Abstract
Background
Recently, accumulating studies have found that ACSL4 dysregulation is related to a great number of malignant tumors. The purpose of the present study was to explore the relationship between ACSL4 expression level and clinical prognosis of hepatocellular carcinoma (HCC) patients.
Material/Methods
The Oncomine and TCGA databases were used to predict the expression of ACSL4 mRNA in HCC and its association with HCC prognosis. Further, immunohistochemistry was performed to verify the ACSL4 protein expression in 116 paired HCC and adjacent normal tissues. Kaplan-Meier and cox analysis were performed to validate the correlation between ACSL4 expression and HCC prognosis.
Results
We first used the Oncomine database to find that ACSL4 mRNA expression level was significantly higher in HCC tissues than that in normal tissues (p all <0.001). The results were consistent with those in the TCGA database. Then, immunohistochemical results demonstrated that the ACSL4 positive expression rate was 70.7% in HCC tissues. ACSL4 differential expression level was significantly related to Edmondson grade (p=0.010), AFP (p=0.001) and TNM stage (p=0.012). Survival analysis revealed that both overall survival (OS) and disease-free survival (DFS) time were remarkably reduced in HCC patients with ACSL4 high expression (p=0.001 and 0.000, respectively). Moreover, Cox multivariate analysis demonstrated that ACSL4 expression was the only independent prognostic factor for both OS and DFS (both p values=0.001).
Conclusions
Taken together, our study demonstrated that ACSL4 was overexpressed in HCC, and it will be a new potential therapeutic target for HCC as an independent adverse prognostic parameter.
MeSH Keywords: Acyl-CoA Oxidase; Carcinoma, Hepatocellular; Prognosis
Background
Hepatocellular carcinoma (HCC) as one of the most common malignancies of the digestive tract, it is the third cause of cancer death worldwide [1], and it ranks second in China’s malignant tumor mortality incidence [2]. Most HCC patients miss the chance of receiving radical resection or liver transplantation due to their disease being diagnosed at a late stage [3]. Therefore, it is helpful to develop diagnostic and therapeutic targets through identification of genes which are differentially expressed in HCC.
Bioinformatics is a powerful tool for mining tumor differential genes. In early studies, it was found that the long chain fatty acyl-CoA ligase 4 (ACSL4) gene was highly expressed in HCC compared with normal tissues. ACSL4, also known as FACL4, was initially identified as its mutation in non-specific X-linked mental retardation in 2002 [4]. Maloberti et al. [5] found that ACSL4 possessed a substrate preference for eicosapentaenoic acid and arachidonic acid (AA). More interestingly, the level of intracellular ACSL4 protein can be altered in turn by the amounts of free AA [6]. So far, it has been proven that the ACSL4 dysregulation is related to a great number of diseases including diabetes [7], atherosclerosis [8], obesity [9], and malignant tumors [10–17]. In a previous study, Sung et al. [10,11] found that ACSL4 was overexpressed in HCC cell lines and tissues compared to normal cell lines. However, the relationship between ACSL4 expression level and clinical prognosis of HCC patients remains largely unclear.
Therefore, in the present study, immunohistochemical staining was done to examine the expressions of ACSL4 in 116 paired HCC and adjacent normal tissue samples. In addition, ACSL4 expression in HCC and its relationships with patients’ clinicopathological factors and prognosis were investigated.
Material and Methods
Bioinformatics prediction
First, the Oncomine database (https://www.oncomine.org/resource/login.html) was used to predict the ACSL4 mRNA expression levels in HCC and normal tissues. Then the Cancer Genome Atlas (TCGA) database was used to predict the relationship between expression levels of ACSL4 mRNA and clinical prognosis of HCC patients.
Patients and samples
One hundred and sixteen cases of HCC patients who had received the curative operation at Anhui Provincial Hospital from January 2009 to June 2013 were selected. We used the tumor-node-metastasis (TNM) classification (sixth edition) of the Union for International Cancer Control (UICC) to evaluate tumor stage. The detailed information on ACSL4 protein expression and other clinicopathological factors (such as gender and age) are listed in Table 1. The study was approved by the Ethics Committee of Anhui Provincial Hospital and all patients signed the written informed consent.
Table 1.
ACSL4 was overexpressed in HCC compared to the adjacent normal tissues.
| ACSL4 expression level | ||
|---|---|---|
| High (n) | Low (n) | |
| HCC tissues | 82 | 34 |
| Adjacent normal tissues | 41 | 65 |
| χ2 | 29.089 | |
| P | 0.000 | |
Immunohistochemistry and analysis
Immunohistochemistry was performed according to the manufacture protocol. After the sections were treated with dewaxing, antigen repair, and serum sealing, ACSL4 antibody (1: 500, ab110007, Abcam, UK) was added at 4°C overnight. The next day, after rewarming for 45 minutes, PBS was used to wash the sections, and then the sections were incubated at room temperature. The staining results were observed by microscope and immunohistochemical scores were calculated as described in a previous report [18].
Statistical analysis
SPSS 19.0 software was used to do statistical analysis. Pearson chi-squared test or Fisher’s test was selected to analyze the correlation between ACSL4 expression and clinicopathological factors. Kaplan-Meier and Cox regression model were employed to analyze the parameters associated to the disease-free survival (DFS) and overall survival (OS) of HCC patients. A p value <0.05 was regarded as statistical significance.
Results
ACSL4 high expression in HCC
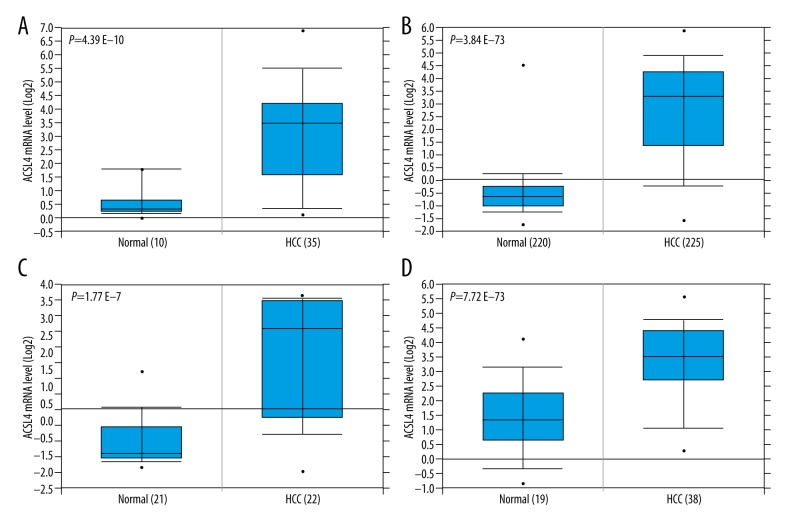
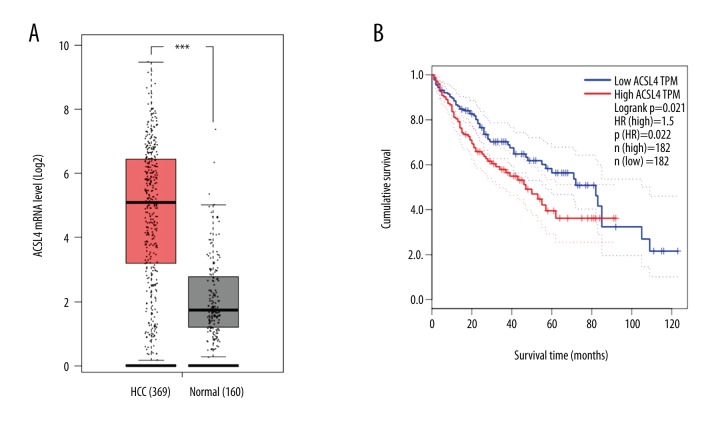
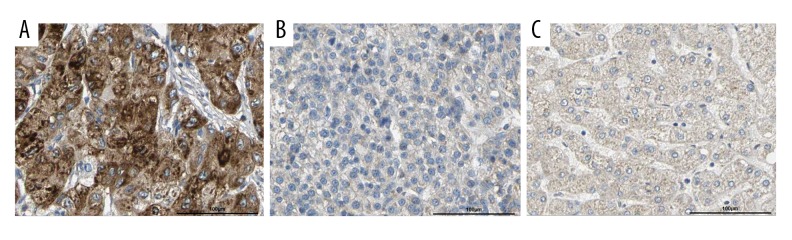
We first used the Oncomine database to find that ACSL4 mRNA expression level was significantly higher in HCC tissues than that in normal tissues (Figure 1, p all <0.001). The results were consistent with those from the TCGA database (Figure 2A). Then, in order to verify the real expression level of ACSL4 in HCC, immunohistochemistry was selected to examine the protein level of ACSL4 in 116 paired HCC and adjacent normal tissues (Figure 3). ACSL4 protein staining was mainly located in the cytoplasm (Figure 3A). ACSL4 positive expression rate was 70.7% (82/116) in HCC tissues (Table 1). The associations of ACSL4 expression with clinicopathological parameters are listed in Table 2. ACSL4 differential expression level was significantly related to Edmondson grade (p=0.010), AFP (p=0.001) and TNM stage (p=0.012).
Figure 1.
High expression levels of ACSL4 mRNA in HCC predicted by the Oncomine database. The Oncomine database mining analysis of ACSL4 mRNA levels in (A) Wurmbach liver (GEO: GSE 6764), (B) Roessler Liver2 (GEO: GSE 14520/GPL3921), (C) Roessler Liver (GEO: GSE 14520/GPL571), and (D) Mas liver (GEO: GSE 14323) grouped by HCC and normal liver.
Figure 2.
Relationship between ACSL4 mRNA and the prognosis of HCC patients predicted by TCGA database. TCGA database mining analysis of (A) ACSL4 mRNA levels grouped by HCC and normal liver and (B) relationship between ACSL4 mRNA and the prognosis of HCC patients.
Figure 3.
Immunochemical staining of ACSL4 in paired HCC and adjacent normal tissues. (A) High expression of ACSL4 in HCC tissues; (B) Low expression of ACSL4 in HCC tissues; (C) Low expression of ACSL4 in adjacent normal tissues.
Table 2.
Relationships among ACSL4 and clinicopathological parameters in HCC patients.
| Variables | Total (n=116) | ACSL4 expression level | ||
|---|---|---|---|---|
| Low (n=34) | High (n=82) | P | ||
| Age | 0.756 | |||
| <60 | 76 | 23 | 53 | |
| ≥60 | 40 | 11 | 29 | |
| Sex | 0.449 | |||
| Male | 71 | 19 | 52 | |
| Female | 45 | 15 | 30 | |
| Tumor Size (cm) | 0.621 | |||
| >5 | 38 | 10 | 28 | |
| ≤5 | 78 | 24 | 54 | |
| Tumor Nodules | 0.967 | |||
| Single | 89 | 26 | 63 | |
| Multiple | 27 | 8 | 19 | |
| Tumor capsula | 0.244 | |||
| Complete | 90 | 24 | 66 | |
| None | 26 | 10 | 16 | |
| Edmondson grade | 0.010* | |||
| I–II | 75 | 28 | 47 | |
| III–IV | 41 | 6 | 35 | |
| HbsAg | 0.430 | |||
| Positive | 97 | 27 | 70 | |
| Negative | 19 | 7 | 12 | |
| Cirrhosis | 0.437 | |||
| Yes | 106 | 30 | 76 | |
| No | 10 | 4 | 6 | |
| Child-Pugh grade | 0.485 | |||
| A | 110 | 33 | 77 | |
| B | 6 | 1 | 5 | |
| AFP (ng/ml) | 0.001* | |||
| >20 | 79 | 14 | 63 | |
| ≤20 | 37 | 18 | 19 | |
| TNM stage | 0.012* | |||
| I–II | 68 | 26 | 42 | |
| III–IV | 48 | 8 | 40 | |
Correlation between ACSL4 expression and survival of HCC patients
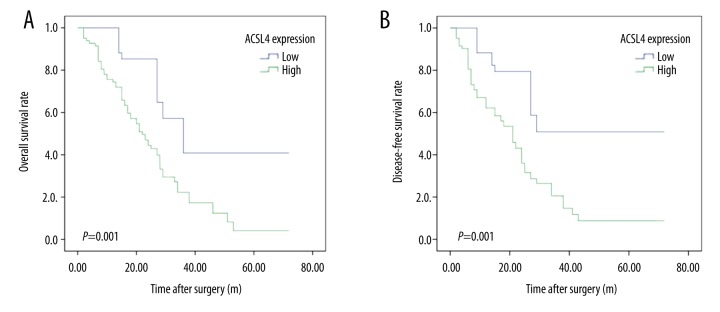
Through mining the TCGA database, we found HCC patients with ACSL4 mRNA high expression had lower survival time than those with ACSL4 mRNA low expression (Figure 2B). Then, our own immunochemical results were consistent with the predictive findings. Survival analysis revealed that ACSL4 expression levels were significantly associated with the survival time of HCC patients. Moreover, compared to those with ACSL4 protein low expression levels, both OS and DFS time were remarkably reduced in HCC patients with ACSL4 high expression levels (p=0.001, Figure 4A and p=0.000, Figure 4B, respectively).
Figure 4.
Kaplan-Meier analysis of overall survival (OS) and disease-free survival (DFS) curves of HCC patients based on ACSL4 expression as high- or low-expression. (A) OS curve of HCC patients based on ACSL4 expression; (B) DFS curve of HCC patients based on ACSL4 expression.
Prognostic value of ACSL4 expression in HCC patients
Initially, we used univariate analysis to reveal that ACSL4 expression, Edmondson grade and TNM stage had statistically prognostic influences on both OS and DFS (Table 3). In addition, Cox multivariate analysis demonstrated that ACSL4 expression was the only independent prognostic factor for both OS and DFS (both p values=0.001, Table 4).
Table 3.
Kaplan-Meir analysis of ACSL4 and other clinicopathological parameters in HCC patients.
| Variable | OS | DFS | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| ACSL4 expression | ||||||
| Low | 0.377 | 0.214–0.665 | 0.001* | 0.358 | 0.203–0.632 | 0.000* |
| High | ||||||
| Age | ||||||
| <60 | 0.744 | 0.467–1.184 | 0.212 | 0.738 | 0.465–1.171 | 0.197 |
| ≥60 | ||||||
| Sex | ||||||
| Male | 1.279 | 0.811–2.108 | 0.289 | 1.224 | 0.777–1.927 | 0.383 |
| Female | ||||||
| Tumor size (cm) | ||||||
| >5 | 0.759 | 0.464–1.241 | 0.272 | 23.268 | 0.518–1.376 | 0.497 |
| ≤5 | ||||||
| Tumor nodules | ||||||
| Single | 0.907 | 0.540–1.526 | 0.714 | 0.931 | 0.554–1.564 | 0.788 |
| Multiple | ||||||
| Tumor capsula | ||||||
| Complete | 0.779 | 0.465–1.307 | 0.345 | 0.785 | 0.472–1.307 | 0.352 |
| None | ||||||
| Edmondson grade | ||||||
| I–II | 0.587 | 0.371–0.928 | 0.022* | 0.566 | 0.358–0.894 | 0.015* |
| III–IV | ||||||
| HbsAg | ||||||
| Positive | 1.154 | 0.643–2.073 | 0.631 | 1.181 | 0.658–2.121 | 0.577 |
| Negative | ||||||
| Cirrhosis | ||||||
| Yes | 0.911 | 0.417–1.989 | 0.814 | 1.108 | 0.508–2.417 | 0.797 |
| No | ||||||
| Child-Pugh grade | ||||||
| A | 0.450 | 0.180–1.124 | 0.087 | 0.601 | 0.242–1.495 | 0.274 |
| B | ||||||
| AFP (ng/ml) | ||||||
| >20 | 0.720 | 0.445–1.167 | 0.183 | 0.784 | 0.484–1.270 | 0.323 |
| ≤20 | ||||||
| TNM stage | ||||||
| I–II | 0.624 | 0.398–0.978 | 0.040* | 0.608 | 0.388–0.951 | 0.029* |
| III–IV | ||||||
Table 4.
Cox regression analysis of ACSL4 and other clinicopathological parameters in HCC patients.
| Variable | OS | DFS | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| ACSL4 expression (low vs. high) | 0.359 | 0.191–0.674 | 0.001* | 0.347 | 0.185–0.650 | 0.001* |
| Age (<60 vs. ≥60) | 0.707 | 0.421–1.187 | 0.190 | 0.714 | 0.430–1.186 | 0.193 |
| Sex (Male vs. Female) | 1.266 | 0.771–2.079 | 0.351 | 1.288 | 0.789–2.104 | 0.312 |
| Tumor size (≤5 vs. >5) | 1.847 | 0.372–1.201 | 0.042* | 0.786 | 0.443–1.392 | 0.408 |
| Tumor nodule (single vs. multiple) | 0.941 | 0.526–1.682 | 0.836 | 0.919 | 0.514–1.641 | 0.774 |
| Edmondson grade (I–II vs. III–IV) | 0.564 | 0.332–0.957 | 0.034* | 0.589 | 0.344–1.008 | 0.053 |
| Tumor capsula (complete vs. none) | 0.584 | 0.325–1.051 | 0.073 | 0.586 | 0.325–1.058 | 0.076 |
| HbsAg (positive vs. negative) | 2.221 | 0.986–5.002 | 0.054 | 1.714 | 0.782–3.757 | 0.178 |
| Cirrhosis (present vs. absent) | 0.521 | 0.168–1.620 | 0.260 | 1.033 | 0.349–3.054 | 0.954 |
| Child-Pugh grade (A vs. B) | 0.698 | 0.252–1.935 | 0.681 | 1.021 | 0.376–2.775 | 0.968 |
| AFP (>20 vs. ≤20) | 1.231 | 0.707–2.144 | 0.463 | 1.297 | 0.752–2.238 | 0.350 |
| TNM stage (I–II vs. III–IV) | 0.898 | 0.539–1.497 | 0.681 | 0.818 | 0.491–1.364 | 0.441 |
Discussion
There has been accumulating evidence from studies demonstrating that ACSL4 is overexpressed in parts of malignant tumors such as liver [10,11], prostate [12,13] and breast cancer [13–16]. In these findings, ACSL4 is reported to perform an oncogene role in promoting tumorigenesis and metastasis. While on the other hand, Ye et al. [17] recently found that ACSL4 may serve as a tumor suppressor gene in gastric cancer possibly involving FAK and P21 signaling. As for HCC, in order to verify the exact role of ACSL4 and explore the relationship between its expression and the prognosis of HCC patients, the present study was designed and completed.
We first used the Oncomine and TCGA databases to reveal that ACSL4 mRNA expression level was significantly higher in HCC tissues than that in normal tissues. Then, in order to validate this phenomenon, 116 cases of paired HCC and normal tissues were selected. Immunochemical results showed that ACSL4 positive expression rate was 70.7% (82/116) in HCC tissues. Compared to those in the adjacent normal tissues, ACSL4 protein expression levels were remarkably higher in HCC tissues. These findings were consistent with the bioinformatics analysis and a report by Sung et al. [10,11]. Moreover, ACSL4 differential expression level was significantly related to Edmondson grade (p=0.010), AFP (p=0.001) and TNM stage (p=0.012). Therefore, the above results suggest a key role for ACSL4 in HCC progression and development.
Though Sung et al. [10,11] reported that ACSL4 was overexpressed in HCC tissues and cell lines, whether it has prognostic significance in HCC has remained unclear. So next, we used the TCGA database to predict that HCC patients with ACSL4 mRNA high expression level had worse OS than those with ACSL4 mRNA low expression level. This finding was validated by our own experimental data. Kaplan-Meir analysis showed that HCC patients with ACSL4 protein high expression had significantly reduced OS and DFS than those with ACSL4 protein low expression. Moreover, both univariate and multivariate analyses demonstrated that ACSL4 was the only independent unfavorable predictor of OS and DFS in HCC patients.
There were several limitations to our study that should be noted. First, this was a retrospective study, possibly resulting in a selective bias. Second, only immunohistochemistry (a semi-quantitative method) was used to detect the ACSL4 protein expression. Finally, we did not explore the exactly underlying molecular mechanisms in this study, which will be elucidated in future studies.
Conclusions
Collectively, our present study demonstrated that ACSL4 was overexpressed in HCC and patients with high expression level of ACSL4 had unfavorable prognosis. ACSL4, as an independent adverse prognostic parameter, will be a new potential therapeutic target for HCC.
Footnotes
Conflicts of interest
None.
Source of support: This work was partly supported by the Anhui Key Laboratory Performance Evaluation Project (1606c08234)
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Zuo T, et al. National cancer incidence and mortality in China, 2012. Chin J Cancer Res. 2016;28:1–11. doi: 10.3978/j.issn.1000-9604.2016.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allaire M, Nault JC. Advances in management of hepatocellular carcinoma. Curr Opin Oncol. 2017;29:288–95. doi: 10.1097/CCO.0000000000000378. [DOI] [PubMed] [Google Scholar]
- 4.Meloni I, Muscettola M, Raynaud M, et al. FACL4, encoding fatty acid-CoA ligase 4, is mutated in nonspecific X-linked mental retardation. Nat Genet. 2002;30:436–40. doi: 10.1038/ng857. [DOI] [PubMed] [Google Scholar]
- 5.Maloberti PM, Duarte AB, Orlando UD, et al. Functional interaction between acyl-CoA synthetase 4, lipooxygenases and cyclooxygenase-2 in the aggressive phenotype of breast cancer cells. PLoS One. 2010;5:e15540. doi: 10.1371/journal.pone.0015540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kan CF, Singh AB, Stafforini DM, et al. Arachidonic acid downregulates acyl-CoA synthetase 4 expression by promoting its ubiquitination and proteasomal degradation. J Lipid Res. 2014;55:1657–67. doi: 10.1194/jlr.M045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Westerbacka J, Kolak M, Kiviluoto T, et al. Genes involved in fatty acid partitioning and binding, lipolysis, monocyte/macrophage recruitment, and inflammation are overexpressed in the human fatty liver of insulin-resistant subjects. Diabetes. 2007;56:2759–65. doi: 10.2337/db07-0156. [DOI] [PubMed] [Google Scholar]
- 8.Askari B, Kanter JE, Sherrid AM, et al. Rosiglitazone inhibits acyl-CoA synthetase activity and fatty acid partitioning to diacylglycerol and triacylglycerol via a peroxisome proliferator-activated receptor-gamma-independent mechanism in human arterial smooth muscle cells and macrophages. Diabetes. 2007;56:1143–52. doi: 10.2337/db06-0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia J, Zhang Y, Xin L, et al. Global transcriptomic profiling of cardiac hypertrophy and fatty heart induced by long-term high-energy diet in bama miniature pigs. PLoS One. 2015;10:e0132420. doi: 10.1371/journal.pone.0132420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sung YK, Hwang SY, Park MK, et al. Fatty acid-CoA ligase 4 is overexpressed in human hepatocellular carcinoma. Cancer Sci. 2003;94:421–24. doi: 10.1111/j.1349-7006.2003.tb01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung YK, Park MK, Hong SH, et al. Regulation of cell growth by fatty acid-CoA ligase 4 in human hepatocellular carcinoma cells. Exp Mol Med. 2007;39:477–82. doi: 10.1038/emm.2007.52. [DOI] [PubMed] [Google Scholar]
- 12.Wu X, Deng F, Li Y, et al. ACSL4 promotes prostate cancer growth, invasion and hormonal resistance. Oncotarget. 2015;6:44849–63. doi: 10.18632/oncotarget.6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monaco ME, Creighton CJ, Lee P, et al. Expression of long-chain fatty Acyl-CoA synthetase 4 in breast and prostate cancers is associated with sex steroid hormone receptor negativity. Transl Oncol. 2010;3:91–98. doi: 10.1593/tlo.09202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, Li Y, Wang J, et al. Long chain fatty Acyl-CoA synthetase 4 is a biomarker for and mediator of hormone resistance in human breast cancer. PLoS One. 2013;8:e77060. doi: 10.1371/journal.pone.0077060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belkaid A, Ouellette RJ, Surette ME. 17β-estradiol-induced ACSL4 protein expression promotes an invasive phenotype in estrogen receptor positive mammary carcinoma cells. Carcinogenesis. 2017;38:402–10. doi: 10.1093/carcin/bgx020. [DOI] [PubMed] [Google Scholar]
- 16.Yen MC, Kan JY, Hsieh CJ, et al. Association of long-chain acyl-coenzyme A synthetase 5 expression in human breast cancer by estrogen receptor status and its clinical significance. Oncol Rep. 2017;37:3253–60. doi: 10.3892/or.2017.5610. [DOI] [PubMed] [Google Scholar]
- 17.Ye X, Zhang Y, Wang X, et al. Tumor-suppressive functions of long-chain acyl-CoA synthetase 4 in gastric cancer. IUBMB Life. 2016;68:320–27. doi: 10.1002/iub.1486. [DOI] [PubMed] [Google Scholar]
- 18.Kudo Y, Ogawa I, Kitajima S, et al. Periostin promotes invasion and anchorage-independent growth in the metastatic process of head and neck cancer. Cancer Res. 2006;66:6928–35. doi: 10.1158/0008-5472.CAN-05-4540. [DOI] [PubMed] [Google Scholar]