Abstract
Squamous cell carcinoma–related oncogene (SCCRO)/DCUN1D1, a component of the neddylation E3 complex, regulates the activity of the cullin–RING–ligase type of ubiquitination E3s by promoting neddylation of cullin family members. Studies have shown that SCCRO regulates proliferation in vitro and in vivo. Here we show that inactivation of SCCRO results in prolonged mitotic time because of delayed and/or failed abscission. The effects of SCCRO on abscission involve its role in neddylation and localization of Cul3 to the midbody. The Cul3 adaptor KLHL21 mediates the effects of SCCRO on abscission, as it fails to localize to the midbody in SCCRO-deficient cells during abscission, and its inactivation resulted in phenotypic changes identical to SCCRO inactivation. Ubiquitination-promoted turnover of Aurora B at the midbody was deficient in SCCRO- and KLHL21-deficient cells, suggesting that it is the target of Cul3KLHL21 at the midbody. Correction of abscission delays in SCCRO-deficient cells with addition of an Aurora B inhibitor at the midbody stage suggests that Aurora B is the target of SCCRO-promoted Cul3KLHL21 activity. The activity of other Cul3-anchored complexes, including Cul3KLHL9/KLHL13, was intact in SCCRO-deficient cells, suggesting that SCCRO selectively, rather than collectively, neddylates cullins in vivo. Combined, these findings support a model in which the SCCRO, substrate, and substrate adaptors cooperatively provide tight control of neddylation and cullin–RING–ligase activity in vivo.
Keywords: cell cycle, inhibitor, mitosis, substrate specificity, ubiquitylation (ubiquitination)
Introduction
Neddylation is a process analogous to ubiquitination in which a tripartite enzymatic cascade results in covalent modification of substrates by the ubiquitin-like protein Nedd8. In contrast to ubiquitination, only a limited number of proteins are subject to neddylation, with the cullin protein family (Cul1, Cul2, Cul3, Cul4, Cul5, and Cul7) the best-characterized targets (1). Cullins serve as the scaffold for assembly of cullin–RING–ligase (CRL)3 type E3 ligases, the most common type of ubiquitination E3s (2, 3). Neddylation of cullins promotes assembly of the CRL complex and optimizes its conformation to allow efficient transfer of ubiquitin from E2 to the substrate protein (4, 5).
Neddylation is thought to regulate the activity of CRLs. However, relatively little is known about the mechanisms by which neddylation is activated or how cullins are selectively neddylated in vivo. Genetic (E1, E2) or pharmaceutical (E1 inhibition with MLN4924) inactivation of core neddylation components typically has broad effects on cullin neddylation in vitro and in vivo, suggesting that they are not regulatory components. We and others identified SCCRO/DCUN1D1 and showed that it functions as a critical component of the neddylation E3 complex (6–10). Biochemical and in vitro analyses show that SCCRO promotes neddylation by enhancing recruitment of E2∼Nedd8 (Ubc12∼Nedd8) thioester to the complex and optimizes the orientation of proteins in the complex to allow efficient transfer of Nedd8 from E2 to the cullin substrates (8). Similar to other core components, SCCRO promotes the neddylation of all cullin family members, albeit with different efficiency (11, 12). Although SCCRO enhances reaction efficacy, it is not required for neddylation in vitro. In contrast, SCCRO plays an essential role in neddylation in vivo by promoting nuclear translocation of cullin–ROC1 complexes, where neddylation is thought to occur (9). Although inactivation of other core components of neddylation in model organisms is lethal, inactivation of SCCRO is not lethal in mice or flies (9, 13). Moreover, the effect of SCCRO on neddylation in vivo appears to be more selective than that of other core components, with the targeted inactivation of SCCRO variably and only partially reducing neddylation of individual cullins. These findings suggest that SCCRO may play a regulatory role in neddylation.
We recently showed that SCCRO affects proliferation in vitro and in vivo (13). Consistent with the involvement of neddylation in proliferation, treatment with MLN2924, a neddylation E1 inhibitor, induces cell cycle arrest in normal and cancerous cells (14–16). We used proliferation as a model to study the effects of SCCRO on neddylation dynamics in vivo. Several different CRLs play roles in cell cycle progression, imparting their effects through ubiquitination of essential regulators. Cul1 (Cul1SKP2, Cul1β-TRCP)-, Cul2-, Cul3-, Cul4A-, and Cul4B-anchored complexes have been shown to play essential roles at various stages of cell cycle progression (17, 18). We found that the effect of SCCRO on proliferation primarily involves its role in cytokinesis during abscission. Furthermore, these effects involve neddylation-based regulation of localization, assembly of the Cul3KLHL21 complex, and ubiquitination-promoted turnover of Aurora B. These findings suggest that SCCRO activity may play a coregulatory role with substrate adaptors to provide specificity to the neddylation pathway.
Results
Targeted disruption of SCCRO results in a defect in mitosis
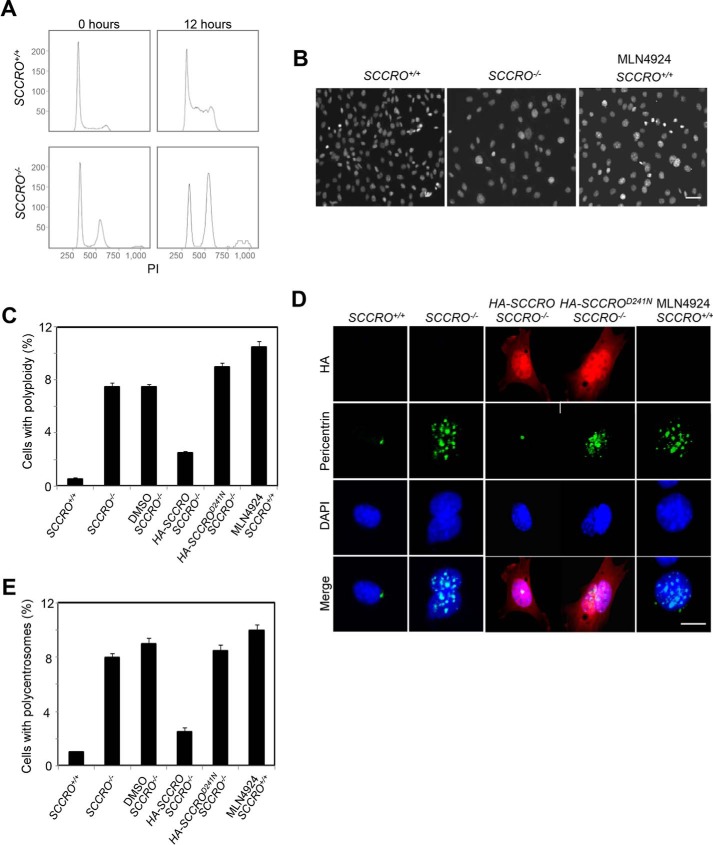
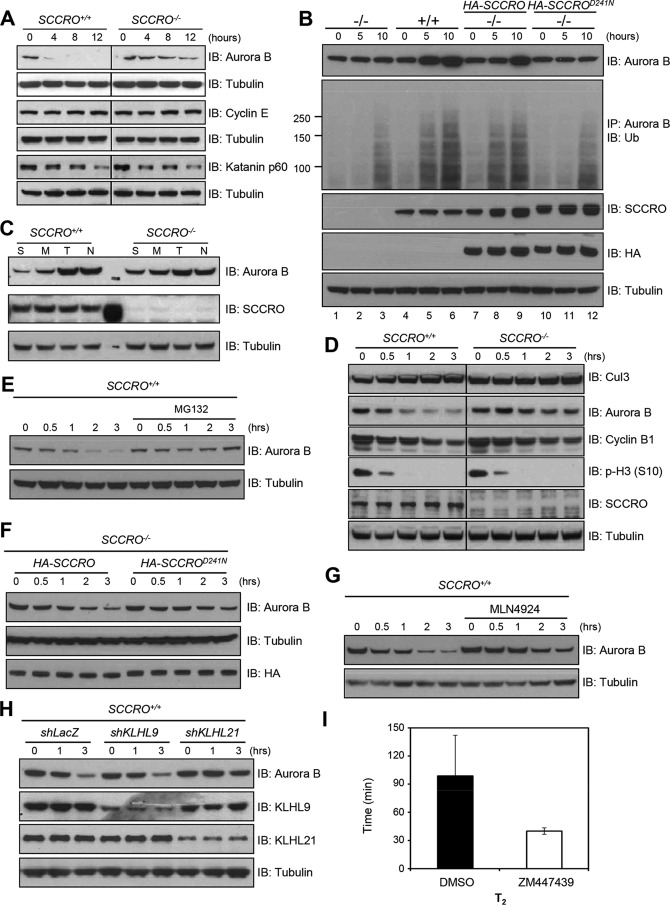
We previously reported that SCCRO−/− mice are runted, which was attributed to a decrease in cell proliferation rather than to alterations in cell size (9). Proliferation defects were observed in SCCRO−/− mouse embryonic fibroblasts (MEFs), which were rescued by re-expression of SCCRO by transfection. Flow cytometry using propidium iodide staining for DNA content indicated an increased >4N fraction in SCCRO−/− MEFs compared with wild-type MEFs (Fig. 1A). Immunostaining with DAPI and anti-pericentrin confirmed the increase in DNA content and disclosed an increase in polycentrosomy in SCCRO−/− MEFs compared with wild-type MEFs (Fig. 1, B–E), suggesting the presence of a mitotic defect. Ploidy and centrosome defects were rescued in SCCRO−/− MEFs by re-expression of SCCRO but not SCCROD241A, a mutant deficient in neddylation activity (Fig. 1, C–E), suggesting that neddylation activity is required for the effect of SCCRO on mitosis. Treatment of SCCRO+/+ MEFs with MLN4924 (Active BioChem) resulted in mitotic delays and phenotypic changes similar to those seen in SCCRO−/− MEFs, confirming the requirement for neddylation activity (Fig. 1, A–E).
Figure 1.
Targeted disruption of SCCRO results in a defect in mitosis. A, flow cytometry analysis (propidium iodide (PI) staining) on MEFs released from synchronization by serum starvation, showing polyploid accumulated in SCCRO−/− MEFs. B, DAPI staining for DNA content confirmed polyploid accumulated in SCCRO−/− MEFs and MLN4924-treated (0.5 μm for 12 h) SCCRO+/+ MEFs. A representative result from staining of SCCRO+/+ MEFs shows an absence of polyploidy. Scale bar = 20 μm. C, quantification of polyploidy in SCCRO+/+ and SCCRO−/− MEFs, SCCRO−/− MEFs infected with a retrovirus carrying SCCRO or SCCROD241N cDNA, and SCCRO+/+ MEFs treated with MLN4924. Polyploidy in SCCRO−/− MEFs could be rescued by overexpression of HA-SCCRO but not the neddylation-dead mutant HA-SCCROD241N. D, immunofluorescence using α-pericentrin antibody, showing normal centrosome numbers and localization in SCCRO+/+ but malpositioned and supernumerary centrosomes in SCCRO−/− MEFs and MLN4924-treated SCCRO+/+ MEFs. The centrosome defect in SCCRO−/− MEFs could be rescued by overexpression of HA-SCCRO but not HA-SCCROD241N. Scale bar = 5 μm. E, quantification of the percentage of polycentrosome cells for C.
SCCRO plays a role in abscission
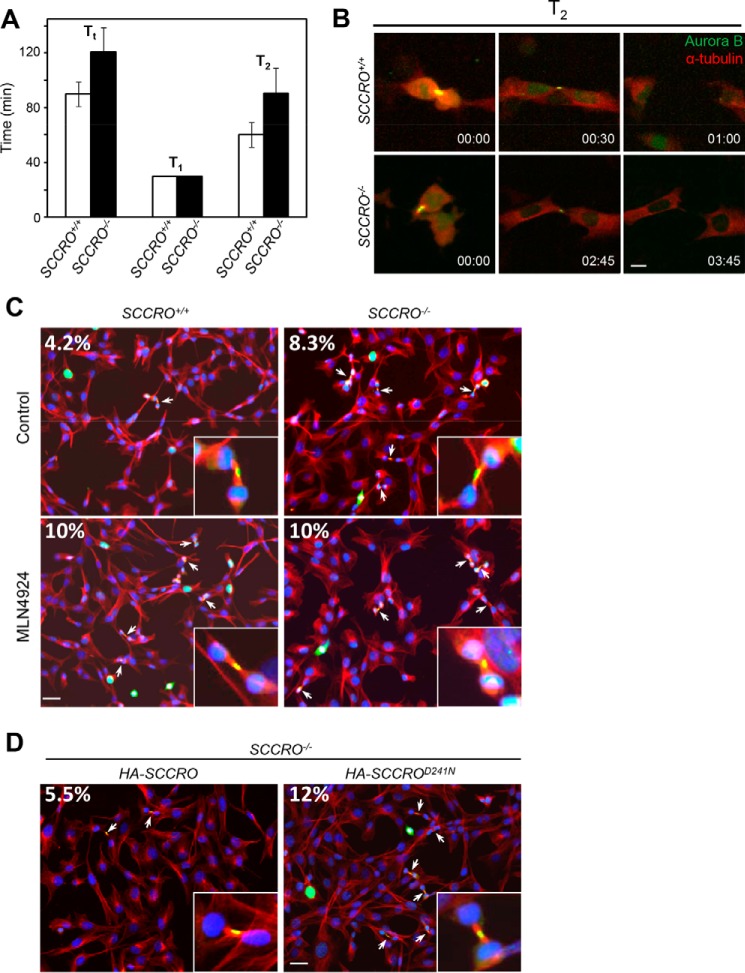
To begin to determine the cause of polyploidy and polycentrosomy in SCCRO−/− cells, we assessed mitotic progression and fidelity using live-cell time-lapse imaging of MEFs stably expressing mCherry–α-tubulin (a major constituent of microtubules) and Aurora B–EGFP (a midbody marker used to assess mitotic progression). Cells were monitored for 24 h using confocal microscopy, and total mitotic duration was assessed as Tt (time from the onset of prophase to the completion of cytokinesis) for individual cells (at least 60 full mitotic events were assessed for each group). We found that Tt was significantly longer in SCCRO−/− MEFs (120.7 ± 18.1 min, mean ± S.D., here and below) than in SCCRO+/+ MEFs (90.0 ± 8.9 min, p < 0.001) (Fig. 2A). When the duration of individual stages of mitosis was assessed, there was no significant difference in the average time from prophase to telophase (T1) between SCCRO−/− and SCCRO+/+ MEFs (30.0 ± 5.0 min for both, p = not significant) (Fig. 2A). In contrast, a significant delay was observed from the onset of late telophase (when the midbody first appears) to the completion of cytokinesis (T2) in SCCRO−/− MEFs (90.7 ± 18.1 min) compared with SCCRO+/+ MEFs (60.0 ± 8.9 min, p < 0.001) (Fig. 2, A and B). Consistent with the presence of a defect in abscission, the number of cells at the midbody stage, identified by immunostaining for α-tubulin and Aurora B, was significantly higher for SCCRO−/− MEFs (8.3%) than for wild-type cells (4.2%, p < 0.001) (Fig. 2C). Re-expression of SCCRO by viral transduction reduced the number of SCCRO−/− MEFs at the midbody stage, whereas re-expression of SCCROD241N had no effect (Fig. 2D). Confirming the importance of neddylation activity, treatment with MLN4924 also increased the number of SCCRO+/+ MEFs at the midbody stage (Fig. 2C). No differences in the morphologic appearance of the intercellular bridge were identifiable in SCCRO+/+ and SCCRO−/− MEFs. Combined, these findings suggest that SCCRO specifically affects abscission and that its effects require its neddylation-promoting activities. In contrast to the isolated defect in abscission in SCCRO−/− MEFs, chemical (MLN4924) or genetic inactivation of core neddylation components has been reported to have broader and pleiotropic effects on mitosis (19–24).
Figure 2.
Depletion of SCCRO in MEFs delays abscission. A, comparison of duration of metaphase and cytokinesis between SCCRO+/+ (white columns) and SCCRO−/− (black columns) MEFs. Note that, although T1 is essentially the same for both cells, SCCRO−/− MEFs have a T2 ∼50% longer than that in SCCRO+/+ MEFs. B, long-term imaging of cell division of SCCRO+/+ and SCCRO−/− MEFs stably expressing Aurora B-EGFP and mCherry–α-tubulin. Compared with SCCRO+/+ MEFs (top row), SCCRO−/− MEFs show delayed abscission (bottom row). Scale bar = 5 μm. C, immunofluorescence using anti-Aurora B (green), anti-α-tubulin (red), and DAPI (blue), showing an increased percentage of midbody cells in SCCRO−/− MEFs compared with SCCRO+/+ MEFs (top row). Treatment with MLN4924 (1 μm for 2 h) increased midbody cells in SCCRO+/+ MEFs to a level similar to that in SCCRO−/− MEFs (bottom row). Insets show a close-up view of midbody cells in each case. The numbers are the percentages of midbody cells. Scale bar = 20 μm. D, the increased percentage of midbody cells seen in SCCRO−/− MEFs was rescued by retroviral introduction of HA-SCCRO but not HA-SCCROD241N.
SCCRO promotes Cul3 neddylation and localization to the midbody
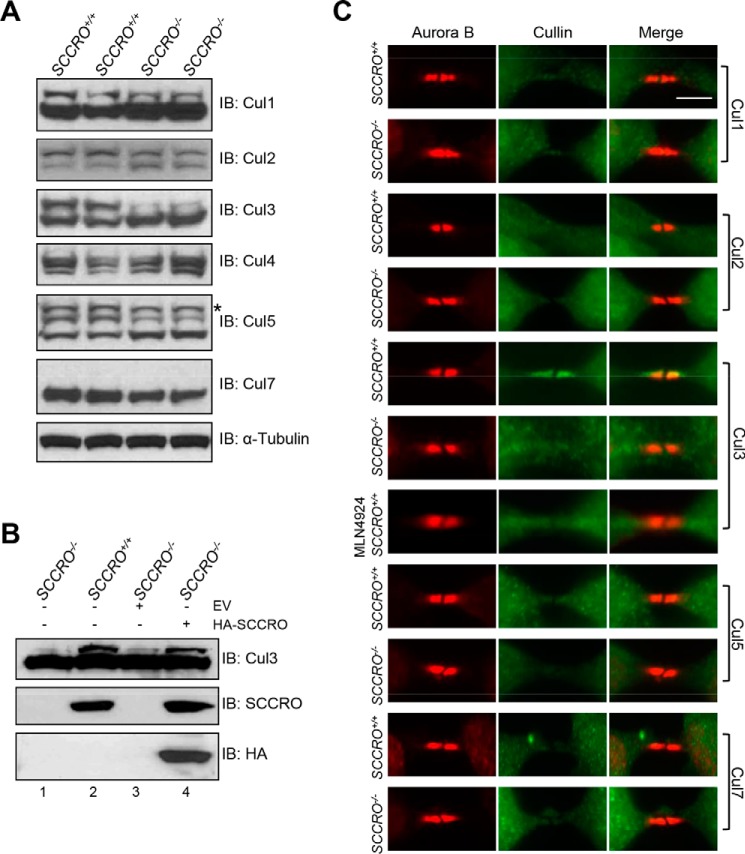
We and others have shown that SCCRO promotes neddylation of several cullin family members, including Cul1, Cul2, Cul3, Cul4, and Cul5, leading to assembly and increased activity of CRL-type ubiquitin ligases and subsequent ubiquitination of CRL substrates (6, 8, 10). To identify protein targets regulated by SCCRO during abscission, we first sought to define specific cullin(s) regulated by SCCRO during cytokinesis. We compared neddylation levels and localization of cullin family members in SCCRO+/+ and SCCRO−/− MEFs by Western blotting and immunostaining, respectively. Although the neddylated fraction of multiple cullins was reduced (Cul1, Cul2, Cul3, and Cul5), the magnitude of reduction was highest for Cul3 in SCCRO−/− MEFs compared with SCCRO+/+ MEFs (Fig. 3A). Defects in neddylation of Cul3 were rescued in SCCRO−/− MEFs by re-expression of SCCRO (Fig. 3B). Immunofluorescence staining showed that Cul3, but not other cullin family members, localized to the midbody in SCCRO+/+ MEFs but failed to localize to the midbody in the majority of SCCRO−/− MEFs (∼83% of cells), suggesting that it may transduce the effects of SCCRO on abscission (Fig. 3C). Consistent with the importance of neddylation activity, treatment with MLN4924 also disrupted the localization of Cul3 to the midbody in SCCRO+/+ MEFs (Fig. 3C, seventh row). Although our findings do not exclude the possibility that SCCRO affects the activity of cullins involved in other stages of mitosis, they suggest that SCCRO-promoted neddylation is required for localization of Cul3 to the midbody during abscission.
Figure 3.
SCCRO promotes Cul3 neddylation and is required for its localization to the midbody. A, Western blot analysis of cell lysates, showing that neddylation of Cul3 was reduced in SCCRO−/− MEFs more than other cullins compared with SCCRO+/+ MEFs. The asterisk denotes possible dineddylated Cul5. IB, immunoblot. B, Western blot analysis of cell lysates of infected SCCRO−/− MEFs, showing that retrovirus-mediated transfection with HA-SCCRO restored SCCRO expression to levels seen in SCCRO+/+ MEFs (center row, compare lanes 2 and 4) and rescued Cul3 neddylation (lane 4). EV = empty vector. C, immunostaining for cullins and Aurora B, showing that only Cul3 localizes to the midbody in SCCRO+/+ MEFs and also showing the absence of Cul3 in the midbody of SCCRO−/− MEFs and MLN4924-treated SCCRO+/+ MEFs. Note that anti-Cul4 did not work for immunostaining. Scale bar = 5 μm.
SCCRO promotes assembly of Cul3KLHL21 at the midbody
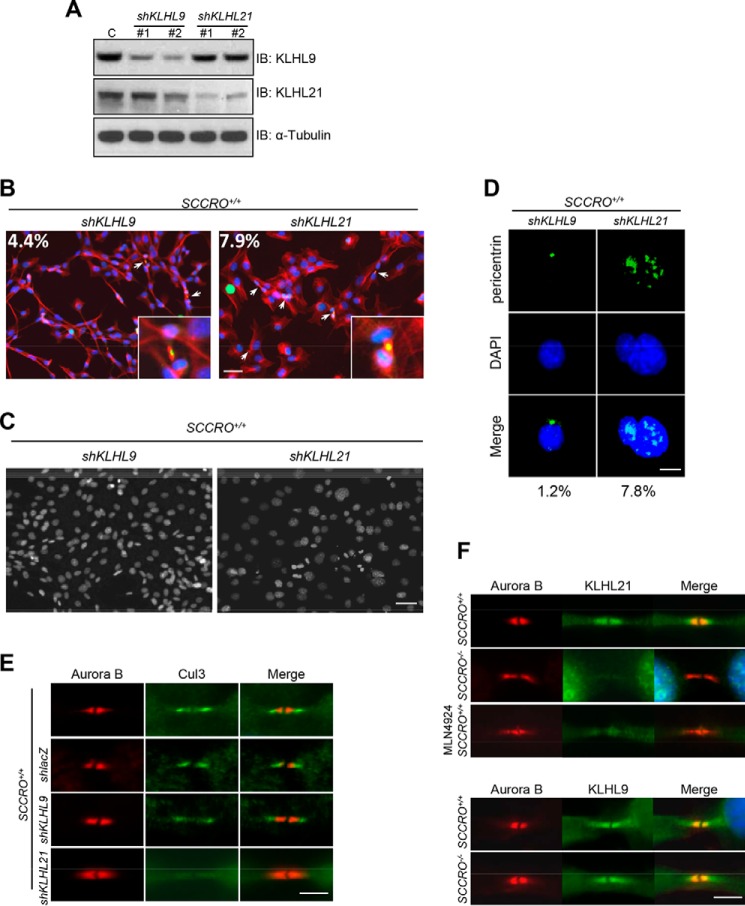
We next sought to identify the substrate adaptor that mediates the interaction of Cul3 and its potential substrate at the midbody. Previously, we reported a newly developed compound that specifically inhibits SCCRO and SCCRO2 activity by interfering with their interactions with UBC12 (25). This novel compound was used to screen for SCCRO-dependent, substrate-specific BTB–Kelch proteins that serve as adaptors in Cul3-anchored CRLs (25). Use of the inhibitor allowed the identification of binding interaction that acutely depends on the neddylation activity of SCCRO. SCCRO inhibitor–dependent binding was observed between Cul3 and several BTB–Kelch proteins, including KLHL23, KLHL9, KLHL18, KLHL13, KLHL26, KLHL20, KLHL25, KLHL7, KLHL21, KLHL22, KLHL12, KLHL42, KLHL24, KLHL11, KLHL15, KLHL36, and KLHL8. Of these, only KLHL9, KLHL13, and KLHL21 have been reported to be involved in cell cycle–related activity (26, 27). To determine whether any of these adaptors is involved in abscission, we knocked down KLHL9 or KLHL21 in SCCRO+/+ MEFs using shRNA (KLHL13 was not tested, as it functions as part of a heterodimer with KLHL9) (Fig. 4A). Phenotypes similar to SCCRO−/− MEFs—including increased numbers of cells at the midbody stage, polyploidy, and polycentrosomy—were seen with KLHL21 knockdown, but not with KLHL9 knockdown, in SCCRO+/+ MEFs (Fig. 4, B–D). In addition, localization of Cul3 to the midbody was lost in SCCRO+/+ MEFs with KLHL21 knockdown but not in those with KLHL9 knockdown (Fig. 4E). Moreover, although both KLHL9 and KLHL21 localized to the midbody, KLHL21 failed to localize to the midbody in SCCRO−/− MEFs (Fig. 4F). KLHL21 also failed to localize to the midbody in SCCRO+/+ MEFs after cells were treated with MLN4924 (Fig. 4F). These findings show that localization of Cul3 and KLHL21 to the midbody requires SCCRO and suggest that Cul3KLHL21 mediates the effects of SCCRO during abscission. Furthermore, because SCCRO−/− MEFs showed no detectable defect in the initiation of cytokinesis or subcellular localization of KLHL9, it is likely that the activity of Cul3KLHL9/KLHL13 was intact in these cells. This suggests that the effects of SCCRO on Cul3 neddylation specifically promote Cul3KLHL21 assembly and activity.
Figure 4.
SCCRO regulates abscission through the Cul3KLHL21 complex. A, Western blot analysis of lysates from SCCRO+/+ MEFs treated with shRNA against KLHL9 or KLHL21 as indicated and probed with KLHL9, KLHL21, and α-tubulin antibodies. C, lacZ shRNA knockdown control; IB, immunoblot. B, KLHL9 (left panel) and KLHL21 (right panel) shRNA-treated SCCRO+/+ MEFs stained for Aurora B (green), α-tubulin (red), and nuclei (blue). KLHL21 shRNA-treated cells show increased levels of midbody cells. Insets, close-up view of midbody cells. The percentages of midbody cells are included inside the images. Scale bar = 20 μm. C, DAPI staining, showing an increased percentage of polyploidy cells in SCCRO+/+ MEFs with KLHL21 knockdown (right panel) compared with those with KLHL9 knockdown (left panel). Scale bar = 20 μm. D, immunofluorescence analysis using anti-pericentrin, showing supernumerary centrosomes in SCCRO+/+ MEFs with KLHL21 knockdown but not in those with KLHL9 knockdown. Scale bar = 5 μm. E, immunostaining for Cul3 and Aurora B, showing an absence of Cul3 from the midbody in SCCRO+/+ MEFs with KLHL21 knockdown (shKLHL21) but not in SCCRO+/+ MEFs with KLHL9 knockdown (shKLHL9) or lacZ knockdown controls (shlacZ). Scale bar = 5 μm. F, immunostaining for KLHL21 and Aurora B, showing an absence of KLHL21 in the midbody of SCCRO−/− MEFs and MLN4924-treated SCCRO+/+ MEFs compared with SCCRO+/+ MEFs (top panel). Note that there is no difference in KLHL9 localization between SCCRO+/+ and SCCRO−/− MEFs (bottom panel).
SCCRO promotes ubiquitination of Aurora B
It was reported previously that Cul3KLHL21 is required for ubiquitination of the mitotic kinase Aurora B (27). During cytokinesis, Aurora B coordinates chromosome segregation with abscission, which occurs only after chromatin is cleared from the cleavage plane (28, 29). Aurora B at the midbody activates the NoCut checkpoint by phosphorylating Shrb/CHMP4C, a subunit of the endosomal sorting complex required for transport (ESCRT-III), to localize it to the Fleming body, which prevents the assembly of a functional abscission complex (30). Subsequent inactivation and degradation of Aurora B is required for abscission to be completed, with prolonged activity resulting in delayed or failed abscission (31). To determine whether levels of Aurora B are regulated by SCCRO through Cul3KLHL21 during cytokinesis, we assessed the levels of total and ubiquitinated Aurora B after chemically blocking protein translation or degradation. Blocking translation by pretreatment with cycloheximide resulted in faster clearance of Aurora B in SCCRO+/+ MEFs than in SCCRO−/− MEFs (Fig. 5A). Proteasome inhibition with MG132 increased accumulation of total and ubiquitinated Aurora B in SCCRO+/+ MEFs compared with SCCRO−/− MEFs. Levels of total and ubiquitinated Aurora B were increased in SCCRO−/− MEFs transfected with HA-SCCRO, but not in those transfected with HA-SCCROD241N, after treatment with MG132 (Fig. 5B). To determine whether the effect of SCCRO on ubiquitination of Aurora B is cell cycle–dependent, we first assessed expression of Aurora B in MEFs synchronized to the G1, S, and M phases. The results of Western blotting showed that levels of Aurora B increased from G0/G1 through G2/M and decreased to their lowest point at the next G0/G1 in SCCRO+/+ MEFs. The decrease in the level of Aurora B protein at G0/G1 was significantly attenuated in SCCRO−/− MEFs (Fig. 5C). To confirm that the level of Aurora B protein decreases during M-to-G1 phase transition, we synchronized cells to G2/M by use of nocodazole and released them into fresh medium. Cells were harvested at different times after release, and lysates were subjected to Western blotting, which showed a more pronounced decrease in levels of Aurora B over time in SCCRO+/+ MEFs than in SCCRO−/− MEFs (Fig. 5D). Addition of MG132 into released cells completely blocked the decrease in the levels of Aurora B during M to G1 phase in SCCRO+/+ MEFs, suggesting that levels of Aurora B could be regulated by ubiquitination-proteasome–mediated degradation (Fig. 5E). Moreover, the decrease in levels of Aurora B correlated with increasing levels of neddylated Cul3 in SCCRO+/+ MEFs, both of which were absent in SCCRO−/− MEFs (Fig. 5D). Consistent with a requirement for neddylation, the defect in Aurora B turnover in SCCRO−/− MEFs was rescued by transfection with SCCRO but not SCCROD241N (Fig. 5F). In addition, when nocodazole-synchronized SCCRO+/+ MEFs were released into medium containing MLN4924, the decrease in levels of Aurora B over time was impaired, which is consistent with the requirement for neddylation activity (Fig. 5G). Degradation of Aurora B was also impaired in SCCRO+/+ MEFs with KLHL21 knockdown but not in those with KLHL9 knockdown (Fig. 5H). Together, these results suggest that degradation of Aurora B at the time of abscission is promoted by Cul3KLHL21 following SCCRO-promoted neddylation of Cul3. It should be noted that the mitotic markers Cyclin B1 and phospho-histone H3 (Ser-10) decrease normally in both SCCRO+/+ and SCCRO−/− MEFs (Fig. 5D), suggesting that SCCRO-deficient cells progress normally through the cell cycle and excluding the possibility that the reduced Aurora B degradation in SCCRO−/− MEFs is an indirect effect of delayed mitotic exit. To confirm the importance of SCCRO-promoted ubiquitination and inactivation of Aurora B during abscission, we assessed the effects of addition of ZM447439 (an Aurora B inhibitor) to the medium when cells entered the midbody stage in SCCRO−/− MEFs by live-cell imaging. The addition of ZM447439 overcame the abscission delay seen in SCCRO−/− MEFs (Fig. 5I) (n = 40 for both DMSO and ZM447439 treatment, p < 0.01). Combined, these results suggest that inactivation of Aurora B by SCCRO-promoted ubiquitination is required for efficient completion of abscission.
Figure 5.
SCCRO promotes ubiquitination of Aurora B. A, Western blot analysis of lysates from MEFs after treatment with cycloheximide at 100 μg/ml for the indicated times, showing a more rapid clearance of Aurora B in SCCRO+/+ MEFs than in SCCRO−/− MEFs. IB, immunoblot. B, Western blot analysis of lysates from MEFs after treatment with MG132 at 25 μm for the indicated times, showing an increase in the levels of Aurora B with time in SCCRO+/+ MEFs compared with SCCRO−/− MEFs (first panel). A similar increase was seen with the expression of HA-SCCRO (lanes 7–9) but not with HA-SCCROD241N (lanes 10–12). The same lysates were also subjected to immunoprecipitation using anti-Aurora B antibody and probed for polyubiquitin chains, showing enrichment of ubiquitinated Aurora B in SCCRO+/+ MEFs and HA-SCCRO–transfected SCCRO−/− MEFs but not in SCCRO−/− MEFs or HA-SCCROD241N–transfected cells (second panel). C, Western blot analysis of lysates from serum-starved (S), mimosine-arrested (M), double thymidine–blocked (T), and nocodazole-arrested (N) MEFs, showing a defect in degradation of Aurora B from M to G1 phase in SCCRO−/− MEFs compared with SCCRO+/+ MEFs. D, Western blot analysis of lysates from MEFs released from nocodazole treatment (100 ng/ml for 12 h), showing an increase of neddylated Cul3 and corresponding degradation of Aurora B in SCCRO+/+ MEFs but not in SCCRO−/− MEFs. E, the degradation of Aurora B seen in SCCRO+/+ MEFs can be reversed by addition of MG132 (50 μm) into medium at the time of release. F, the defect in Aurora B degradation observed in SCCRO−/− MEFs was rescued by retroviral introduction of HA-SCCRO but not HA-SCCROD241N. G, nocodazole-treated SCCRO+/+ MEFs released into medium containing 1 μm MLN4924 exhibited a similar defect in Aurora B degradation as that seen in SCCRO−/− MEFs. H, Western blot analysis of lysate from MEFs released from nocodazole arrest, showing a defect in degradation of Aurora B in SCCRO+/+ MEFs with KLHL21 knockdown compared with SCCRO+/+ MEFs with KLHL9 knockdown. I, duration of T2 in SCCRO−/− MEFs in the presence of either DMSO or ZM447439. DMSO or ZM447439 was added to the culture medium when cells reached the midbody stage (n = 40 for both DMSO and ZM447439 treatment, p < 0.01).
Defective abscission in SCCRO−/− MEFs leads to delay and/or failure of cytokinesis
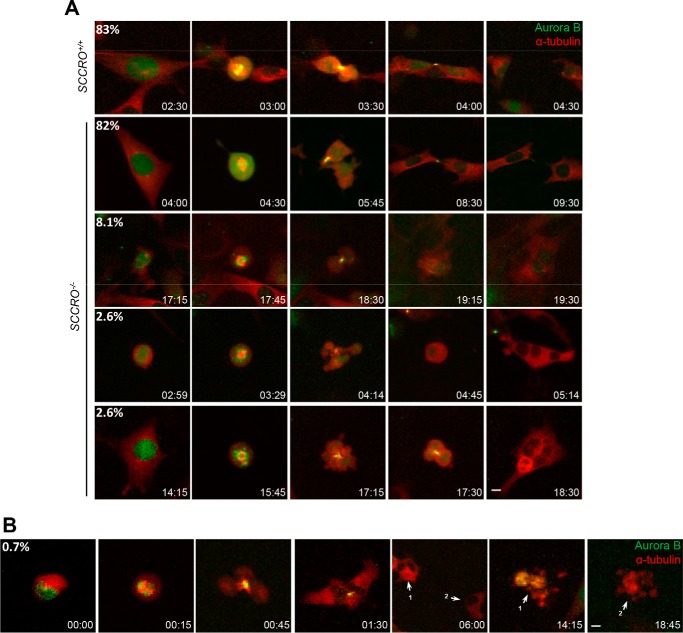
To assess the effects of aberrant localization of Cul3 and KLHL21 to the midbody in SCCRO−/− MEFs, we performed live-cell imaging and monitored Aurora B localization in SCCRO+/+ and SCCRO−/− MEFs stably expressing mCherry–α-tubulin and Aurora B–EGFP. We found that Aurora B localized normally during mitosis in SCCRO−/− cells, to the centromeres at metaphase, to the mitotic spindle midzone at anaphase, and to the midbody at late telophase. However, in contrast to SCCRO+/+ cells—in which Aurora B was removed from the midbody, after which abscission was completed—in SCCRO−/− cells, it persisted at the midbody and was associated with a significant delay in abscission (Fig. 6A, first and second rows, and Movies S1 and S2). Moreover, tetraploid cells with two centrosomes were observed in SCCRO−/− MEFs after failed abscission and regression of abscission furrows (Fig. 6A, third row, and Movie S3). Tetraploidy and the accompanying gain in the number of centrosomes increased the potential for multipolar spindle formation and inappropriate kinetochore-microtubule attachments at metaphase, both of which can lead to chromosome missegregation and aneuploidy. In many cases, this led to the development of polyploidy giant cells because of another failed abscission (Fig. 6A, fourth and fifth rows, and Movies S4 and S5) or asymmetric division (Fig. 6B and Movie S6). These aneuploid cells eventually underwent apoptosis (Fig. 6B and Movie S6). Our findings suggest that SCCRO-promoted neddylation of Cul3 is required for the localization and activity of Cul3KLHL21 at the midbody, which allows ubiquitination-promoted turnover of Aurora B and subsequent completion of cytokinesis. These results also suggest that KLHL21- and SCCRO-promoted neddylation of Cul3 may cooperate to promote localization of the Cul3KLHL21 complex to the midbody but that neither is sufficient on its own. It is still unclear whether the Cul3KLHL21 complex assembles before or after localization to the midbody.
Figure 6.
Defective abscission in SCCRO−/− MEFs leads to a delay in and failure of abscission. A, long-term imaging of cell division of SCCRO+/+ and SCCRO−/− MEFs stably expressing Aurora B-EGFP and mCherry–α-tubulin. Compared with SCCRO+/+ MEFs (first row, 83% of cells completed abscission in 60 min or less), defects in SCCRO−/− MEFs include delayed abscission (second row, 82% of cells took >75 min to complete abscission), regression of abscission furrow (third row), multipolar spindle formation (fourth row), and development of polyploidy giant cells (fifth row). Scale bar = 5 μm. B, asymmetric division and, eventually, apoptosis of SCCRO−/− MEFs. 1 and 2 denote the two daughter cells resulting from asymmetric division.
Discussion
Neddylation of cullins serves as a key regulator of CRL activity. Neddylation promotes the release of the stoichiometric inhibitory effects of CAND1 to allow assembly of CRLs (32–35). In addition, neddylation enhances CRL activity by inducing conformational change in cullins, which causes the enzymatic component of the complex (ROC1) to come into proximity with E2 to facilitate transfer of ubiquitin from E2 to the substrate (36). Although it is well established that neddylation regulates the activity of CRL complexes, it remains unclear how specificity is achieved in the neddylation reaction. Previous studies have shown that core components of neddylation have indiscriminate effects on cullin neddylation, suggesting that they may not play a regulatory role (19–23). Accumulating evidence suggests that substrates and substrate adaptors serve as primary regulators of CRL activity. The addition of substrate adaptors to in vitro reactions is sufficient to promote cullin neddylation and CRL assembly, suggesting that the effect of substrate adaptors on CRL activity involves regulation of neddylation (37). Consistent with this, mutations in cullins in the binding region of substrate adaptors or substrate recognition subunits result in reduced neddylation (38). Although these findings suggest that the substrates and substrate adaptors may regulate and provide specificity for cullin neddylation, it remains to be determined whether other regulators exist and how the signal is transduced to the core neddylation machinery.
SCCRO is a component of E3 for neddylation. Several studies have shown that SCCRO forms stable stoichiometric complexes with cullins and CAND1 (6, 9, 10, 12). In this complex, SCCRO is not sufficient to overcome the inhibitory effects of CAND1 on cullin neddylation in vitro (10). The addition of testis lysates from SCCRO+/+ mice to in vitro reactions overcomes the inhibition of neddylation of recombinant Cul1 resulting from binding to CAND1. This suggests that factors in the lysate are required to release the inhibitory effects of CAND1 on cullin neddylation. Interestingly, the addition of testis lysates from SCCRO−/− mice is not able to overcome CAND1 inhibition of recombinant Cul1 under identical conditions, suggesting that SCCRO is also required (10). Keuss et al. (12) found that the addition of substrate adaptor (KLHL3) overcame the inhibition of CAND1, allowing SCCRO to promote cullin neddylation in vitro (Cul3). This suggests that binding of the substrate adaptor to the SCCRO–cullin–ROC1 complex is required to promote neddylation. Heir et al. reported that binding of substrate (HIF1α) to the substrate receptor von Hippel-Lindau (VHL) promotes binding of SCCRO to VHL and cullin (Cul2) (39). This led to the suggestion that SCCRO functions as a “substrate sensor switch,” with binding of substrate to SCCRO via the substrate adaptor being required to trigger cullin neddylation.
Our findings show that SCCRO plays a role in regulating cytokinesis in vivo. A panoply of spatial and biochemical stimuli regulate the timing of cytokinesis, with Aurora B playing an essential role. Drosophila cells lacking Aurora B and mammalian cells treated with Aurora B inhibitors do not undergo cytokinesis, leading to regression of the cleavage furrow and polyploidy (40, 41). Interestingly, overexpression of Aurora B also results in polyploidy, reflecting the differential activities of Aurora B during cytokinesis (42). The published data suggest that CRL-promoted ubiquitination of Aurora B plays an important role in regulating cytokinesis. Aurora B ubiquitination is decreased, and its clearance from the midbody is delayed in SCCRO-deficient cells, leading to abscission delay and failure. Rescue of delayed abscission in SCCRO-deficient cells by pharmacological inhibition of Aurora B in cells at the midbody stage strongly implicates Aurora B as a key target of SCCRO. Two different Cul3-anchored CRLs (Cul3KLHL9/KLHL13 and Cul3KLHL21) have been reported to target Aurora B for ubiquitination during cytokinesis (26, 27, 43, 44). Ubiquitination of Aurora B by the Cul3KLHL9/KLHL13 complex regulates its removal from mitotic chromosomes to initiate cytokinesis; in contrast, Cul3KLHL21 mediates ubiquitination of Aurora B after the spindle assembly checkpoint has been satisfied. Although cytokinesis is not initiated in cells depleted of KLHL9 and/or KLHL13 by RNAi, KLHL21-depleted cells initiate but fail to complete cytokinesis, suggesting that these complexes act independently and sequentially. Even though there is a significant decrease in global levels of neddylated Cul3 in SCCRO-deficient cells, only the activity of selected Cul3-anchored CRL complexes is affected. SCCRO-deficient cells initiate cytokinesis normally, suggesting that the activity of the Cul3KLHL9/KLHL13 complex is intact (26). Interestingly, RNAi knockdown of KLHL21, but not KLHL9, results in phenotypic changes identical to those seen in SCCRO-deficient cells. This is consistent with the fact that the activity of the Cul3KLHL21 complex, but not the Cul3KLHL9/KLHL13 complex, is regulated by SCCRO.
In our study, both Cul3 and KLHL21 failed to localize to the midbody in SCCRO-deficient cells. Interestingly, Cul3 also failed to localize to the midbody in cells with KLHL21 knockdown. These findings suggest that both SCCRO and KLHL21 are required for proper localization of Cul3KLHL21 to the midbody. It is unclear whether midbody localization of Cul3 and KLHL21 occurs before or after assembly of the complex. The previous findings that neddylation occurs in the nucleus and that unneddylated cullins are unstable suggest that CRL complexes assemble before translocation to the site of activity (9). Moreover, SCCRO was not detected at the midbody under any conditions (data not shown). This leads to a model where the substrate adaptor and SCCRO coactivate neddylation of the substrate in the nucleus and translocate as part of the CRL complex to the site of activity. The role of the substrate and the factors governing temporal and spatial sequences with which SCCRO, substrates, and substrate adaptors impart their effects remain to be determined.
Inactivation of an SCCRO orthologue (DCN1) in yeast and Caenorhabditis elegans leads to lethality. In contrast, SCCRO knock-out mice and flies are viable. Bioinformatics analysis shows that SCCRO has four paralogues in mammals, which can be classified into three subgroups on the basis of their phylogeny and N-terminal sequences: SCCRO and SCCRO2 (DCUN1D2) contain an ubiquitin-associated domain, SCCRO3 (DCUN1D3) contains a myristoyl sequence, and SCCRO4 (DCUN1D4) and SCCRO5 (DCUN1D5) contain a nuclear localization signal in the N terminus (12, 45, 46). We have shown that SCCRO paralogues have both independent and overlapping activities in regulating cullin neddylation in higher organisms (13). It is likely that SCCRO paralogues function to diversify the neddylation signal to provide tight control of CRL activity.
Finally, SCCRO is amplified and overexpressed in a variety of human cancers. The increased proliferation resulting from SCCRO overexpression in cellular and animal models (9, 13), along with the high prevalence of polyploidy in tumors with SCCRO amplification, suggests the possibility that the cancer-promoting activity of SCCRO may result in premature removal of Aurora B, accelerated abscission, and aneuploidy and may contribute to genetic instability. Moreover, although proteasome and neddylation inhibitors have been shown to have therapeutic efficacy in humans, their broad-based activity induces severe side effects in a significant number of patients. The selective effects of SCCRO in neddylation, combined with an “oncogene addiction” phenotype associated with its overexpression in human cancer, suggest that SCCRO may be an excellent therapeutic target.
Experimental procedures
Immunofluorescence and live imaging analysis
MEFs were stained with anti-α-tubulin (Calbiochem), anti-β-tubulin (Sigma), anti-pericentrin (Abcam), anti-Aurora B (BD Transduction Laboratories), anti-KLHL9 (Abcam), anti-KLHL21 (GeneTex), anti-Cul1 (Invitrogen), anti-Cul2 (Novus), anti-Cul3 (Santa Cruz Biotechnology), anti-Cul4 (Santa Cruz Biotechnology), anti-Cul5 (Santa Cruz Biotechnology), anti-Cul7 (Bethyl), and MitoTracker Red (Invitrogen). Whole slides were scanned using the Mirax scanner (Carl Zeiss) with a 20 × 0.8 numerical aperture objective. Confocal imaging was performed using a Leica TCS SP2 system with 20 × 0.7 NA and 63 × 1.2 NA water immersion objectives. Live-cell imaging was performed using a Zeiss LSM 5 Live microscope equipped with an incubation chamber (37 °C, humidified, 5% CO2) and a 63 × 1.4 NA Plan-Apochromat water immersion objective (Zeiss). Sample illumination was generally kept to a minimum and had no adverse effect on cell division and proliferation. Image analysis was performed by using Metamorph software. Linear contrast adjustments were applied, with constant settings for different experimental conditions.
Cell synchronization
Cells were synchronized at M/G1, late G1, G1/S, or G2/M by serum starvation, mimosine arrest, double thymidine block, or nocodazole arrest, as described elsewhere (47).
KLHL9 and KLHL21 knockdown shRNA plasmids were purchased from Sigma. The sequences were 5′-CCGGCACGCACAGTTCGGTTGTATTCTCGAGAATACAACCGAACTGTGCGTGTTTTTG-3′ and 5′-CCGGCCCTGTTCTAACCTAATATAACTCGAGTTATATTAGGTTAGAACAGGGTTTTTG-3′ for KLHL9 and 5′-CCGGTGTGCCTAGTATTGATCTATACTCGAGTATAGATCAATACTAGGCACATTTTTG-3′ and 5′-CCGGACTGCGTGTGGAGATACAATTCTCGAGAATTGTATCTCCACACGCAGTTTTTTG-3′ for KLHL21.
Western blotting
For Western blotting, protein was separated by use of SDS/PAGE, transferred to PVDF membranes (Whatman), and probed with different antibodies. Anti-SCCRO monoclonal antibody was produced and used as described elsewhere (10). Anti-Tex14 was purchased from Abcam. Other antibodies used were anti-HA (Covance), anti-Cyclin E (Santa Cruz Biotechnology), anti-Katanin p60 (Santa Cruz Biotechnology), anti-α-tubulin (Calbiochem), anti-Aurora B (BD Transduction Laboratories), anti-KLHL9 (Abcam), anti-KLHL21 (GeneTex), and anti-Cul3 (BD Transduction Laboratories).
Immunoprecipitation
Immunoprecipitations were performed essentially as described elsewhere (48). In brief, lysates from MEFs were incubated with 4 μg of anti-Aurora B (Cell Signaling Technology) and 20 μl of protein A + protein G-agarose beads by gentle rocking at 4 °C overnight. The beads were washed three times with lysis buffer and once with PBS. Bound proteins were eluted with 2× Laemmli buffer, resolved on SDS-PAGE gels, and analyzed by Western blotting.
Author contributions
B. S. conceived and coordinated the study and experiments. B. S. and G. H. wrote the manuscript. G. H. and A. J. K. designed, performed, and analyzed the experiments shown in Figs. 1–6. K. X. and K. M. provided technical assistance and contributed to the preparation of the figures. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Sho Fujisawa for help with preparing Fig. 6.
This work was supported in part by NCI, National Institutes of Health Cancer Center Support Grant P30 CA008748 and by a research grant from the Hackers for Hope. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Movies S1–S6.
- CRL
- cullin–RING–ligase
- SCCRO
- squamous cell carcinoma–related oncogene
- MEF
- mouse embryonic fibroblast
- EGFP
- enhanced GFP
- Tt
- time from the onset of prophase to the completion of cytokinesis
- T1
- time from prophase to telophase
- T2
- time from the onset of late telophase to the completion of cytokinesis.
References
- 1. Xirodimas D. P. (2008) Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem. Soc. Trans. 36, 802–806 [DOI] [PubMed] [Google Scholar]
- 2. Cardozo T., and Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 3. Petroski M. D., and Deshaies R. J. (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 4. Duda D. M., Borg L. A., Scott D. C., Hunt H. W., Hammel M., and Schulman B. A. (2008) Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saha A., and Deshaies R. J. (2008) Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell 32, 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kurz T., Chou Y. C., Willems A. R., Meyer-Schaller N., Hecht M. L., Tyers M., Peter M., and Sicheri F. (2008) Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol. Cell 29, 23–35 [DOI] [PubMed] [Google Scholar]
- 7. Kurz T., Ozlü N., Rudolf F., O'Rourke S. M., Luke B., Hofmann K., Hyman A. A., Bowerman B., and Peter M. (2005) The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435, 1257–1261 [DOI] [PubMed] [Google Scholar]
- 8. Scott D. C., Monda J. K., Grace C. R., Duda D. M., Kriwacki R. W., Kurz T., and Schulman B. A. (2010) A dual E3 mechanism for Rub1 ligation to Cdc53. Mol. Cell 39, 784–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang G., Kaufman A. J., Ramanathan Y., and Singh B. (2011) SCCRO (DCUN1D1) promotes nuclear translocation and assembly of the neddylation E3 complex. J. Biol. Chem. 286, 10297–10304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim A. Y., Bommeljé C. C., Lee B. E., Yonekawa Y., Choi L., Morris L. G., Huang G., Kaufman A., Ryan R. J., Hao B., Ramanathan Y., and Singh B. (2008) SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J. Biol. Chem. 283, 33211–33220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Monda J. K., Scott D. C., Miller D. J., Lydeard J., King D., Harper J. W., Bennett E. J., and Schulman B. A. (2013) Structural conservation of distinctive N-terminal acetylation-dependent interactions across a family of mammalian NEDD8 ligation enzymes. Structure 21, 42–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keuss M. J., Thomas Y., Mcarthur R., Wood N. T., Knebel A., and Kurz T. (2016) Characterization of the mammalian family of DCN-type NEDD8 E3 ligases. J. Cell Sci. 129, 1441–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fu W., Sun J., Huang G., Liu J. C., Kaufman A., Ryan R. J., Ramanathan S. Y., Venkatesh T., and Singh B. (2016) Squamous cell carcinoma-related oncogene (SCCRO) family members regulate cell growth and proliferation through their cooperative and antagonistic effects on cullin neddylation. J. Biol. Chem. 291, 6200–6217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han K., Wang Q., Cao H., Qiu G., Cao J., Li X., Wang J., Shen B., and Zhang J. (2016) The NEDD8-activating enzyme inhibitor MLN4924 induces G2 arrest and apoptosis in T-cell acute lymphoblastic leukemia. Oncotarget 7, 23812–23824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y., Shi C. C., Zhang H. P., Li G. Q., and Li S. S. (2016) MLN4924 suppresses neddylation and induces cell cycle arrest, senescence, and apoptosis in human osteosarcoma. Oncotarget 7, 45263–45274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li L., Liu B., Dong T., Lee H. W., Yu J., Zheng Y., Gao H., Zhang Y., Chu Y., Liu G., Niu W., Zheng S., Jeong L. S., and Jia L. (2013) Neddylation pathway regulates the proliferation and survival of macrophages. Biochem. Biophys. Res. Commun. 432, 494–498 [DOI] [PubMed] [Google Scholar]
- 17. Nakayama K. I., and Nakayama K. (2006) Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 18. Sumara I., Maerki S., and Peter M. (2008) E3 ubiquitin ligases and mitosis: embracing the complexity. Trends Cell Biol. 18, 84–94 [DOI] [PubMed] [Google Scholar]
- 19. Lin J. J., Milhollen M. A., Smith P. G., Narayanan U., and Dutta A. (2010) NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 70, 10310–10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Milhollen M. A., Narayanan U., Soucy T. A., Veiby P. O., Smith P. G., and Amidon B. (2011) Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 71, 3042–3051 [DOI] [PubMed] [Google Scholar]
- 21. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., et al. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
- 22. Singer J. D., Gurian-West M., Clurman B., and Roberts J. M. (1999) Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 13, 2375–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tateishi K., Omata M., Tanaka K., and Chiba T. (2001) The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J. Cell Biol. 155, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leck Y. C., Choo Y. Y., Tan C. Y., Smith P. G., and Hagen T. (2010) Biochemical and cellular effects of inhibiting Nedd8 conjugation. Biochem. Biophys. Res. Commun. 398, 588–593 [DOI] [PubMed] [Google Scholar]
- 25. Scott D. C., Hammill J. T., Min J., Rhee D. Y., Connelly M., Sviderskiy V. O., Bhasin D., Chen Y., Ong S.-S., Chai S. C., Goktug A. N., Huang G., Monda J. K., Low J., Kim H. S., et al. (2017) Blocking an N-terminal acetylation–dependent protein interaction inhibits an E3 ligase. Nature Chem. Biol. 10.1038/nchembio.2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sumara I., Quadroni M., Frei C., Olma M. H., Sumara G., Ricci R., and Peter M. (2007) A Cul3-based E3 ligase removes Aurora B from mitotic chromosomes, regulating mitotic progression and completion of cytokinesis in human cells. Dev. Cell 12, 887–900 [DOI] [PubMed] [Google Scholar]
- 27. Maerki S., Olma M. H., Staubli T., Steigemann P., Gerlich D. W., Quadroni M., Sumara I., and Peter M. (2009) The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J. Cell Biol. 187, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eggert U. S., Mitchison T. J., and Field C. M. (2006) Animal cytokinesis: from parts list to mechanisms. Annu. Rev. Biochem. 75, 543–566 [DOI] [PubMed] [Google Scholar]
- 29. Glotzer M. (2005) The molecular requirements for cytokinesis. Science 307, 1735–1739 [DOI] [PubMed] [Google Scholar]
- 30. Carlton J. G., Caballe A., Agromayor M., Kloc M., and Martin-Serrano J. (2012) ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science 336, 220–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mathieu J., Cauvin C., Moch C., Radford S. J., Sampaio P., Perdigoto C. N., Schweisguth F., Bardin A. J., Sunkel C. E., McKim K., Echard A., and Huynh J. R. (2013) Aurora B and cyclin B have opposite effects on the timing of cytokinesis abscission in Drosophila germ cells and in vertebrate somatic cells. Dev. Cell 26, 250–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu K., Chen A., and Pan Z. Q. (2000) Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275, 32317–32324 [DOI] [PubMed] [Google Scholar]
- 33. Liu J., Furukawa M., Matsumoto T., and Xiong Y. (2002) NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol. Cell 10, 1511–1518 [DOI] [PubMed] [Google Scholar]
- 34. Zheng J., Yang X., Harrell J. M., Ryzhikov S., Shim E. H., Lykke-Andersen K., Wei N., Sun H., Kobayashi R., and Zhang H. (2002) CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol. Cell 10, 1519–1526 [DOI] [PubMed] [Google Scholar]
- 35. Goldenberg S. J., Cascio T. C., Shumway S. D., Garbutt K. C., Liu J., Xiong Y., and Zheng N. (2004) Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119, 517–528 [DOI] [PubMed] [Google Scholar]
- 36. Kawakami T., Chiba T., Suzuki T., Iwai K., Yamanaka K., Minato N., Suzuki H., Shimbara N., Hidaka Y., Osaka F., Omata M., and Tanaka K. (2001) NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20, 4003–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bornstein G., Ganoth D., and Hershko A. (2006) Regulation of neddylation and deneddylation of cullin1 in SCFSkp2 ubiquitin ligase by F-box protein and substrate. Proc. Natl. Acad. Sci. U.S.A. 103, 11515–11520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chew E. H., and Hagen T. (2007) Substrate-mediated regulation of cullin neddylation. J. Biol. Chem. 282, 17032–17040 [DOI] [PubMed] [Google Scholar]
- 39. Heir P., Sufan R. I., Greer S. N., Poon B. P., Lee J. E., and Ohh M. (2013) DCNL1 functions as a substrate sensor and activator of cullin 2-RING ligase. Mol. Cell. Biol. 33, 1621–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hauf S., Cole R. W., LaTerra S., Zimmer C., Schnapp G., Walter R., Heckel A., van Meel J., Rieder C. L., and Peters J. M. (2003) The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Giet R., and Glover D. M. (2001) Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J. Cell Biol. 152, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Giet R., Petretti C., and Prigent C. (2005) Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 15, 241–250 [DOI] [PubMed] [Google Scholar]
- 43. Nguyen H. G., Chinnappan D., Urano T., and Ravid K. (2005) Mechanism of Aurora-B degradation and its dependency on intact KEN and A-boxes: identification of an aneuploidy-promoting property. Mol. Cell. Biol. 25, 4977–4992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stewart S., and Fang G. (2005) Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res. 65, 8730–8735 [DOI] [PubMed] [Google Scholar]
- 45. Huang G., Stock C., Bommeljé C. C., Weeda V. B., Shah K., Bains S., Buss E., Shaha M., Rechler W., Ramanathan S. Y., and Singh B. (2014) SCCRO3 (DCUN1D3) antagonizes the neddylation and oncogenic activity of SCCRO (DCUN1D1). J. Biol. Chem. 289, 34728–34742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bommeljé C. C., Weeda V. B., Huang G., Shah K., Bains S., Buss E., Shaha M., Gönen M., Ghossein R., Ramanathan S. Y., and Singh B. (2014) Oncogenic function of SCCRO5/DCUN1D5 requires its Neddylation E3 activity and nuclear localization. Clin. Cancer Res. 20, 372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jackman J., and O'Connor P. M. (2001) Methods for synchronizing cells at specific stages of the cell cycle. Curr. Protoc. Cell Biol. Chapter 8, Unit 8.3 [DOI] [PubMed] [Google Scholar]
- 48. Sarkaria I., O-charoenrat P., Talbot S. G., Reddy P. G., Ngai I., Maghami E., Patel K. N., Lee B., Yonekawa Y., Dudas M., Kaufman A., Ryan R., Ghossein R., Rao P. H., Stoffel A., et al. (2006) Squamous cell carcinoma related oncogene/DCUN1D1 is highly conserved and activated by amplification in squamous cell carcinomas. Cancer Res. 66, 9437–9444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.