskaya EE, Pougach K, Logacheva M, Wiedenheft B, Davidson AR, Severinov K, et al. 2015. Foreign DNA acquisition by the I-F CRISPR-Cas system requires all components of the interference machinery. Nucleic Acids Res 43: 10848–10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li J, Zhao H, Sheng G, Wang M, Yin M, Wang Y. 2015. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 163: 840–853. [DOI] [PubMed] [Google Scholar]
- Wei Y, Terns RM, Terns MP. 2015. Cas9 function and host genome sampling in Type II-A CRISPR-Cas adaptation. Genes Dev 29: 356–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. 2012a. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu Rev Genet 46: 311–339. [DOI] [PubMed] [Google Scholar]
- Westra ER, van Erp PB, Kunne T, Wong SP, Staals RH, Seegers CL, Bollen S, Jore MM, Semenova E, Severinov K, et al. 2012b. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, van Houte S, Oyesiku-Blakemore S, Makin B, Broniewski JM, Best A, Bondy-Denomy J, Davidson A, Boots M, Buckling A. 2015. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr Biol 25: 1043–1049. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, et al. 2011. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci 108: 10092–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Seetharam AS, Musharova O, Severinov K, Brouns SJ, Severin AJ, Sashital DG. 2015. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res 43: 10831–10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Whitis NR, Sashital DG. 2016. Conformational control of cascade interference and priming activities in CRISPR immunity. Mol Cell 64: 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. 2012. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 40: 5569–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Shitrit D, Goren MG, Burstein D, Pupko T, Qimron U. 2013. DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc Natl Acad Sci 110: 14396–14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.