ABSTRACT
Glioblastoma (GBM) is the most prevalent and malignant brain tumor, displaying notorious resistance to conventional therapy, partially due to molecular and genetic heterogeneity. Understanding the mechanisms for gliomagenesis, tumor stem/progenitor cell propagation and phenotypic diversity is critical for devising effective and targeted therapy for this lethal disease. The basic helix-loop-helix transcription factor OLIG2, which is universally expressed in gliomas, has emerged as an important player in GBM cell reprogramming, genotoxic resistance, and tumor phenotype plasticity. In an animal model of proneural GBM, elimination of mitotic OLIG2+ progenitors blocks tumor growth, suggesting that these progenitors are a seeding source for glioma propagation. OLIG2 deletion reduces tumor growth and causes an oligodendrocytic to astrocytic phenotype shift, with PDGFRα downregulation and reciprocal EGFR signaling upregulation, underlying alternative pathways in tumor recurrence. In patient-derived glioma stem cells (GSC), knockdown of OLIG2 leads to downregulation of PDGFRα, while OLIG2 silencing results in a shift from proneural-to-classical gene expression pattern or a proneural-to-mesenchymal transition in distinct GSC cell lines, where OLIG2 appears to regulate EGFR expression in a context-dependent manner. In addition, post-translational modifications such as phosphorylation by a series of protein kinases regulates OLIG2 activity in glioma cell growth and invasive behaviors. In this perspective, we will review the role of OLIG2 in tumor initiation, proliferation and phenotypic plasticity in animal models of gliomas and human GSC cell lines, and discuss the underlying mechanisms in the control of tumor growth and potential therapeutic strategies to target OLIG2 in malignant gliomas.
KEYWORDS: classical, GBM stem cells, glioblastoma, glioma model, mesenchymal, OLIG2, proneural, transcriptome analysis, tumor subtype, tumor phenotype plasticity
Introduction
Glioblastoma multiforme (GBM) is the most common and aggressive malignant primary brain tumor in adults. GBMs are resistant to conventional radiotherapy and chemotherapies, resulting in an average life expectancy of less than 14 months.1,2 These tumors display significant biologic and genetic heterogeneity, which contributes to treatment challenges faced by clinicians.3,4
Much of the tumor heterogeneity of GBMs can be attributed to their distinct genetic alterations, expression programs, tumor cell stages and compositions within the tumors.2,5 GBM may arise from neural stem/progenitor cells (NPCs), astrocytes, oligodendrocyte precursor cells (OPCs), and even neurons, further contributing to its biologic diversity.6−11 Based on gene expression profiles in tumor tissues, GBMs can be classified into 4 distinct molecular subtypes: proneural [oligodendrocyte progenitor (OPC) signature], classical (astrocytic signature), neural (neuronal signature), and mesenchymal (reactive astrocyte and microglia signature).12 The proneural GBM subtype displays characteristic genetic alterations including platelet-derived growth factor receptor α (PDGFRA) amplification, mutations in the tumor suppressor TP53 (p53), and/or isocitrate dehydrogenases (IDH).2,12 The classical subtype is characterized by mutational activation of the epidermal growth factor receptor (EGFR) or by extra copies of EGFR and/or loss of CDKN2A locus.12,13 The mesenchymal subtype is characterized by NF1 deletion, elevated CHI3L1, TLR2/4, and TRADD, and high expression levels of the genes in the TNF-α and NF-κB pathways.12 The GBM phenotype can be both tumor-cell intrinsic, and glioma-cell extrinsic, in which case the tumor microenvironment contributes to the expression signature. Several recent studies suggest that the neural and mesenchymal genetic signature may be attributable to the normal brain tissue and tumor stromal components, respectively, as opposed to the tumor cells themselves.14-17 A mesenchymal microenvironment (i.e. macrophages/microglia) or contamination of glioma samples with stromal tissues could be sufficient to lead to classification as the mesenchymal subtype17 (https://t.co/3sEaUE3yFx). GSCs generated from most mesenchymal GBM tumor tissues, which is composed of a variety of cellular compositions including necrotic components, lose their mesenchymal footprint and acquire a proneural glioma cell signature even in early passages.18,19
Recent single cell transcriptome analysis further reveals various degrees of genetic subtype heterogeneity within a single tumor.4,20 Individual cells within a tumor exhibit diverse transcriptional programs including oncogenic pathways, immune components, and stemness-related expression profiles. Notably, GBM subtype classifiers based on bulk tumor tissues are expressed in different cell populations within a tumor at a variable level,4 indicating intra-tumor heterogeneity in GBM. At present, however, the molecular and cellular mechanisms underlying the tumor phenotype diversity and plasticity are not fully understood.
OLIG2 is a central nervous system (CNS) restricted transcription factor that plays a critical role in glial progenitor proliferation, OPC specification from neural progenitors or their primitive progenitors (pri-OPC) and the OPC-astrocyte fate switch by inhibiting astrocytic differentiation in the developing brain.21−25 OLIG2 is also ubiquitously expressed in gliomas,26,27 and has an important role in gliomagenesis and tumor phenotype plasticity.28−31 Recently, OLIG2 has been identified as one of the core transcription factors together with SOX2, POU3F2 and SALL2 that can reprogram differentiated GBM cells into the glioma stem cells.32 These observations suggest that OLIG2 may sustain tumor growth and regulate phenotypic plasticity, both of which contribute to GBM treatment resistance.
OLIG2+ propagating cells as a seeding source for initiation of proneural-like glioma
In human brain tumors, OLIG2 is present, to various extents, in all grades of pediatric and adult diffuse gliomas including astrocytomas, oligodendrogliomas, and GBMs.26,27,33 It is particularly enriched in the proneural GBM subtype, which possesses a gene expression profile resembling that of OPCs,12 although lower levels of expression are seen in other GBM subtypes.12,34 Notably, a large population of OLIG2+ cells in human gliomas, particularly proneural GBMs, has been found to express a proliferative marker Ki67 and a stem-cell marker CD133, suggesting that proliferative OLIG2+ cells are tumor-propagating cells.28-30
In a glioma mouse model carrying Trp53/Nf1 mutations, mosaic analysis with double markers reveals an expansion of OLIG2+ progenitors or OPCs during glioma initiation,7 suggesting that OLIG2+ OPCs are the primary source of tumor-propagating cells in this mouse model. Consistently, deletion of Nf1/Trp53/Pten or Nf1/Trp53 in adult NG2+ OPCs induced formation of gliomas, which mainly occur in the ventral brain and potentially represent a subset of GBM.11,35 In addition, in a murine model of proneural GBM harboring Pten/Trp53 mutations and PDGFB expression,36,37 the majority of proliferative cells in tumor tissues are OLIG2-positive.28 These observations suggest that OLIG2+ cells are highly proliferative tumor propagating cells in proneural GBMs.12,28,29
To assess whether Olig2+ proliferating cells are required for glioma initiation, an in vivo cell suicide approach has been used to deplete mitotic OLIG2+ progenitors in glioma-forming mice by carrying Olig2-TK, where a ganciclovir (GCV)-inducible suicide gene HSV-TK is knocked in at the Olig2 locus. Elimination of Olig2+ mitotic progenitor cells by GCV treatment in the murine proneural GBM harboring Pten/Trp53 mutations and PDGFB expression at an early phase of gliomagenesis essentially blocks glioma initiation and growth28 (Fig. 1A). In addition, GCV treatment at a late phase of tumorigenesis delays tumor progression and extends the survival rate in the glioma-forming mice. This study provides the first evidence that Olig2+ mitotic progenitors are essential for glioma cell initiation, and progression in a proneural GBM-like animal model. Recent studies indicate that a nestin-expressing quiescent progenitor cell population propagates glioblastoma growth after chemotherapy,38 it would be interesting to determine whether OLIG2+ cells also represent a subset of quiescent progenitors that lead to tumor recurrence after treatment.
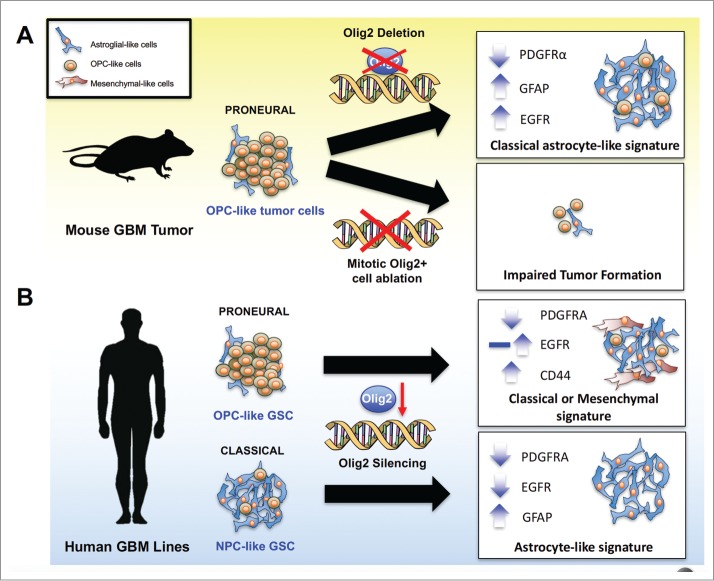
Figure 1.
OLIG2+ mitotic cells and OLIG2 function in glioma cell growth (A) In a murine model of proneural GBM, Olig2 knockout delays tumor growth, and changes the gene expression profile from the proneural to the astrocyte-associated classical phenotype, leading to increased EGFR expression and sensitivity to EGFR inhibitors. In addition, the induced suicide of mitotic OLIG2+ cells blocks tumorigenesis, suggesting an essential role of these cells in glioma initiation. (B) In both proneural and classical human GBM cell lines, OLIG2 knockdown downregulates PDGFRA expression, whereas the effect of OLIG2 silencing on full-length EGFR expression differs by genetic background. In proneural OPC-like GSCs, OLIG2 knockdown may exhibit upregulation or no change in the of EGFR expression, resulting in a classical or mesenchymal phenotype shift. In contrast, silencing of OLIG2 in classical NPC-like GSCs leads to EGFR downregulation and GFAP upregulation, manifesting an astrocyte signature.
The role of OLIG2 in tumor cell growth in animal models of gliomas
The requirement of OLIG2 in gliomagenesis is context-dependent. Expression of a constitutively active EGFRvIII from retrovirus is able to induce neural progenitors from Cdkn2a−/− (a.k.a. Ink4 a/Arf−/−) mouse embryos to form gliomas in allografts in immunodeficient SCID mice, however, neural progenitors from Olig1/2 null Cdkn2a−/− neural progenitors transduced by EGFRvIII do not form gliomas.29 This study suggests that Olig2 is required for glioma formation from neural stem cells under the genetic background of Cdkn2a−/− and EGFRvIII overexpression in allografts. In contrast, gliomas generated endogenously in a mouse model of proneural GBM carrying Trp53 and Pten deletions with PDGFB overexpression, Olig2 deletion slows but does not prevent tumor cell growth.28 The phenotypic differences in tumor growth after Olig2-deletion may be attributable to distinct genetic backgrounds representing classical and proneural subtypes (Cdkn2a−/−; EGFRvIII overexpression vs. Trp53/Pten deletion/PDGFB overexpression) and different animal models (allografts vs. endogenous tumors). The function of OLIG2 in brain tumorigenesis has been shown to be dependent on the mutation status of p53,39 which is frequently mutated in GBM patients.
OLIG2 appears to be critical for the maintenance of tumor-propagating neurospheres or self-renewal with Trp53/Pten deletions/PDGFB overexpression. Tumor cells eventually form an adherent monolayer following Olig2 deletion,28,29 suggesting that OLIG2 is required for tumor stem/progenitor cell maintenance under this context. Consistently, expression of OLIG2, together with SOX2, POU3F2 and SALL2, can reprogram differentiated human GBM cells into stem-like tumor propagating cells (TPCs),32 suggesting that OLIG2 is important for maintaining GSC stemness. Strikingly, despite the lack of tumorsphere formation, Olig2-deleted tumor cells with Trp53/Pten deletions/PDGFB overexpression are able to proliferate in a monolayer culture, display clonogenicity, and form secondary tumors after orthotopic transplantation in immunodeficient mice at a reduced rate.28 These studies suggest a context-dependent OLIG2 function in glioma formation and their growth rate, with alternative pathways sustaining tumor growth in the absence of OLIG2.
OLIG2 regulates oncogenic pathways and PDGFRA signaling for tumor cell growth
Gene expression profiling and genomic occupancy analyses indicate that, besides targeting PDGFRa, OLIG2 targets the enhancers of the cell cycle and oncogenic regulatory genes including Cdca8, Cdc20, Cdc25 c, along with cell proliferation-promoting genes e.g. Aurka, Pim1 and proto-oncogenes c-Myc and c-Jun, much greater in the tumor cells from proneural GBM mouse models than in normal neural stem cells.28 OLIG2 may therefore function as a pioneer factor to regulate chromatin remodeling events in transformed cells, which render the E box accessible to OLIG2 and promote tumor cell growth.40 Besides promoting cell proliferation activators, OLIG2 also appears to oppose the tumor suppressor p53 by direct transcriptional repression of p53-induced cell cycle inhibitor p21,29 which may underlie blunting of the p53-mediated response to genotoxic damage to sustain tumor cell growth.39
Consistent with these murine model observations, OLIG2 expression is positively correlated with PDGFRA among proneural GBMs in the Cancer Genome Atlas (TCGA) data set.12 Interestingly, phylogenic modeling of GBM samples in the TCGA has identified gain of chromosome 7, which harbors a PDGFRA ligand, PDGFA, as a potential driving event in human proneural-like GBMs without GCIMP during tumor evolution.41
In multiple human proneural GSC cell lines, OLIG2-knockdown leads to downregulation of PDGFRα signaling.28,31 Similarly, Olig2-deleted tumors in a murine proneural GBM model exhibit downregulation of PDGFRα expression both in vitro and in vivo.28 This suggests a critical role of OLIG2-activated PDGFRα signaling in proneural glioma growth, consistent with the developmental role of OLIG2 in PDGFRα signaling and maintenance of the OPC state.21,22
OLIG2 regulates tumor phenotype plasticity
Deletion of Olig2 in the murine proneural GBM model carrying p53/Pten deletion and PDGFB expression leads to a gene expression signature shift from the proneural to the classical tumor pattern based on transcriptome profiling analysis, manifested by an elevation of astrocytic signature genes and EGFR, and a decrease in OPC-related proneural signature genes28 (Fig. 1A). Thus, despite downregulated PDGFRα signaling, Olig2-deleted tumor cells appear to reciprocally utilize EGFR signaling as an alternative pathway to maintain tumor cell growth. Upregulation of EGFR is in keeping with the role of OLIG2 in the OPC-astrocyte fate switch,21,24,42 wherein OLIG2 suppresses expression of the astrocyte-enriched genes like GFAP and NFIa.42−45 It is worth noting that Olig2 deletion does not necessarily cause a glioma subtype conversion. Rather, Olig2 deletion leads to a transcriptional program shift with apparent upregulation of astrocytic classical-like GBM signature, while downregulation of OPC-associated proneural GBM signature in the animal glioma model.28 However, overexpression of activated EGFR has been shown to promote neural stem/progenitor cells or OPC expansion during development46-48 and cancer10,49,50 with no clear links to astrocytic development; thus, it is also possible that Egfr may be upregulated in in NPC-like tumor cells or pre-OPCs in tumors with Trp53/Pten deletions and PDGFB overexpression following Olig2 deletion.
Silencing or deletion of OLIG2 with shRNA or genomic deletion in an array of human proneural GBM lines also results in upregulation of EGFR and the astrocytic signature28 (Kosty and Lu, unpublished) (Fig. 1B). Consistent with the murine glioma models carrying Trp53/Pten deletion,28 the gene set expression analyses suggest that OLIG2 deletion engenders a tumor phenotype shift from a proneural to an astrocytic gene expression pattern, including upregulation of EGFR and GFAP. Thus, OLIG2 has a role in maintaining the OPC-signature associated proneural phenotype in GSCs.
Recently, a feed-forward loop between OLIG2 and EGFR was observed to sustain pro-mitogenic functions in tumor cell growth in neural progenitor cells from a murine glioma model with EGFRvIII overexpression and Cdkn2 anull background.31,51 Lentivirus-mediated knockdown of OLIG2 in human classical GSC lines led to a downregulation of EGFR (Fig. 1B). In contrast, the full-length EGFR transcript was expressed at low levels in proneural GSC lines at baseline, and exhibited little change with OLIG2 knockdown.31 It is noteworthy that EGFR expression can be detected in a variety of GSCs without OLIG2 expression32,52 (J.K. Q.L., unpublished observations). Some GSC lines with no or minimal OLIG2 expression express high levels of EGFR and are able to proliferate rapidly (J.K, Q.L., R.K., and S.M. unpublished observations),19,35 suggesting EGFR expression is not fully dependent on OLIG2. Conversely, even though RTK signaling is required for the activation of core transcriptional regulators such as OLIG2 and SOX2 in established GBM, once activated the maintenance of their expression becomes independent of the RTK signaling51 suggesting that relationship between expression of RTKs and OLIG2 may be stage- or cellular context-dependent, and is far more complex than previously thought.
Intriguingly, OLIG2 knockdown in a set of proneural GSC lines results in upregulation of the mesenchymal marker CD44 as well as an increase in a set of the markers associated with the mesenchymal GBM subtype (e.g., TGFB1, STAT3, FN1, CTGF, and CHI3L1).31 These data suggest that absence of OLIG2 leads to a proneural-to-mesenchymal transition in the GBM cells.31 It is worth noting that an array of markers such as CD44, CHI3L1, STAT3 and CTGF examined after OLIG2 knockdown are also highly enriched in the astrocyte lineage,45 which may be representative of reactive astrocytic features within the tissue of mesenchymal subtype tumors12 and in keeping with upregulation of astrocytic signature expression pattern in the Olig2-ablated glioma carrying Pten;Trp53 deletion and PDGFB overexpression.28 Nonetheless, whole transcriptome analysis of gene expression patterns would need to further define the tumor subtype change in the absence of OLIG2 in GSCs.
Discrepancies in the relationship between OLIG2 and EGFR in different GSC cell lines and animal models may in part be due to different mutations and cell types examined. In murine glioma models, upregulated EGFR expression is detected in Olig2-deleted tumors derived from the progenitors in the adult white matter carrying Pten;Trp53 deletion and PDGFB overexpression, wherein the tumor cells are the OPC-enriched proneural subtype. The tumors formed in the absence of Olig2 in this model potentially represent astrocytes converted or transformed from OPCs. In contrast, EGFR appears to be downregulated in Olig2 null NPCs isolated from the embryonic ganglionic eminences with or without the Cdkn2a−/−;EGFRvIII mutation (a model for classical GBM). While Olig2 can be detected in uncommitted SVZ progenitors of the developing forebrain, it has been recently demonstrated that many of these co-express both EGFR and OLIG2 in the germinal matrix of the developing human brain.53 As a discrete analysis of these populations following Olig2 embryonic deletion has never been completed, it would be interesting to know if Olig2 is also essential for EGFR expression in the NPCs of embryonic eminence regions during brain development. Recent single-cell RNA-seq studies54 has further demonstrated that atop the cellular hierarchy of oligodendroglioma resides cells with an expression profile reminiscent of embryonic radial glia, notably, these also express high amounts of OLIG2 and both RTKs (EGFR, PDGFRA).
A systematic survey of OLIG2 requirement for maintaining EGFR expression remains to be defined in brain tumor cell types. On a genomic level, OLIG2, along with a histone mark H3K27ac, binds to Egfr regulatory elements in murine NPCs,55 murine proneural tumors,28 human GSCs,32 and human fetal-derived NSCs (R.K., S.M. unpublished). Notably, levels of the activating histone mark H3K27ac on the multiple enhancer elements of the Egfr locus55 are elevated in Olig2-knockout tumors compared with control OLIG2+ tumors,28 suggesting a repressor function of OLIG2 on Egfr expression in murine proneural tumor cells carrying Pten;Trp53 deletion.
Collectively, the disparate observations suggest a context-dependent function of OLIG2 regarding EGFR expression, while OLIG2 is consistently required for PDGFRα expression in tumor cells regardless of tumor subtype. Further analysis of a larger cohort of GBM cells lines is required to address the effect of the OLIG2 loss on tumor phenotype transition and plasticity.
It is worth noting that the proneural to mesenchymal transition is often associated with more aggressive GBMs with regard to cell invasiveness and drug resistance.12,34,56,57 The capacity for the OLIG2-knockdown GSC cells to exhibit these mesenchymal characteristics, such as increased invasion and drug resistance, remain to be determined, although OLIG2 knockdown leads to a reduction of tumor growth rate in these human GSC cell lines.28,31 Nonetheless, multiple unaccounted-for heterogeneities in tumor stem cell properties and their fitness with a tumor tissue with diverse cellular compositions and genetic mutations may be contributable to subtype conversion and phenotypic changes in response to the OLIG2 state.
Modification of Olig2 activity for glioma therapy
Olig2 deletion in murine glioma carrying Pten/Trp53 mutations and PDGFB expression leads to a delay in the onset and progression of tumors, however, the tumor cells are able to grow eventually. Transcriptome profiling analysis identifies an upregulation of EGFR signaling pathway in the tumor cells in the absence of Olig2, despite the downregulation of PDGFRα signaling. Consistently, treatment of Olig2-deleted tumor cells with EGFR inhibitors blocked tumor cell proliferation and induced apoptosis, suggesting that Olig2 loss sensitizes glioma cells to EGFR inhibition.28 Inhibition of OLIG2 expression may therefore be useful to enhance the efficacy of EGFR inhibitors. In addition, OLIG2 appears to oppose p53 responses to genotoxic damage in malignant gliomas by transcriptionally repressing a p53-regulated effector p2129,39, suggesting that OLIG2 confers radiation and chemo-resistance in glioma cells. These observations provide a proof of principle to stratify therapies among distinct subtypes of malignant gliomas by inducing differential patient responses to a treatment through targeting OLIG2.
Phosphorylation of a highly conserved triple serine motif in the N-terminus of OLIG2 at S10, S13, and S14 has also been shown to be important for regulation of glioma cell proliferation and invasion.30,58,59 Hyper-phosphorylated OLIG2, mimicked by a triple phosphomimetic (TPM) variant, results in greater cell proliferation, while a triple phosphonull (TPN) variant results in impaired cell proliferation.58 This is partially due to attenuation of p53-mediated responses by both wildtype and phosphomimetic forms of OLIG2.58 Although the TPN mutant OLIG2 results in decreased cell proliferation, it is associated with increased invasiveness of murine neural stem cells carrying a Cdkn2 a deletion and EGFRvIII overexpression and human GSC cells.30 Similarly, human GSC cells with low levels of OLIG2 phosphorylation (pOLIG2low) exhibit an increase in tumor invasion both in culture and in xenografts compared with those with OLIG2 hyper-phosphorylation. Tumors generated from OLIG2TPN mutants and pOLIG2low GSC cells show an increase in expression of genes associated with invasion including TWIST1, CD33, TFGβ2, CREB, and ZEB1.
A recent series of elegant experiments, 3 serine/threonine protein kinases, glycogen synthase kinase 3α/β (GSK3α/β), casein kinase 2 (CK2) and cyclin-dependent kinase 1/2 (CDK1/2), have been identified as necessary and sufficient to phosphorylate the S10, S13, and S14 residues in OLIG2, respectively.59 In the proposed sequentially priming model, phosphorylation of S13 and S10 by GSK3 and CK2, respectively, is contingent upon phosphorylation of S14 by CDK1/2, which initiates a phosphorylation cascade that also includes 3 additional serine residues, S3, S6, and S9, hence forming a hexa-phosphoserine “acid blob,” which can oppose the actions of p53. Inhibition of GSK3α/β, CK2, and CDK1/2 in murine NPCs by treatment with small molecule inhibitors results in elevation of a p53-target p21, consistent with a repressive function of OLIG2 on p21 expression. Interestingly, peripheral administration of CK2 inhibitor CX-4945, capable of blood-brain barrier penetration, to a pediatric astrocytoma mouse model harboring BRAFV600E and deletion of Ink4 a/ARF, decreases the level of phosphorylated OLIG2. Glioma cells treated CX-4945 exhibit significantly lower cell proliferation rates. In addition, concurrent treatment of CX-4945 with a BRAF antagonist PLX-4720 improved the survival of orthotopically transplanted animals harboring BRAFV600E and Ink4 a/ARF deletion. It would be interesting to determine if the inhibition of OLIG2 phosphorylation by these upstream kinases would also alter tumor cell invasiveness. Blockade of OLIG2 expression or activity though kinase inhibition, together with other oncogenic pathway inhibitors, may provide a promising avenue for the development or discovery of novel therapeutic agents in the treatment of GBM.
Conclusions
Much work is still needed to fully understand the biologic events that lead to the development, progression and drug resistance of glioma stem/initiating cells. OLIG2 likely plays an important role in the growth of at least a subset of GBMs, though the relationship between OLIG2 and tumor growth and phenotypes in primary and recurrent GBM may be context-dependent. Further studies are needed to elucidate the mechanisms underlying OLIG2 activity and its posttranslational modifications or interacting partners in the regulation of GBM formation, growth, and invasion. Given that OLIG2 is critical for glioma initiation and progression, modulation of OLIG2 expression or activity by its upstream kinases, in conjunction with radiotherapy and targeted therapeutics, may potentially improve the treatment efficacy in malignant gliomas.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to Ravinder Verma for providing unpublished information, Rohit Rao for contribution in initial manuscript preparation, and Edward Hurlock for critical comments.
Funding
This study was funded in part by grants from the Mayfield Education and Research Foundation to JK and Molecular Therapeutics Program to QRL, and the US National Institutes of Health (R01 NS078092) to QRL and (R01 NS088648) to SM.
References
- [1].Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9:717-26. doi: 10.1016/S1474-4422(10)70105-8. PMID:20610347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al.. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462-77. doi: 10.1016/j.cell.2013.09.034. PMID:24120142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al.. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807-12. doi: 10.1126/science.1164382. PMID:18772396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, et al.. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396-401. doi: 10.1126/science.1254257. PMID:24925914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al.. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318-25. doi: 10.1038/nature08712. PMID:20032975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080-4. doi: 10.1126/science.1226929. PMID:23087000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, et al.. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146:209-21. doi: 10.1016/j.cell.2011.06.014. PMID:21737130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005;8:119-30. doi: 10.1016/j.ccr.2005.07.004. PMID:16098465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15:45-56. doi: 10.1016/j.ccr.2008.12.006. PMID:19111880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Persson AI, Petritsch C, Swartling FJ, Itsara M, Sim FJ, Auvergne R, Goldenberg DD, Vandenberg SR, Nguyen KN, Yakovenko S, et al.. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18:669-82. doi: 10.1016/j.ccr.2010.10.033. PMID:21156288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Galvao RP, Kasina A, McNeill RS, Harbin JE, Foreman O, Verhaak RGW, Nishiyama A, Miller CR, Zong H. Transformation of quiescent adult oligodendrocyte precursor cells into malignant glioma through a multistep reactivation process. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4214-E23. doi: 10.1073/pnas.1414389111. PMID:25246577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al.. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98-110. doi: 10.1016/j.ccr.2009.12.020. PMID:20129251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hayden EC. Genomics boosts brain-cancer work. Nature. 2010;463:278. doi: 10.1038/463278a. PMID:20090720 [DOI] [PubMed] [Google Scholar]
- [14].Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, Mellano A, Senetta R, Cassenti A, Sonetto C, et al.. Stromal contribution to the colorectal cancer transcriptome. Nat Genet. 2015;47:312-9. doi: 10.1038/ng.3224. PMID:25706627 [DOI] [PubMed] [Google Scholar]
- [15].Martinez E, Yoshihara K, Kim H, Mills GM, Trevino V, Verhaak RG. Comparison of gene expression patterns across 12 tumor types identifies a cancer supercluster characterized by TP53 mutations and cell cycle defects. Oncogene. 2015;34:2732-40. doi: 10.1038/onc.2014.216. PMID:25088195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, Sevillano M, Palomo-Ponce S, Tauriello DV, Byrom D, et al.. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320-9. doi: 10.1038/ng.3225. PMID:25706628 [DOI] [PubMed] [Google Scholar]
- [17].Kim H, Verhaak RG. Transcriptional mimicry by tumor-associated stroma. Nat Genet. 2015;47:307-9. doi: 10.1038/ng.3255. PMID:25814306 [DOI] [PubMed] [Google Scholar]
- [18].Eidel O, Burth S, Neumann JO, Kieslich PJ, Sahm F, Jungk C, Kickingereder P, Bickelhaupt S, Mundiyanapurath S, Baumer P, et al.. Tumor Infiltration in Enhancing and Non-Enhancing Parts of Glioblastoma: A Correlation with Histopathology. PLoS One. 2017;12:e0169292. doi: 10.1371/journal.pone.0169292. PMID:28103256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bhat KP, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, et al.. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331-46. doi: 10.1016/j.ccr.2013.08.001. PMID:23993863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Meyer M, Reimand J, Lan X, Head R, Zhu X, Kushida M, Bayani J, Pressey JC, Lionel AC, Clarke ID, et al.. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci U S A. 2015;112:851-6. doi: 10.1073/pnas.1320611111. PMID:25561528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75-86. doi: 10.1016/S0092-8674(02)00678-5. PMID:11955448 [DOI] [PubMed] [Google Scholar]
- [22].Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61-73. doi: 10.1016/S0092-8674(02)00677-3. PMID:11955447 [DOI] [PubMed] [Google Scholar]
- [23].Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157-63. doi: 10.1016/S0960-9822(02)00926-0. PMID:12121626 [DOI] [PubMed] [Google Scholar]
- [24].Zhu X, Zuo H, Maher BJ, Serwanski DR, LoTurco JJ, Lu QR, Nishiyama A. Olig2-dependent developmental fate switch of NG2 cells. Development. 2012;139:2299-307. doi: 10.1242/dev.078873. PMID:22627280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhang L, He X, Liu L, Jiang M, Zhao C, Wang H, He D, Zheng T, Zhou X, Hassan A, et al.. Hdac3 Interaction with p300 Histone Acetyltransferase Regulates the Oligodendrocyte and Astrocyte Lineage Fate Switch. Dev Cell. 2016;36:316-30. doi: 10.1016/j.devcel.2016.01.002. PMID:26859354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lu QR, Park JK, Noll E, Chan JA, Alberta J, Yuk D, Alzamora MG, Louis DN, Stiles CD, Rowitch DH, et al.. Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc Natl Acad Sci U S A. 2001;98:10851-6. doi: 10.1073/pnas.181340798. PMID:11526205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ligon KL, Alberta JA, Kho AT, Weiss J, Kwaan MR, Nutt CL, Louis DN, Stiles CD, Rowitch DH. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63:499-509. doi: 10.1093/jnen/63.5.499. PMID:15198128 [DOI] [PubMed] [Google Scholar]
- [28].Lu F, Chen Y, Zhao C, Wang H, He D, Xu L, Wang J, He X, Deng Y, Lu EE, et al.. Olig2-Dependent Reciprocal Shift in PDGF and EGF Receptor Signaling Regulates Tumor Phenotype and Mitotic Growth in Malignant Glioma. Cancer Cell. 2016;29:669-83. doi: 10.1016/j.ccell.2016.03.027. PMID:27165742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, Bachoo RM, Kane M, Louis DN, Depinho RA, et al.. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53:503-17. doi: 10.1016/j.neuron.2007.01.009. PMID:17296553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Singh SK, Fiorelli R, Kupp R, Rajan S, Szeto E, Lo Cascio C, Maire CL, Sun Y, Alberta JA, Eschbacher JM, et al.. Post-translational Modifications of OLIG2 Regulate Glioma Invasion through the TGF-beta Pathway. Cell Rep. 2016;16:950-66. doi: 10.1016/j.celrep.2016.06.045. PMID:27396340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kupp R, Shtayer L, Tien AC, Szeto E, Sanai N, Rowitch DH, Mehta S. Lineage-Restricted OLIG2-RTK Signaling Governs the Molecular Subtype of Glioma Stem-like Cells. Cell Rep. 2016;16:2838-45. doi: 10.1016/j.celrep.2016.08.040. PMID:27626655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, Rabkin SD, Riggi N, Chi AS, Cahill DP, Nahed BV, et al.. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580-94. doi: 10.1016/j.cell.2014.02.030. PMID:24726434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Otero JJ, Rowitch D, Vandenberg S. OLIG2 is differentially expressed in pediatric astrocytic and in ependymal neoplasms. J Neurooncol. 2011;104:423-38. doi: 10.1007/s11060-010-0509-x. PMID:21193945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et al.. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157-73. doi: 10.1016/j.ccr.2006.02.019. PMID:16530701 [DOI] [PubMed] [Google Scholar]
- [35].Alcantara Llaguno SR, Wang Z, Sun D, Chen J, Xu J, Kim E, Hatanpaa KJ, Raisanen JM, Burns DK, Johnson JE, et al.. Adult Lineage-Restricted CNS Progenitors Specify Distinct Glioblastoma Subtypes. Cancer Cell. 2015;28:429-40. doi: 10.1016/j.ccell.2015.09.007. PMID:26461091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lei L, Sonabend AM, Guarnieri P, Soderquist C, Ludwig T, Rosenfeld S, Bruce JN, Canoll P. Glioblastoma models reveal the connection between adult glial progenitors and the proneural phenotype. PLoS One. 2011;6:e20041. doi: 10.1371/journal.pone.0020041. PMID:21625383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sonabend AM, Bansal M, Guarnieri P, Lei L, Amendolara B, Soderquist C, Leung R, Yun J, Kennedy B, Sisti J, et al.. The transcriptional regulatory network of proneural glioma determines the genetic alterations selected during tumor progression. Cancer Res. 2014;74:1440-51. doi: 10.1158/0008-5472.CAN-13-2150. PMID:24390738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522-6. doi: 10.1038/nature11287. PMID:22854781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mehta S, Huillard E, Kesari S, Maire CL, Golebiowski D, Harrington EP, Alberta JA, Kane MF, Theisen M, Ligon KL, et al.. The central nervous system-restricted transcription factor Olig2 opposes p53 responses to genotoxic damage in neural progenitors and malignant glioma. Cancer Cell. 2011;19:359-71. doi: 10.1016/j.ccr.2011.01.035. PMID:21397859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, et al.. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248-61. doi: 10.1016/j.cell.2012.12.006. PMID:23332759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, Charles N, Michor F, Holland EC. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26:288-300. doi: 10.1016/j.ccr.2014.06.005. PMID:25117714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR. A crucial role for Olig2 in white matter astrocyte development. Development. 2007;134:1887-99. doi: 10.1242/dev.02847. PMID:17428828 [DOI] [PubMed] [Google Scholar]
- [43].Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953-68. doi: 10.1016/j.neuron.2006.11.019. PMID:17178400 [DOI] [PubMed] [Google Scholar]
- [44].Glasgow SM, Zhu W, Stolt CC, Huang TW, Chen F, LoTurco JJ, Neul JL, Wegner M, Mohila C, Deneen B. Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci. 2014;17:1322-9. doi: 10.1038/nn.3790. PMID:25151262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, et al.. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Aguirre A, Dupree JL, Mangin JM, Gallo V. A functional role for EGFR signaling in myelination and remyelination. Nat Neurosci. 2007;10:990-1002. doi: 10.1038/nn1938. PMID:17618276 [DOI] [PubMed] [Google Scholar]
- [47].Aguirre A, Rubio ME, Gallo V. Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature. 2010;467:323-7. doi: 10.1038/nature09347. PMID:20844536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gonzalez-Perez O, Romero-Rodriguez R, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Epidermal growth factor induces the progeny of subventricular zone type B cells to migrate and differentiate into oligodendrocytes. Stem Cells. 2009;27:2032-43. doi: 10.1002/stem.119. PMID:19544429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sugiarto S, Persson AI, Munoz EG, Waldhuber M, Lamagna C, Andor N, Hanecker P, Ayers-Ringler J, Phillips J, Siu J, et al.. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell. 2011;20:328-40. doi: 10.1016/j.ccr.2011.08.011. PMID:21907924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ivkovic S, Canoll P, Goldman JE. Constitutive EGFR signaling in oligodendrocyte progenitors leads to diffuse hyperplasia in postnatal white matter. J Neurosci. 2008;28:914-22. doi: 10.1523/JNEUROSCI.4327-07.2008. PMID:18216199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Singh DK, Kollipara RK, Vemireddy V, Yang XL, Sun Y, Regmi N, Klingler S, Hatanpaa KJ, Raisanen J, Cho SK, et al.. Oncogenes Activate an Autonomous Transcriptional Regulatory Circuit That Drives Glioblastoma. Cell Rep. 2017;18:961-76. doi: 10.1016/j.celrep.2016.12.064. PMID:28122245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Caren H, Stricker SH, Bulstrode H, Gagrica S, Johnstone E, Bartlett TE, Feber A, Wilson G, Teschendorff AE, Bertone P, et al.. Glioblastoma Stem Cells Respond to Differentiation Cues but Fail to Undergo Commitment and Terminal Cell-Cycle Arrest. Stem Cell Reports. 2015;5:829-42. doi: 10.1016/j.stemcr.2015.09.014. PMID:26607953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Tome-Garcia J, Tejero R, Nudelman G, Yong RL, Sebra R, Wang H, Fowkes M, Magid M, Walsh M, Silva-Vargas V, et al.. Prospective Isolation and Comparison of Human Germinal Matrix and Glioblastoma EGFR+ Populations with Stem Cell Properties. Stem Cell Reports. 2017;8:1421-9. doi: 10.1016/j.stemcr.2017.03.019. PMID:28434940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K, Fisher JM, Rodman C, Mount C, Filbin MG, et al.. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309-13. doi: 10.1038/nature20123. PMID:27806376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Mateo JL, van den Berg DL, Haeussler M, Drechsel D, Gaber ZB, Castro DS, Robson P, Crawford GE, Flicek P, Ettwiller L, et al.. Characterization of the neural stem cell gene regulatory network identifies OLIG2 as a multifunctional regulator of self-renewal. Genome Res. 2015;25:41-56. doi: 10.1101/gr.173435.114. PMID:25294244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol. 2014;232:165-77. doi: 10.1002/path.4282. PMID:24114756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Piao Y, Liang J, Holmes L, Henry V, Sulman E, de Groot JF. Acquired resistance to anti-VEGF therapy in glioblastoma is associated with a mesenchymal transition. Clin Cancer Res. 2013;19:4392-403. doi: 10.1158/1078-0432.CCR-12-1557. PMID:23804423 [DOI] [PubMed] [Google Scholar]
- [58].Sun Y, Meijer DH, Alberta JA, Mehta S, Kane MF, Tien AC, Fu H, Petryniak MA, Potter GB, Liu Z, et al.. Phosphorylation state of Olig2 regulates proliferation of neural progenitors. Neuron. 2011;69:906-17. doi: 10.1016/j.neuron.2011.02.005. PMID:21382551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhou J, Tien AC, Alberta JA, Ficarro SB, Griveau A, Sun Y, Deshpande JS, Card JD, Morgan-Smith M, Michowski W, et al.. A Sequentially Priming Phosphorylation Cascade Activates the Gliomagenic Transcription Factor Olig2. Cell Rep. 2017;18:3167-77. doi: 10.1016/j.celrep.2017.03.003. PMID:28355568 [DOI] [PMC free article] [PubMed] [Google Scholar]