Abstract
A remarkable property of the machinery for import of peroxisomal matrix proteins is that it can accept already folded proteins as substrates. This import involves binding of newly synthesized proteins by cytosolic peroxisomal biogenesis factor 5 (PEX5) followed by insertion of the PEX5–cargo complex into the peroxisomal membrane at the docking/translocation module (DTM). However, how these processes occur remains largely unknown. Here, we used truncated PEX5 molecules to probe the DTM architecture. We found that the DTM can accommodate a larger number of truncated PEX5 molecules comprising amino acid residues 1–197 than full-length PEX5 molecules. A shorter PEX5 version (PEX5(1–125)) still interacted correctly with the DTM; however, this species was largely accessible to exogenously added proteinase K, suggesting that this protease can access the DTM occupied by a small PEX5 protein. Interestingly, the PEX5(1–125)–DTM interaction was inhibited by a polypeptide comprising PEX5 residues 138–639. Apparently, the DTM can recruit soluble PEX5 through interactions with different PEX5 domains, suggesting that the PEX5–DTM interactions are to some degree fuzzy. Finally, we found that the interaction between PEX5 and PEX14, a major DTM component, is stable at pH 11.5. Thus, there is no reason to assume that the hitherto intriguing resistance of DTM-bound PEX5 to alkaline extraction reflects its direct contact with the peroxisomal lipid bilayer. Collectively, these results suggest that the DTM is best described as a large cavity-forming protein assembly into which cytosolic PEX5 can enter to release its cargo.
Keywords: peroxisome, protein import, protein sorting, receptor recycling, ubiquitylation (ubiquitination), PEX14, PEX5, docking/translocation module
Introduction
Peroxisomal matrix proteins are encoded in nuclear genes, synthesized on cytosolic ribosomes, and post-translationally targeted to the organelle (1) via one of two peroxisomal targeting signals (PTSs),4 the PTS type 1 (PTS1) and the PTS type 2 (PTS2), respectively. The PTS1 is a small C-terminally located peptide frequently ending with the sequence SKL present in most peroxisomal matrix proteins (2, 3). The PTS2 is a degenerated nonapeptide present at the N termini of just a few mammalian proteins (4, 5). The machinery that recognizes these proteins and promotes their translocation across the peroxisome membrane is rather complex. In mammals, it comprises at least 10 peroxins plus a few other proteins mostly involved in ubiquitination/deubiquitination events (for a review, see Ref. 6). These components can be grouped into four sets: 1) the shuttling receptors PEX5 and the PEX5–PEX7 complex, which recognize PTS1 and PTS2 proteins, respectively (7–12); 2) the peroxisomal membrane docking/translocation module (DTM) comprising PEX13, PEX14, and the three “Really Interesting New Gene” (RING) finger peroxins PEX2, PEX10, and PEX12 (13, 14); 3) the receptor export module (REM) comprising the two “ATPases associated with diverse cellular activities” (AAA), the mechanoenzymes PEX1 and PEX6, and their membrane anchor PEX26 (15); and 4) a group of soluble proteins involved in ubiquitination/deubiquitination of PEX5 (i.e. E2D1/2/3 and USP9X (16–18)) and recognition of monoubiquitinated PEX5 by the REM (AWP1 (19)).
According to current models (6, 20–23), sorting of proteins to the peroxisomal matrix starts with their recognition in the cytosol by the shuttling receptors PEX5 or PEX5–PEX7. The receptor–cargo complex then interacts with the peroxisomal membrane DTM, an event ultimately leading to the insertion of the receptor into the organelle membrane with the concomitant translocation of the cargo into the peroxisome matrix (24–27). At this stage, PEX5 behaves as a transmembrane protein (28). Indeed, biochemical experiments have shown that DTM-bound PEX5 exposes the majority of its mass into the organelle matrix, whereas a small N-terminal domain of ∼2 kDa remains exposed to the cytosolic milieu (29). Interestingly, binding of cytosolic cargoes by the corresponding receptors, translocation of these cargo proteins across the peroxisomal membrane, and their release into the organelle matrix are all NTP hydrolysis-independent events (24, 26, 30). Apparently, the driving force for peroxisomal protein import relies on the strong protein–protein interactions that are established between PEX5 on one side and DTM components on the other side (30, 31). Several short linear motifs found at the N-terminal half of PEX5 (eight in the human protein), the so-called pentapeptide motifs (see below), are crucial for the PEX5–DTM interaction. All of them interact with the N-terminal domain of PEX14 quite strongly (dissociation constants in the nanomolar range), whereas motifs 2–4 also interact with PEX13 (Refs. 32–34; see Fig. 1).
Figure 1.
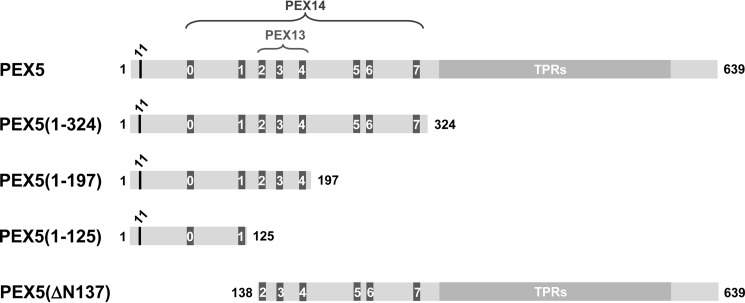
PEX5 molecules used in this study. Full-length and truncated versions of the large isoform of human PEX5 are schematically represented. The conserved cysteine residue at position 11 (black), which was replaced by an alanine or a lysine in many of the proteins used in this work; the eight pentapeptide motifs responsible for the interaction with PEX13 and PEX14 (numbered from 0 to 7; dark gray); and the structured C-terminal half comprising seven TPRs (light gray) are indicated.
After cargo release, the receptors are recycled back into the cytosol. This is the only part of the protein transport cycle that requires energy from ATP hydrolysis (30). Extraction of receptors from the DTM involves two steps. First, PEX5 is monoubiquitinated at a conserved cysteine residue (Cys-11 in the mammalian protein) (18, 35). Subsequently, monoubiquitinated PEX5 (Ub-PEX5) is extracted back into the cytosol in an ATP-dependent manner by the REM, a step that also triggers the release of PEX7 from the DTM (27, 36, 37). After removal of ubiquitin in the cytosol, PEX5 then engages in a new protein transport cycle (17, 38, 39).
A remarkable property of the peroxisomal matrix protein import machinery (PIM) is its capacity to accept already folded proteins as substrates. Actually, in some cases, even oligomeric proteins can be imported into peroxisomes, although the import efficiency of this type of cargoes is probably low (Refs. 40–42; for a review, see Ref. 43). How the PIM accomplishes this feat while at the same time ensuring that matrix proteins are retained in the organelle is currently a central question in the field of peroxisome biogenesis. Some models have been proposed (see “Discussion”), but the data supporting each of these different perspectives are still scarce. In this work, we revisited the hitherto intriguing resistance of DTM-bound PEX5 to alkaline extraction, a property that has been used to suggest that PEX5 is a pore-forming protein. In addition, we used an established cell-free in vitro system and truncated versions of PEX5 to probe the architecture of the DTM. Collectively, our results suggest that the DTM is conceptually best described as a gated large cavity-containing protein assembly in which soluble PEX5 enters to release its cargo.
Results
The PEX5–PEX14 interaction is resistant to alkaline pH
As stated above, PEX5 acquires a transmembrane topology during its transient passage through the DTM (29). Strikingly, DTM-embedded PEX5 cannot be extracted from peroxisomes by alkaline (pH 11.5) solutions, a property generally attributed to proteins that interact directly with the lipid bilayer of biological membranes (10, 28, 44, 45). This biochemical behavior of peroxisomal PEX5 is intriguing because its N-terminal half (the domain necessary and sufficient for insertion of the receptor into the peroxisome membrane) lacks phylogenetically conserved hydrophobic or amphipathic regions that might support a direct contact of the protein with the lipid bilayer of the peroxisomal membrane (25, 46, 47). Actually, a recombinant protein comprising this PEX5 domain alone is highly soluble and shows no tendency to precipitate or aggregate even upon boiling, a property that stems from its intrinsically disordered nature (48). Despite this inconsistency, the striking alkaline resistance of peroxisomal PEX5 has led some authors to propose that PEX5 may function as a pore-forming protein (Refs. 49 and 50; see also “Discussion”). There is, however, a simpler explanation for the peculiar biochemical behavior of peroxisomal PEX5: the protein–protein interactions involving PEX5 and DTM components may be alkaline pH-resistant. We decided to test this possibility.
There are only two DTM components known to interact with the N-terminal half of PEX5. These are PEX13 and PEX14 (32–34, 51, 52), respectively. The PEX13–PEX5 interaction is rather weak and actually difficult to capture in in vitro binding assays (33, 53–55). PEX14, by contrast, interacts very strongly with PEX5 (13, 14, 52, 56). The main interaction involves the first 80 amino acid residues of PEX14, a domain that is probably embedded in the peroxisomal membrane or even exposed into the organelle matrix, and any of the eight pentapeptide motifs present in the N-terminal half of PEX5 (32, 52, 56–59). We focused on the latter interaction.
Two strategies were used to assess whether or not the PEX5–PEX14 interaction is alkaline pH-resistant. In the first strategy, we subjected recombinant PEX5, a protein comprising the first 80 amino acid residues of PEX14 (hereafter referred to as NDPEX14), and a mixture of both proteins to pH 11.5 polyacrylamide gel electrophoresis. The results in Fig. 2A show that indeed this protein interaction is stable at pH 11.5. In the second strategy, we subjected the same recombinant proteins to size-exclusion chromatography performed in the presence of 0.1 m sodium carbonate, the solution generally used to extract biological membranes. As shown in Fig. 2B, protein complexes between PEX5 and NDPEX14 were easily detected. Thus, there is no need to assume that the resistance of peroxisomal PEX5 to alkaline extraction reflects an interaction of the protein with the lipid bilayer of the organelle; the properties of the PEX5–PEX14 interaction fully explain the striking biochemical behavior of DTM-embedded PEX5.
Figure 2.
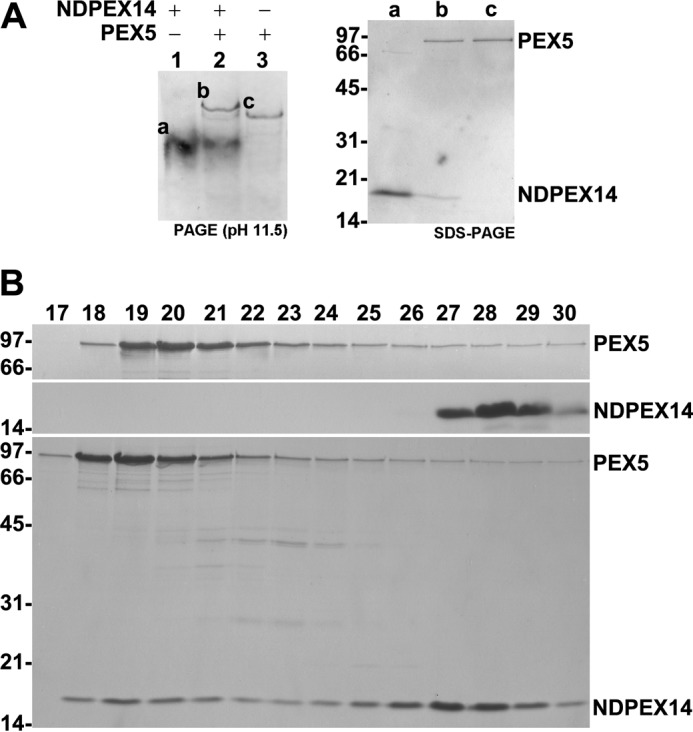
The PEX5–PEX14 interaction is resistant to alkaline pH. A, recombinant NDPEX14 (lane 1), PEX5 (lane 3), and a mixture of both proteins (lane 2) were subjected to PAGE under alkaline conditions (50 mm phosphate buffer, pH 11.5), blotted onto a nitrocellulose membrane, and stained with Ponceau S (left panel). Bands marked “a–c” were excised from the membrane, and the proteins were eluted with Laemmli sample buffer and analyzed by SDS-PAGE/Coomassie Blue staining (right panel). B, recombinant PEX5 (top panel), NDPEX14 (middle panel), and a mixture of both proteins (bottom panel) were subjected to size-exclusion chromatography under alkaline conditions (0.12 m sodium carbonate). The void volume of the column was in fraction 14. Fractions were collected and analyzed by SDS-PAGE/Coomassie Blue staining. In A and B, numbers to the left indicate the molecular mass (kDa) of protein standards.
The DTM can accommodate more molecules of a truncated PEX5 species than full-length PEX5
The very low abundance of PEX5 and DTM components, even in peroxisome-rich cells, together with the lability of detergent-solubilized PEX5–DTM complex during standard biochemical procedures has greatly hampered a structural characterization of the peroxisomal protein translocon (13, 14). There are, nevertheless, indirect approaches, such as in vitro binding analyses, that can provide valuable data on the architecture/mechanism of protein complexes (60). We applied such a strategy to the PEX5–DTM complex.
We first asked whether or not C-terminally truncated PEX5 molecules can interact with the peroxisomal membrane at the same stoichiometry of full-length PEX5. For this purpose, we used an established postnuclear supernatant (PNS)-based in vitro system programmed with in vitro synthesized 35S-labeled PEX5 proteins and assessed insertion of the different PEX5 species into the peroxisome membrane by treating organelle suspensions with proteinase K (PK) (29, 61). This assay explores the fact that soluble PEX5 is extremely sensitive to proteolysis due to the natively unfolded nature of its N-terminal half, whereas PEX5 inserted at the DTM is resistant to PK (29, 48). Two populations of DTM-embedded PEX5 can be discerned after protease treatment. These are the so-called stage 2 and stage 3 PEX5 (29, 35). Stage 2 PEX5 corresponds to non-ubiquitinated PEX5; this species is clipped by PK, losing a domain of ∼2 kDa from its N terminus. Stage 3 PEX5 is completely resistant to the protease and represents monoubiquitinated PEX5. Besides full-length PEX5 possessing an alanine instead of a cysteine at position 11 (PEX5(C11A); see below), two other in vitro synthesized 35S-labeled PEX5 proteins were used in these initial experiments, namely PEX5(1–324;C11A) and PEX5(1–197;C11A) comprising amino acid residues 1–324 and 1–197 of PEX5, respectively (see Fig. 1). Once at the DTM, these C11A mutants cannot be monoubiquitinated and exported (38), a property that should allow us to reach saturation of PEX5-binding sites at the peroxisome membrane more easily. For practical reasons (see “Experimental procedures”), these proteins were synthesized in vitro from pET-28-based plasmids. Thus, they all contain a histidine tag at their N termini (see supplemental Fig. S1A). The in vitro assays were performed at 37 °C and in the presence of ATP, conditions that lead to the export of endogenous rat PEX5 from the peroxisome and to a very low occupation of DTMs by this functional PEX5 species (see supplemental Fig. S2).
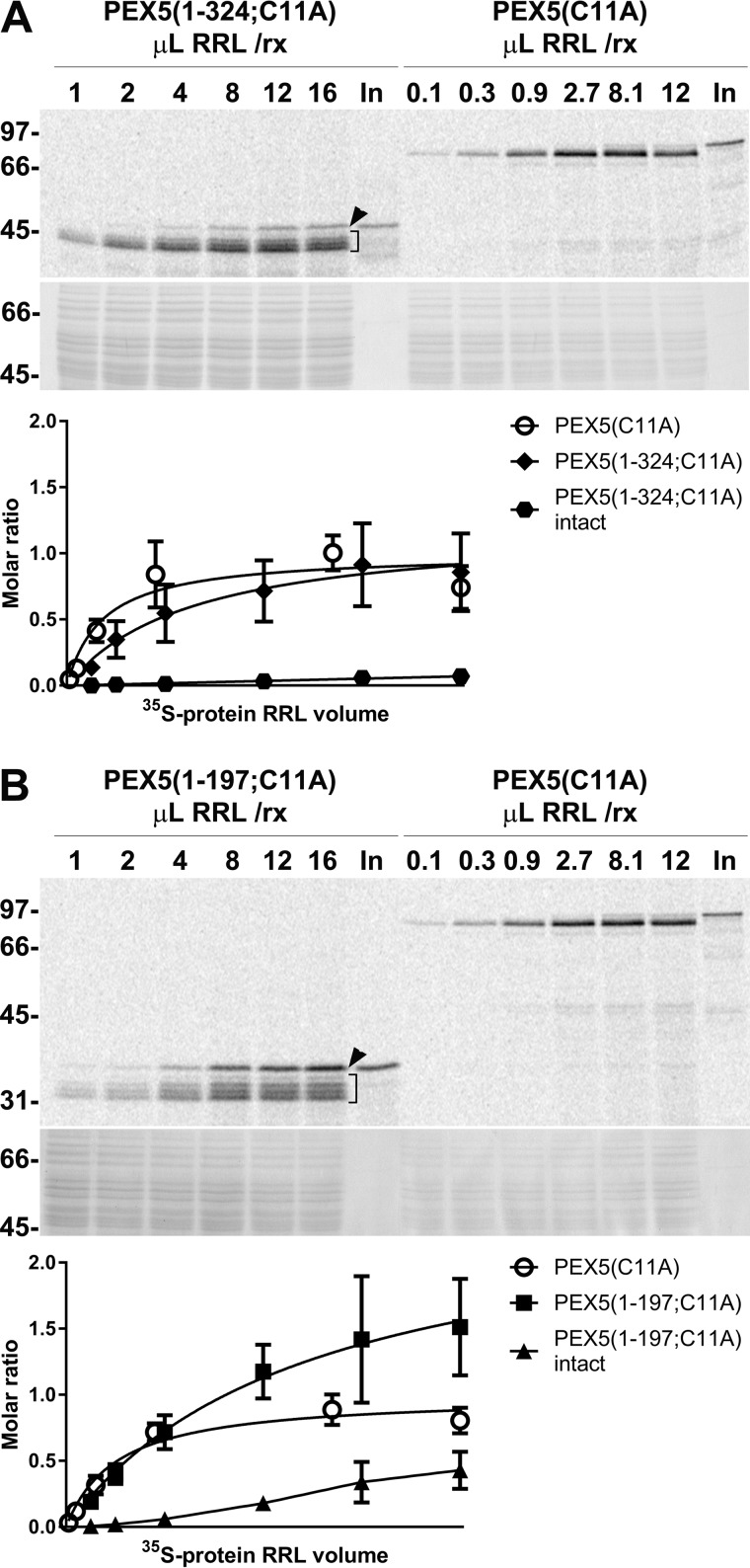
As shown in Fig. 3, we were able to reach saturation or near-saturation conditions for many of the PK-resistant species detected in these assays (A and B, upper panels). Interestingly, both protease-protected PEX5(1–324;C11A) and PEX5(1–197;C11A) displayed an unexpected heterogeneity in these experiments. Indeed, in addition to a set of partially cleaved species, a protein band corresponding to intact radiolabeled protein was also detected, particularly in the PEX5(1–197;C11A) assays (Fig. 3, A and B, indicated with brackets and arrowheads, respectively).
Figure 3.
The DTM can accommodate more PEX5(1–197;C11A) molecules than full-length PEX5(C11A). A, increasing volumes of RRLs (volumes in μl indicated at the top of each lane) containing either PEX5(1–324;C11A) or PEX5(C11A) were used in PNS-based in vitro reactions (“rx”) performed in the presence of ATP for 45 min. Organelle suspensions were then treated with PK and analyzed by SDS-PAGE/phosphorimaging. Total organelle samples (PEX5(1–324;C11A)) or one-third of them (PEX5(C11A)) were loaded onto the gel. The phosphorimage (upper panel) and a section of the corresponding Coomassie Blue-stained gel (middle panel) are shown. Numbers to the left indicate the molecular mass (kDa) of protein standards. Lower panel, data from the phosphorimaging quantitation were normalized for protein loads and methionine number in each radiolabeled protein (see supplemental Fig. S1 for details), fitted to a dose-response one-site-specific binding curve, and divided by the Bmax value obtained for full-length PEX5(C11A). Thus, values in the ordinate represent molar ratios of PEX5(1–324;C11A) to PEX5(C11A) (averages and standard deviations (error bars) from four replicates are shown). Bmax values for PEX5(1–324;C11A) and PEX5(C11A) are 1.21 (95% confidence interval (CI) = 0.88–1.99) and 1.00 (95% CI = 0.85–1.18), respectively. Note that the abscissa scale is different for the two radiolabeled proteins. B, exactly as in A but using radiolabeled PEX5(1–197;C11A) and PEX5(C11A). Bmax values for PEX5(1–197;C11A) and PEX5(C11A) are 2.52 (95% CI = 1.82–4.46) and 1.00 (95% CI = 0.90–1.11), respectively. In A and B, lanes In, RRL containing the indicated 35S-labeled proteins. Brackets and arrowheads indicate the PK-cleaved and intact PK-resistant 35S-labeled proteins, respectively. Note that intact PEX5(1–324;C11A) and intact PEX5(1–197;C11A) data cannot be fitted to the same dose-response curve (“ambiguous fit”); in these cases, the lines simply connect averages. Note also that for some points the error bars are shorter than the height of the symbols.
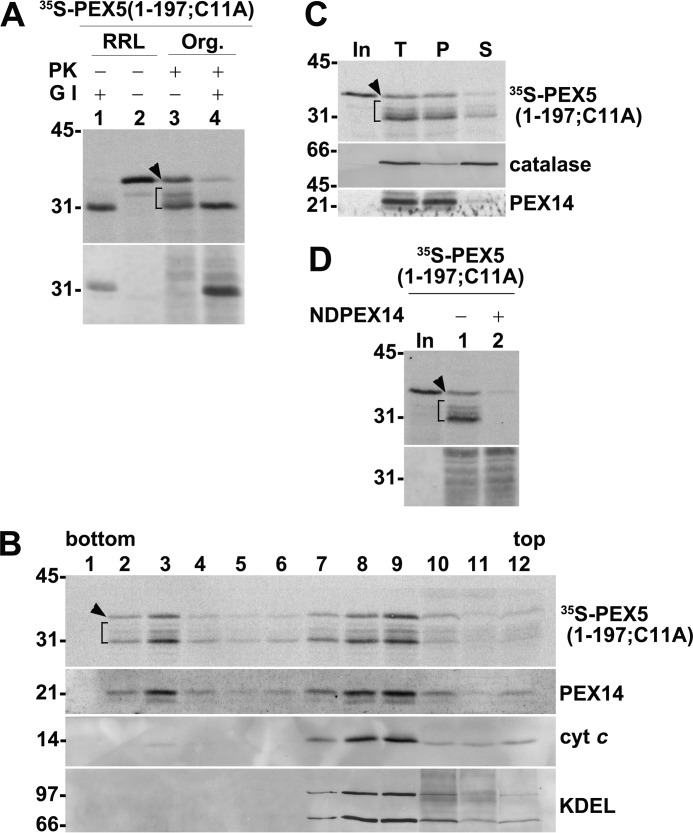
The partially cleaved species represent membrane-embedded molecules exposing their N termini into the cytosol, i.e. at the stage 2 level, as determined by digesting the radiolabeled proteins with Genenase I (29). Genenase I is an engineered subtilisin from Bacillus amyloliquefaciens (62) that cleaves PEX5 near its N terminus immediately after Phe-24 (Ref. 29; see supplemental Figs. S3 and S4). As shown in Fig. 4A, Genenase I converted all PK-resistant PEX5(1–197;C11A) species into a single 31-kDa protein, which co-migrates with the smallest PEX5(1–197;C11A) fragment generated by PK.
Figure 4.
Characterization of protease-protected PEX5(1–197;C11A): part I. A, RRL containing PEX5(1–197;C11A) (lanes 1 and 2) or PK-treated organelles (Org.) from an in vitro assay programmed with PEX5(1–197;C11A) (lanes 3 and 4) were incubated or not with Genenase I (G I) for 30 min at 23 °C as indicated. Samples were analyzed by SDS-PAGE/Western blotting/autoradiography. The autoradiograph (upper panel) and the corresponding Ponceau S-stained membrane (lower panel) are shown. B, a PK-treated in vitro reaction performed with PEX5(1–197;C11A) was loaded onto the top of a Histodenz gradient and centrifuged. Twelve fractions were then collected from the bottom of the gradient and analyzed by SDS-PAGE/Western blotting/autoradiography. An autoradiograph (upper panel) showing the distribution of PK-resistant 35S-labeled PEX5(1–197;C11A) species and Western blots probed with antibodies directed to PEX14 (peroxisomes), cytochrome c (cyt c; mitochondria), and the retention signal KDEL (endoplasmic reticulum) are presented. Note that PEX14 is converted into a small fragment upon PK digestion (59). C, PK-treated organelles from an in vitro assay programmed with radiolabeled PEX5(1–197;C11A) were disrupted by sonication. Half of the sample was kept on ice (lane T), and the other half was centrifuged to separate membrane (lane P) and soluble (lane S) fractions. Samples were analyzed by SDS-PAGE/Western blotting/autoradiography. The autoradiograph (upper panel) and blots showing the distribution of catalase (a peroxisomal matrix protein) and PEX14 (an intrinsic membrane protein) are presented. D, radiolabeled PEX5(1–197;C11A) was subjected to a PNS-based in vitro assay in the absence (lane 1) or presence (lane 2) of 10 μm NDPEX14. PK-treated organelles were analyzed by SDS-PAGE/autoradiography. The autoradiograph (upper panel) and the corresponding Ponceau S-stained membrane (lower panel) are shown. In C and D, lanes In, RRL containing 35S-labeled PEX5(1–197;C11A). A–D, brackets and arrowheads indicate the PK-cleaved and PK-resistant intact 35S-labeled proteins, respectively. Numbers to the left indicate the molecular mass (kDa) of protein standards.
The heterogeneity of the PK-cleaved PEX5 species detected in these experiments together with the distribution of methionine residues at the N terminus of PEX5 raises some uncertainty in the calculation of relative molar ratios (see supplemental Fig. S1). Thus, three different possibilities for the distribution of PK cleavage sites in the N termini of PEX5 truncated species were considered (see supplemental Fig. S1F for details). The data from the most likely possibility (scenario 1; see supplemental Fig. S1F) are shown in Fig. 3, A and B (lower panels). The results suggest that, at saturation, the approximate ratio of PEX5-binding sites in peroxisomes for PEX5(1–197;C11A), PEX5(1–324;C11A), and PEX5(C11A) is 2.5:1.2:1, respectively. In the other two possibilities, the PK cleavage sites were assumed to be at positions leading to the largest (scenario 2) and smallest (scenario 3) PEX5(1–197;C11A):PEX5(1–324;C11A):PEX5(C11A) ratios, respectively: values of 3.2:1.4:1 and 1.8:1:1 were obtained for each of these scenarios (see supplemental Fig. S1F). Thus, regardless of the actual location of PK cleavage sites in the N termini of PEX5 truncated species, it is clear that peroxisomes can accommodate more PEX5(1–197;C11A) molecules than full-length PEX5. Actually, these ratios are probably larger, particularly for PEX5(1–197;C11A), because PK-resistant intact PEX5(1–197;C11A) is also specifically bound to peroxisomes as shown below.
The finding that a non-ubiquitinable PEX5 species can acquire an organelle-associated status remaining completely resistant to PK was particularly striking because up to now the only PEX5 species known to display this behavior was DTM-embedded Ub-PEX5. Because of the potential mechanistic implications of this finding, we analyzed the properties of this species in detail.
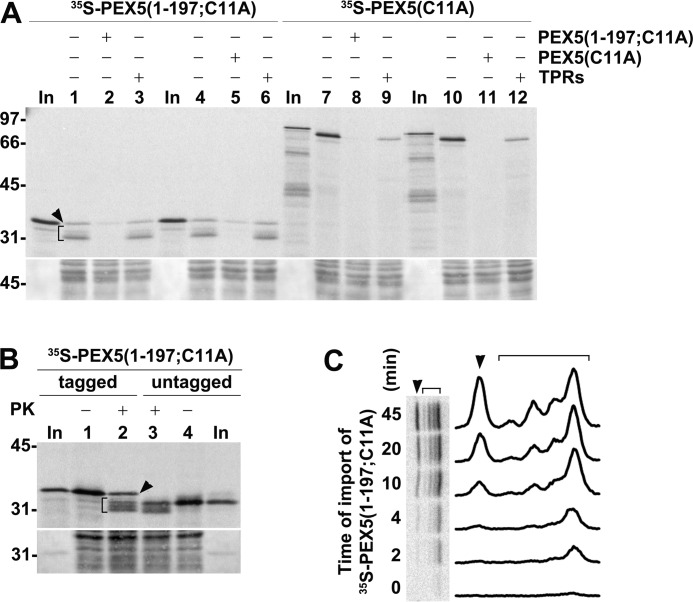
Considering that unusually large amounts of radiolabeled proteins were used in these assays (see “Experimental procedures”), we first addressed the possibility that intact protease-resistant PEX5(1–197;C11A) might result from some nonspecific event (e.g. encapsulation of a small fraction of the radiolabeled protein into some membrane vesicles during sample processing). However, we found that this species co-purifies with peroxisomes upon gradient centrifugation as does stage 2 PEX5(1–197;C11A) (Fig. 4B). Also, similarly to stage 2 species, intact protease-resistant PEX5(1–197;C11A) cannot be extracted from the peroxisomal membrane by sonication in a low-ionic strength buffer, conditions that lead to the release of catalase, a peroxisomal matrix protein (Fig. 4C). Thus, intact protease-resistant PEX5(1–197;C11A) represents a peroxisome membrane-bound species. Furthermore, its insertion into the organelle membrane is blocked when the in vitro assays are performed in the presence of NDPEX14 (Fig. 4D). As stated above, NDPEX14 binds strongly to the pentapeptide motifs present in PEX5 (33, 52, 56), thus blocking insertion of the shuttling receptor into the peroxisome membrane (24, 61). Importantly, the amount of protease-resistant intact PEX5(1–197;C11A) is largely decreased when the in vitro assays are performed in the presence of recombinant (full-length) PEX5(C11A) (Fig. 5A, compare lanes 4 and 5), and conversely, recombinant PEX5(1–197;C11A) blocks insertion of radiolabeled PEX5(C11A) into the peroxisome membrane (Fig. 5A, compare lanes 7 and 8). Thus, both truncated and full-length PEX5 species compete for the same PEX5-binding sites at the peroxisome membrane.
Figure 5.
Characterization of protease-protected PEX5(1–197;C11A): part II. A, PNS-based in vitro assays showing that PEX5(1–197;C11A) competes with PEX5(C11A) and vice versa. PNS in import buffer containing ATP was preincubated for 5 min at 37 °C in the absence or presence of the indicated recombinant proteins (15 μg of PEX5(1–197;C11A), PEX5(C11A), or TPRs). Radiolabeled PEX5(1–197;C11A) and PEX5(C11A) were then added as specified, and the reactions were incubated for 30 min. PK-treated organelles were then subjected to SDS-PAGE/Western blotting/autoradiography. The autoradiograph (upper panel) and a portion of the Ponceau S-stained membrane (lower panel) are shown. Lanes In, RRL containing radiolabeled PEX5(1–197;C11A) and PEX5(C11A) as indicated. Stage 2 PEX5(1–197;C11A) (bracket) and PK-resistant intact PEX5(1–197;C11A) (arrowhead) are also indicated. B, PEX5(1–197;C11A) lacking an N-terminal histidine tag also yields PK-resistant intact species in in vitro assays. RRL (16 μl) containing radiolabeled untagged PEX5(1–197;C11A) was used in a PNS-based in vitro assay. The reaction was divided into two halves (lanes 3 and 4), and one-half was treated with PK as indicated. For comparison, an identical assay performed with histidine-tagged PEX5(1–197;C11A) (lanes 1 and 2) is also shown. Organelle pellets were then subjected to SDS-PAGE/Western blotting/autoradiography. The autoradiograph (upper panel) and a portion of the Ponceau S-stained membrane (lower panel) are shown. Lanes In, RRL containing histidine-tagged or untagged PEX5(1–197;C11A). In A and B, numbers to the left indicate the molecular mass (kDa) of protein standards. C, radiolabeled PEX5(1–197;C11A) was incubated with PNS in import buffer containing ATP. At the indicated time points, aliquots were withdrawn and treated with PK, and the organelles were analyzed by SDS-PAGE/phosphorimaging. The brackets and arrowheads indicate stage 2 and PK-resistant intact PEX5(1–197;C11A), respectively. A densitometry profile of each lane is also shown.
The presence of a histidine tag at the N terminus of PEX5(C11A) does not change its behavior upon PK treatment. As shown in Fig. 3, essentially all protease-protected PEX5(C11A) is clipped by the protease into a slightly shorter protein as previously observed for untagged PEX5 (e.g. see Fig. 1 in Ref. 35). Nevertheless, aiming at better understanding the nature of PK-resistant intact PEX5(1–197;C11A), we still considered that this might not be so for truncated PEX5 species. Thus, we performed an in vitro assay using a large volume of a rabbit reticulocyte lysate (RRL) containing an untagged version of PEX5(1–197;C11A). As shown in Fig. 5B, similar abnormal protease-resistant species were detected with this radiolabeled protein as well. Apparently, intact PK-resistant species are generated when large amounts of truncated PEX5 molecules are used in these assays.
Finally, we analyzed the import kinetics of intact PK-resistant PEX5(1–197;C11A). The results in Fig. 5C suggest that this species displays a slower kinetics than partially accessible PK-resistant PEX5 species. Seemingly, the peroxisomal PEX5-binding site occupied by this abnormal PEX5 population is kinetically distinct from the one occupied by the partially protease-accessible species as the saturation binding experiments shown in Fig. 3B might already suggest.
In summary, the data above suggest that PEX5(1–197;C11A) and PEX5(C11A) bind to the same sites at the peroxisome membrane. However, they do so with different stoichiometries. Apparently, many of the sites available to bind a full-length PEX5(C11A) molecule can accommodate more than one PEX5(1–197;C11A) molecule.
DTM-bound PEX5(1–125;C11A/K) is accessible to PK
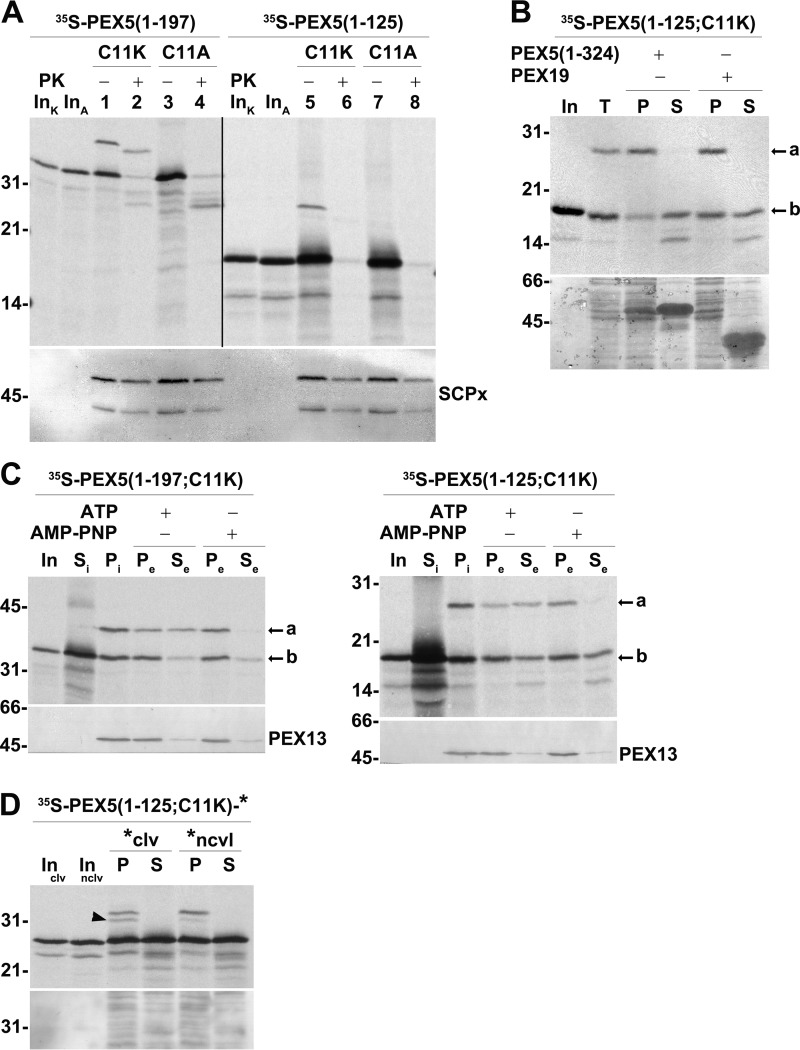
In addition to PEX5(1–197;C11A) and PEX5(1–324;C11A), one other C-terminally truncated species comprising just the first 125 amino acid residues of PEX5 (PEX5(1–125;C11A)) was characterized in this work. Our initial aim was to use this protein in the saturation binding experiments described above and determine whether the DTM could accommodate an even larger number of a smaller PEX5 species. However, this was not possible because PEX5(1–125;C11A) is mostly accessible to PK in the in vitro assays (Fig. 6A, compare lanes 3 and 4 with lanes 7 and 8). Strikingly, when a ubiquitinable version of this protein, PEX5(1–125;C11K), possessing a lysine at position 11 was used in in vitro assays supplemented with AMP-PNP (conditions that allow monoubiquitination of PEX5 at the peroxisome but not its export by the REM (17)), a monoubiquitinated species was clearly detected (Fig. 6A, lane 5; see also supplemental Fig. S5). Interestingly and similarly to PEX5(1–125;C11A), this monoubiquitinated species (Ub-PEX5(1–125;C11K)) is also accessible to PK (Fig. 6A, compare lanes 1 and 2 with lanes 5 and 6).
Figure 6.
DTM-bound PEX5(1–125;C11K/A) is accessible to PK. A, PEX5(1–125;C11K) is correctly monoubiquitinated but does not acquire a PK-protected status. A primed PNS (see “Experimental procedures”) was used in AMP-PNP-supplemented in vitro assays programmed with radiolabeled PEX5(1–197;C11K) (lanes 1 and 2; C11K), PEX5(1–197;C11A) (lanes 3 and 4; C11A), PEX5(1–125;C11K) (lanes 5 and 6; C11K), or PEX5(1–125;C11A) (lanes 7 and 8; C11A). One-half of each reaction was treated with PK as indicated. Organelle fractions were analyzed by SDS-PAGE/Western blotting/autoradiography. The autoradiograph (upper panel) and the corresponding nitrocellulose membrane probed with an antibody directed to sterol carrier protein x (SCPx; lower panel) to assess intactness of peroxisomes (86) are shown. The exposure time of the PEX5(1–125;C11K/A) panel was 4-fold longer than that of PEX5(1–197;C11K/A) to obtain similar intensities of the ubiquitinated species. Note that PEX5(1–197;C11K/A) and PEX5(1–125;C11K/A) have the same number of methionines. Lanes InK and InA, RRL containing the C11K and C11A versions of the indicated 35S-proteins, respectively. B, Ub-PEX5(1–125;C11K), but not PEX5(1–125;C11K), is tightly bound to organelles. Radiolabeled PEX5(1–125;C11K) was incubated with a primed PNS in AMP-PNP-containing import buffer for 30 min at 37 °C. The organelles were then recovered by centrifugation, resuspended in import buffer, and divided into three tubes. One tube was kept on ice (lane T), and the other two tubes were incubated for 15 min at 37 °C in the presence of 10 μg of either recombinant PEX5(1–324) or PEX19 as indicated. Organelles (P) and the corresponding supernatants (S) were separated by centrifugation and analyzed by SDS-PAGE/autoradiography. Lane In, RRL containing the radiolabeled protein. The autoradiograph (upper panel) and a portion of the corresponding Ponceau S-stained membrane (lower panel) are shown. C, Ub-PEX5(1–125;C11K) is a substrate for the REM. Radiolabeled PEX5(1–197;C11K) (left panels) or PEX5(1–125;C11K) (right panels) was incubated with a primed PNS in import buffer supplemented with ubiquitin aldehyde and AMP-PNP. The reactions were then centrifuged to separate supernatant fraction (Si) from organelles (Pi). The organelles were resuspended in an ATP- or AMP-PNP-containing import buffer and further incubated for 15 min at 37 °C. The organelle suspensions were again centrifuged to obtain a supernatant (Se) and an organelle pellet (Pe). Samples were analyzed by SDS-PAGE/Western blotting/autoradiography. The autoradiographs (upper panels) and the behavior of endogenous PEX13 (lower panels) are shown. Si, equivalent to 50 μg of PNS; Pi, Pe, and Se, equivalent to 600 μg of PNS. Lanes In, RRL containing the radiolabeled protein. In B and C, a and b indicate monoubiquitinated and non-ubiquitinated PEX5 species, respectively. D, radiolabeled PEX5(1–125;C11K)-clv is partially processed in the PNS-based in vitro assay. Radiolabeled PEX5(1–125;C11K)-clv and PEX5(1–125;C11K)-nclv were subjected to PNS-based in vitro assays in the presence of AMP-PNP for 60 min. The reactions were then centrifuged to separate organelles (lanes P) from soluble proteins (lanes S). Organelles and soluble fractions from 600 and 100 μg of PNS, respectively, were subjected to SDS-PAGE/Western blotting/autoradiography. Lanes Inclv and Innclv, RRL containing the indicated 35S-labeled proteins. The autoradiograph (upper panel) and the corresponding Ponceau S-stained membrane (lower panel) are shown. The cleaved species is indicated by an arrowhead. Numbers to the left indicate the molecular mass (kDa) of protein standards.
Interaction of PEX5 with the peroxisomal DTM occurs in two steps: 1) a reversible docking event in which PEX5 remains protease-accessible (24) and 2) insertion of PEX5 into the DTM, an essentially irreversible step (in the absence of ATP) that yields protease-resistant PEX5 (29). Given the results described above for PEX5(1–125;C11A/K), we hypothesized that this protein might be unable to enter the DTM remaining trapped at the docking step and thus protease-accessible. To test this, organelles from an in vitro assay programmed with radiolabeled PEX5(1–125;C11K) and performed in the presence of AMP-PNP were resuspended in import buffer and incubated with a vast excess of recombinant PEX5(1–324), which competes with PEX5 at the docking step (24), or PEX19, a protein involved in a different aspect of peroxisome biogenesis and used here as a negative control (63). After centrifugation to separate organelle-associated proteins from soluble proteins, samples were analyzed by SDS-PAGE/autoradiography. Note that because no protease treatment step can be included in these assays, radiolabeled protein nonspecifically adsorbed to the organelles is not destroyed, thus explaining the relatively large background observed in this type of experiment (see Ref. 24). Despite this limitation, the results in Fig. 6B show that a considerable amount of unmodified PEX5(1–125;C11K) was specifically extracted from the organelles upon incubation with recombinant PEX5(1–324) as expected for a PEX5 species at the docking step (24). By contrast, essentially all Ub-PEX5(1–125;C11K) remained in the organelle pellet. Seemingly, Ub-PEX5(1–125;C11K) is beyond the docking step.
Data showing that Ub-PEX5(1–125;C11K) is actually upstream of the export step were obtained in a two-step import/export experiment (17). Briefly, an organelle pellet from an in vitro assay programmed with radiolabeled PEX5(1–125;C11K) and performed in the presence of AMP-PNP (to accumulate Ub-PEX5(1–125;C11K) at the DTM) was resuspended in import buffer and incubated in the presence of either AMP-PNP (to maintain the block of the REM) or ATP (to activate the REM). Organelle-bound and soluble proteins were then analyzed by SDS-PAGE/autoradiography. As shown in Fig. 6C (right panel), no Ub-PEX5(1–125;C11K) was released into the supernatant in the reaction containing AMP-PNP as expected. In contrast, a large fraction of Ub-PEX5(1–125;C11K) was found in the supernatant from the ATP-containing export reaction. Exactly the same results were obtained with PEX5(1–197;C11K), which was used in this assay as a positive control (Fig. 6C, left panel). Thus, Ub-PEX5(1–125;C11K) is a substrate for the REM.
Finally, to better characterize PEX5(1–125;C11K), we asked whether a portion of its polypeptide chain becomes exposed into the peroxisomal matrix during its passage through the DTM. For this, we adapted an experimental strategy previously used to show that a portion of the polypeptide chains of PEX5 and PEX7 reach the peroxisome matrix (27, 64). Specifically, we produced two almost identical PEX5(1–125;C11K) proteins, both having at their C termini an extension of about 5 kDa. In the first protein, named PEX5(1–125;C11K)-clv, the extension comprises a cleavable but otherwise non-functional PTS2 derived from pre-thiolase presequence (64) followed by 19 amino acid residues corresponding to the N terminus of human sterol carrier protein-2. The second protein, PEX5(1–125;C11K)-nclv, is virtually identical to PEX5(1–125;C11K)-clv with the exception that it lacks the last two residues of the non-functional pre-thiolase presequence. This deletion was designed based on previous work showing that a similar deletion in pre-phytanoyl-CoA hydroxylase dramatically decreased its processing efficiency at the peroxisomal matrix (65). Thus, if the C terminus of PEX5(1–125;C11K)-clv becomes exposed into the peroxisomal matrix, a 2-kDa shorter protein should be generated by the action of Tysnd1, the peroxisomal matrix protease that processes PTS2-containing proteins (66), whereas no or very little cleavage should be observed for PEX5(1–125;C11K)-nclv. The results of an in vitro assay performed with these two proteins revealed precisely this behavior, although the fraction of cleaved PEX5(1–125;C11K)-clv is relatively small (Fig. 6D). We conclude that at least a fraction of PEX5(1–125;C11K)-clv acquires a transmembrane topology during its transient passage through the DTM.
Taken together, the data shown in Fig. 6 suggest that a small protein comprising just the first 125 amino acid residues of PEX5 can enter the DTM, yielding a correct substrate for the RING finger peroxins and to the REM, but remain largely accessible to PK added from the cytosolic side of the peroxisomal membrane.
Two non-overlapping PEX5 fragments can interact with the DTM in a competitive manner
We have previously shown that a truncated PEX5 lacking the first 110 amino acid residues of the receptor can still become inserted into the DTM in a cargo-dependent manner (67). In this work, larger N-terminal truncations were characterized. The aim was to obtain a functional N-terminally truncated PEX5 species displaying no sequence overlap with PEX5(1–125) so that we could better understand the architecture/function of the DTM.
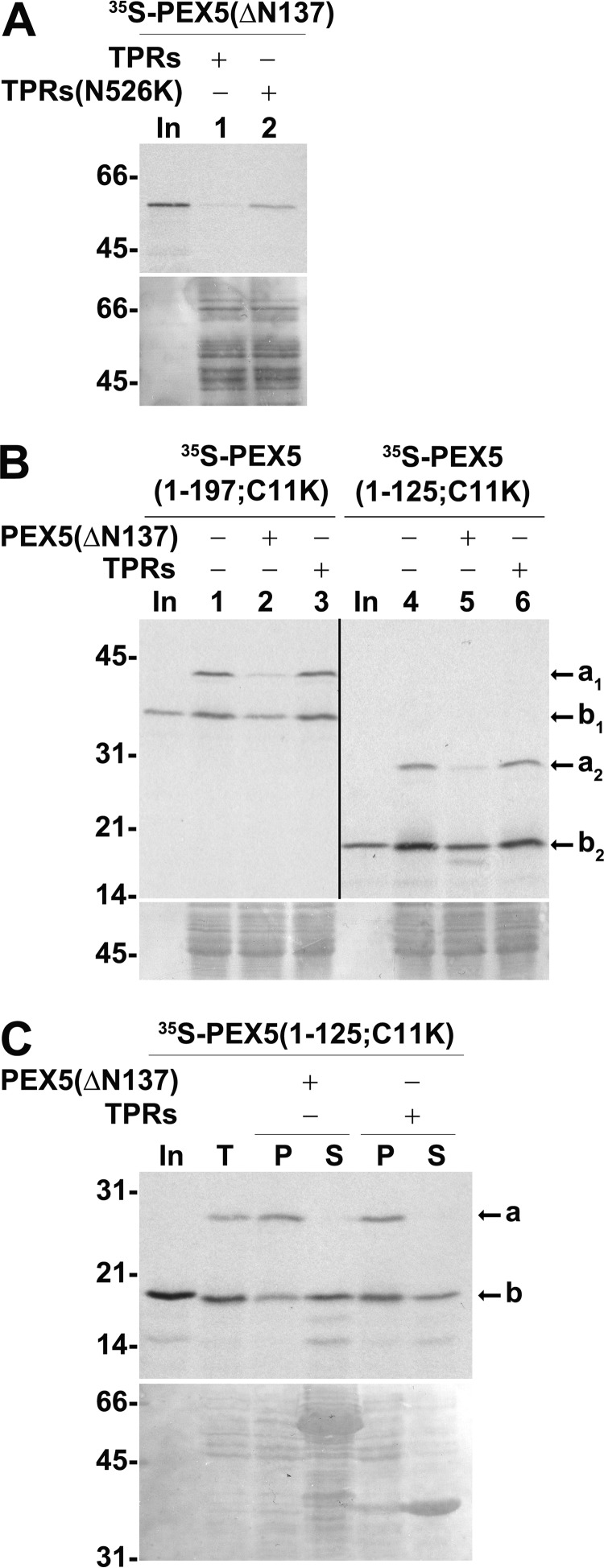
We found that truncating the first 137 amino acid residues of PEX5 resulted in a species (PEX5(ΔN137)) that is still active in the in vitro assays as demonstrated by the acquisition of an organelle-associated, protease-protected status in a cargo-dependent manner (see Fig. 7A). No import capacity was observed for a slightly shorter protein lacking amino acid residues 1–148 (data not shown).
Figure 7.
Two non-overlapping PEX5 fragments interact with the DTM in a competitive manner. A, PNS-based in vitro assays were performed with radiolabeled PEX5(ΔN137) in the presence of ATP and 1 μm recombinant TPRs or TPRs(N526K). Note that the N526K mutation abolishes the PTS1 binding activity of TPRs (87). PK-treated organelles were then analyzed by SDS-PAGE/Western blotting/autoradiography. Lane In, RRL containing 35S-labeled PEX5(ΔN137). B, radiolabeled PEX5(1–197;C11K) or PEX5(1–125;C11K) was subjected to PNS-based in vitro assays in the presence of AMP-PNP and in the absence or presence of 1 μm recombinant TPRs or PEX5(ΔN137) as indicated. Organelle fractions were analyzed by SDS-PAGE/Western blotting/autoradiography. The exposure time of the PEX5(1–125;C11K) panel was 4-fold longer than that of PEX5(1–197;C11K). Lanes In, RRL containing the indicated 35S-proteins. C, radiolabeled PEX5(1–125;C11K) was incubated with a primed PNS in AMP-PNP-containing import buffer for 30 min. The organelles were then isolated by centrifugation, resuspended in import buffer, and divided into three tubes. One tube remained on ice (lane T). The other tubes were incubated for 15 min at 37 °C in the presence of 10 μg of either recombinant PEX5(ΔN137) or TPRs as indicated. Organelles (P) and soluble proteins (S) were separated by centrifugation and analyzed by SDS-PAGE/Western blotting/autoradiography. Lane In, RRL containing 35S-labeled PEX5(1–125;C11K). In B and C, a and b indicate monoubiquitinated and non-ubiquitinated PEX5 species, respectively. A–C, autoradiographs (upper panels) and corresponding Ponceau S-stained membranes (lower panels) are shown. Numbers to the left indicate the molecular mass (kDa) of protein standards.
We then used recombinant PEX5(ΔN137) in competition experiments in the PNS-based in vitro assays. As might be expected for two proteins that share some of their DTM-interacting regions (see Fig. 1), recombinant PEX5(ΔN137) competes with PEX5(1–197;C11K) for insertion into the DTM as shown by the large decrease in the amount of monoubiquitinated PEX5(1–197;C11K) in organelle pellets (Fig. 7B, lanes 1–3). Importantly, the same result was observed for PEX5(1–125;C11K) (Fig. 7B, lanes 4–6).
Competition between PEX5(ΔN137) and PEX5(1–125;C11K) could occur at the docking step or only at the insertion step. To clarify this, we repeated the experiment presented in Fig. 6B but this time using recombinant PEX5(ΔN137) as a competitor of organelle-associated PEX5(1–125;C11K). The results in Fig. 7C suggest that PEX5(ΔN137) competes with PEX5(1–125;C11K) at the docking step.
Discussion
Protein translocons with the capacity to accept already folded proteins as substrates are not unique to peroxisomes. Indeed, several other protein transport systems found in both eukaryotes and prokaryotes have this ability. These include the nuclear import/export machinery; the chloroplast; bacterial twin-arginine translocators; a vast number of bacterial protein secretion machineries such as the type II, type IV, and some type V secretion systems; and pore-forming proteins of both prokaryotic and eukaryotic origin (68–73). Although the diversity of architectures and compositions of these machineries is overwhelming, they can be coarsely grouped into three mechanistic classes: 1) those that comprise membrane proteins with the capacity to transiently assemble into a “custom”-size transmembrane pore every time a cargo has to be transported (i.e., the twin-arginine translocator); 2) those built of proteins that are initially soluble but that undergo oligomerization and major conformational changes enabling them to insert into a membrane, creating pores of variable dimensions (pore-forming proteins/toxins); and 3) those that comprise a gated pore of fixed geometry and composition through which folded and soluble cargoes are transported (e.g. the nuclear pore complex and the bacterial type II and IV secretion systems) (68–73). Which of these three mechanisms, if any, best describes the PIM has been a major question in the field.
We have previously proposed that newly synthesized folded peroxisomal matrix proteins are translocated across the organelle membrane by PEX5 itself when the soluble receptor interacts with and becomes inserted into the DTM (29, 31). It is explicitly assumed in this model that soluble cargo-loaded PEX5 is recruited into a proteinaceous pore/channel formed by DTM components. The transmembrane topology of DTM-embedded PEX5 (29), the absence of any obvious phylogenetically conserved hydrophobic or amphipathic domains in PEX5, the fact that the DTM-interacting domain of PEX5 (i.e. its N-terminal half) is a natively unfolded domain that remains in solution even upon boiling (48), and the membrane topology of PEX14, which has its strongest PEX5-interacting domain embedded in the peroxisomal membrane or even exposed into the organelle matrix (59), are some of the facts supporting this idea (see also Ref. 47). The resistance of DTM-embedded PEX5 to alkaline extraction, although striking, has been interpreted within the context of all the other PEX5 data as probably meaning that there is some unusually strong protein–protein interaction involving PEX5 on one side and members of the DTM on the other side (28, 44, 45, 47).
A different perspective is adopted in the so-called transient pore model (49), which uses the alkaline resistance of peroxisomal PEX5 as its main supporting fact. We note that in its essence the transient pore model is a derivative of the one described above; i.e. it also proposes that cytosolic cargo proteins are pushed across the organelle membrane by PEX5 itself when the receptor interacts with the DTM. The novelty of this model is the idea that PEX5 becomes integrated into the lipid bilayer of the peroxisomal membrane, thus acting as a pore-forming toxin-like protein (49, 74). The data in this work provide a much simpler explanation for the alkaline resistance of DTM-embedded PEX5 and therefore reconcile the discrepancies between the biochemical properties of peroxisomal PEX5 and the primary structure and biochemical behavior of its N-terminal half, which were never fully explained by the transient pore model. Clearly, there is no need to assume that PEX5 is a pore-forming toxin-like protein because the resistance of its peroxisomal pool to alkaline extraction can now be explained by the properties of the PEX5–PEX14 interaction. Although additional biochemical work and detailed structural data will be necessary to understand how the PIM transports newly synthesized proteins across the organelle membrane, the data presented here for the PEX5 truncated species can be used to infer some of its properties (see Fig. 8).
Figure 8.
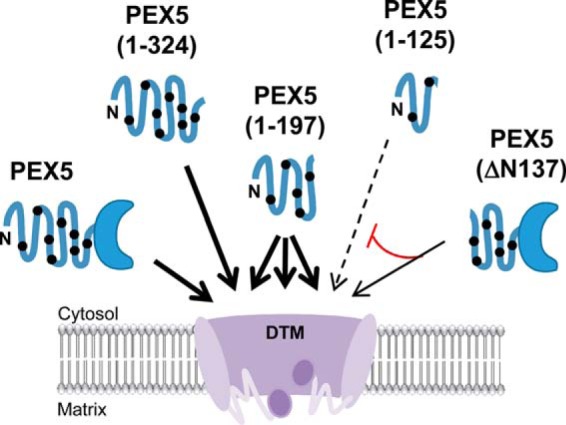
Model of the peroxisomal matrix protein translocon (DTM). The DTM, comprising the transmembrane proteins PEX14, PEX13, and RING finger peroxins PEX2, PEX1,0 and PEX12, is a large cavity-forming protein assembly into which soluble PEX5 enters to release its cargo. Each site at the DTM available to bind one full-length PEX5 molecule can accommodate one PEX5(1–324) molecule or two to three PEX5(1–197) molecules (thick arrows). The small PEX5(1–125) species also interacts correctly with DTM, albeit with low efficiency (dashed arrow); interestingly, it remains largely accessible to proteinase K added from the cytosolic side of the membrane. Furthermore, PEX5(ΔN137) competes with PEX5(1–125) in the interaction with the DTM (red line), a finding suggesting that the DTM can recruit cytosolic PEX5 through different pentapeptide motifs. The putative disorder of DTM components (purple ovals with wavy lines) might contribute to this interaction flexibility. The C-shaped form and the thick blue line represent the PTS1-binding domain and the intrinsically disordered N-terminal half of PEX5, respectively. The black dots indicate the pentapeptide motifs involved in the interactions with PEX14 and PEX13.
The finding that the DTM can accommodate more molecules of PEX5(1–197;C11A) than full-length PEX5 is particularly interesting. First, it suggests that each site at the DTM available to bind a single full-length PEX5 molecule contains more than one PEX5-interacting domain. Second, because several PEX5(1–197;C11A) molecules can enter into a single DTM site, at least some of these PEX5-interacting domains are probably equivalent; i.e. they can bind to the same PEX5 sequence(s). We tried to extend these findings by using an even smaller PEX5 protein, PEX5(1–125;C11A/K), harboring just two pentapeptide motifs in our in vitro import/export assays. Our hypothesis was that at saturating conditions an even larger number of PEX5(1–125) molecules should be found at the DTM. However, this was not possible because PEX5(1–125;C11A/K) is inefficient in these in vitro assays (e.g. see Fig. 6A), and more importantly, peroxisome associated PEX5(1–125;C11A/K) is largely accessible to PK. Despite this limitation, several conclusions can be drawn from the PEX5(1–125;C11K) data. First, despite representing just of full-length PEX5, PEX5(1–125;C11K) is still correctly monoubiquitinated at the DTM and exported by the REM. Clearly, amino acid residues 126–639 of PEX5 convey no essential information to the ubiquitination/export components of the PIM. Another interesting property of PEX5(1–125;C11K) is that its entry into the DTM can be competed by PEX5(ΔN137). We do not know presently whether this competition is due to steric hindrance or whether the two proteins bind the same DTM component(s); but regardless, the fact that two non-overlapping PEX5 fragments can enter into the DTM suggests that this membrane complex can engage in interactions with PEX5 using different binding paths. In other words, it is possible that the first set of interactions that are established between a single PEX5 molecule and the DTM during the docking/insertion steps, most of them likely involving PEX14 (75, 76), do not occur necessarily through a predefined order of events. If true, this would suggest that at least some of the PEX5–DTM interactions are polymorphic in nature and probably better described by the principles that rule fuzzy protein–protein interactions (see Refs. 77 and 78). The data discussed above for PEX5(1–197) and the fact that the N-terminal half of PEX5 is a natively unfolded domain interacting with DTM components mainly (if not exclusively) through several small motifs together with an abundance of intrinsic disorder in many DTM components (e.g. 40 and 60% of human PEX13 and PEX14 sequences, respectively, are predicted to be intrinsically disordered by the PONDR®-VLXT algorithm (79, 80), Molecular Kinetics, Inc.) are surely compatible with this idea.
Finally, the fact that PEX5(1–125;C11K) enters into the DTM where it is monoubiquitinated, yielding the correct substrate for the REM, and yet remains accessible to PK added from the cytosolic side of the peroxisomal membrane is also interesting. In principle, this could simply suggest that PEX5(1–125;C11K) remains trapped at the docking step and that docking at the DTM would already be sufficient to position this PEX5 fragment in the correct orientation to be monoubiquitinated at the DTM and exported by the REM. However, we found that the interaction of Ub-PEX5(1–125;C11K) with the DTM, in contrast to the PEX5–DTM interaction at the docking step, is essentially irreversible. Furthermore, the data shown in Fig. 6D suggest that the C terminus of at least a fraction of PEX5(1–125;C11K)-clv reaches the matrix side of the peroxisome membrane. Thus, it is more likely that PEX5(1–125;C11A/K) becomes inserted into the DTM as all the other PEX5 proteins used in this work do, but because of its small size there is still sufficient space inside the DTM to provide access to PK. Altogether, the data presented in this work reveal novel aspects of the PEX5–DTM interaction and provide additional evidence to support the idea that the DTM comprises the transmembrane hydrophilic channel in which PEX5 enters to deliver its cargoes into the peroxisome matrix.
Experimental procedures
Plasmids
The oligonucleotides used to generate the plasmids described below are listed in supplemental Table S1. The cDNA encoding the large isoform of human PEX5 (10), hereafter referred to as PEX5, was obtained by PCR amplification of the plasmid pQE-30-PEX5 (53) and cloned into the NdeI/SalI restriction sites of pET-28c (Novagen), originating pET-28-PEX5. To generate the plasmid encoding full-length PEX5 possessing an alanine instead of a cysteine at position 11, the 0.3-kb NcoI/SdaI fragment of pET-28-PEX5(1–324;C11A) (27) was inserted into NcoI/SdaI-digested pET-28-PEX5, originating pET-28-PEX5(C11A). The cDNAs encoding the first 125 and 197 amino acid residues of PEX5 possessing an alanine at position 11 were obtained by PCR amplification of the plasmid pET-28-PEX5(1–324;C11A) and cloned into the NdeI/HindIII and XbaI/HindIII sites of pET-28a vector (Novagen), originating plasmids pET-28-PEX5(1–125;C11A) and pET-28-PEX5(1–197;C11A), respectively. The plasmid encoding the first 324 amino acid residues of PEX5 possessing a lysine at position 11 (pET-28PEX5(1–324;C11K)) was obtained with the QuikChange® site-directed mutagenesis kit (Agilent Technologies) using pET-28-PEX5(1–324) as template (16) and the primers described elsewhere (38). To generate plasmids encoding the first 125 and 197 amino acid residues of PEX5 possessing a lysine at position 11, the EcoRV/SdaI 1.5-kb fragment of pET-28PEX5(1–324;C11K) was inserted into EcoRV/SdaI-digested pET-28-PEX5(1–125;C11A) and pET-28-PEX5(1–197;C11A), originating plasmids pET-28-PEX5(1–125;C11K) and pET-28-PEX5(1–197;C11K), respectively. The cDNA encoding amino acids 138–639 of PEX5 (PEX5(ΔN137)) was obtained by PCR amplification from the plasmid pGEM4-PEX5(C11K) (38) and cloned into the NdeI/SalI restriction sites of pET-23a (Novagen), originating pET-23-PEX5(ΔN137). The NdeI/SalI insert of this plasmid was then cloned into the NdeI/SalI sites of pET-28a; this plasmid was then digested with NdeI, dephosphorylated with calf intestine phosphatase (New England Biolabs), and ligated to an adaptor encoding the recognition sequence of the tobacco etch virus (TEV) protease, originating pET-28-TEV-PEX5(ΔN137). A synthetic gene encoding a fusion protein comprising the first 125 amino acids of human PEX5 with the C11K mutation and possessing at its C terminus the polypeptide MQRRQVVLGHLRGPADSGWMPQAAPC*LSGAGFPEAASSFRTHQVSAAPT was codon-optimized for expression in RRL, synthesized, and cloned into the NdeI/BamHI sites of pET-28a by Genscript. The resultant plasmid, pET-28-PEX5(1–125;C11K)-clv, encodes the N-terminally histidine-tagged PEX5(1–125;C11K) protein fused to amino acid residues 1–30 of human pre-thiolase harboring the L4R mutation, which abolishes its PTS2 function (64), followed by the first 19 residues of the human sterol carrier protein-2 precursor (italic). The cleavage site for the peroxisomal matrix protease Tysnd1 is marked with an asterisk (4, 66). A nearly identical plasmid encoding a fusion protein lacking the −2 and −1 residues of the Tysnd1 cleavage site (Pro-25 and Cys-26; numbering of full-length human pre-thiolase; underlined in sequence above) was constructed in the exact same way, yielding pET-28-PEX5(1–125;C11K)-nclv. For simplicity, the two proteins encoded by these plasmids are referred to as PEX5(1–125;C11K)-clv and PEX5(1–125;C11K)-nclv, respectively.
Expression and purification of recombinant proteins
Recombinant histidine-tagged PEX5, PEX5(C11A), and PEX5(ΔN137) (53); a protein comprising amino acid residues 315–639 of PEX5 (referred to as tetratricopeptide repeats (TPRs) (48)); TPRs containing the missense mutation N526K (TPRs(N526K); numbering of full-length PEX5 (81)); NDPEX14 (48); PEX19 (82); glutathione S-transferase-ubiquitin fusion protein (35); and PEX5(1–324) and PEX5(1–197;C11A) (61) were purified as described before. The N-terminal histidine tag of PEX5(ΔN137) was removed using histidine-tagged TEV protease in 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA-NaOH, pH 8.0, 1 mm DTT overnight at 4 °C. The protein solution was then incubated with HIS-Select Nickel Affinity Gel beads (Sigma) for 2 h at 4 °C, and PEX5(ΔN137) was recovered in the non-bound fraction. The protein was concentrated by repeated centrifugation using Vivaspin® 2 sample concentrators as described before (61).
PAGE at pH 11.5
Recombinant PEX5 (2 μg), NDPEX14 (5 μg), and a mixture of both proteins were incubated for 15 min at 23 °C in a final volume of 2 μl. After dilution with 8 μl of 25 mm phosphate buffer, pH 11.5, 2.5 mm DTT, 18% (w/v) glycerol, 0.05% (w/v) bromphenol blue, the samples were loaded onto a 9% polyacrylamide gel (16.5 cm × 14.5 cm × 0.75 mm) made in 50 mm Na2HPO4, pH 11.5, with 5 m KOH. The gel was run in the same pH 11.5 buffer at 4 °C for 5 h 30 min with power limited to 3 watts. Proteins were transferred onto a nitrocellulose membrane and stained with Ponceau S. Proteins were eluted from the membrane by incubating the excised bands in SDS-PAGE sample buffer as described previously (83).
Size-exclusion chromatography
Recombinant PEX5 (180 μg), NDPEX14 (300 μg), and a mixture of both proteins were incubated in 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA-NaOH, pH 8.0, 1 mm DTT (final volume, 50 μl) for 15 min at 23 °C. Samples were then diluted with 200 μl of 0.15 m Na2CO3, pH 10.9 after dilution, and incubated on ice for 30 min, and a 200-μl aliquot was injected into a SuperoseTM 12 10/300 GL column running with 0.12 m Na2CO3, 0.5 mm DTT, pH 11.6, at 4 °C at a flow rate of 0.5 ml/min. Fractions of 0.5 ml were collected, and 30-μl aliquots were analyzed by SDS-PAGE/Coomassie Blue staining.
PNS-based in vitro assays
Liver PNS was prepared from overnight-fasted Wistar Han male rats 6–10 weeks of age as described (61). PNS-based in vitro reactions were performed as described recently (61) using 600 μg of total PNS protein and 0.1–16 μl of RRL containing the indicated 35S-labeled protein per 100-μl of reaction volume in import buffer (0.25 m sucrose, 20 mm MOPS-KOH, pH 7.2, 50 mm KCl, 3 mm MgCl2, 2 μg/ml E-64, 96 μg/ml methionine). Reactions were typically supplemented with 2 mm reduced l-glutathione, 10 μm bovine ubiquitin, and 5 mm ATP or 5 mm AMP-PNP. In the latter situation, the PNS was first primed for import by incubation for 5 min at 37 °C in the presence of 0.3 mm ATP. Ubiquitin aldehyde was used at 1 μm final concentration. Unless otherwise indicated, in vitro reactions were incubated for 45 min at 37 °C and treated with 400 μg/ml (final concentration) PK for 40 min on ice followed by PK inactivation with 500 μg/ml (final concentration) PMSF. Organelles were then processed for SDS-PAGE as described (61). In most experiments, gels were blotted onto a nitrocellulose membrane (GE Healthcare) for autoradiography. For quantitative autoradiography, the gels were instead stained with 0.2% (w/v) Coomassie, 50% (v/v) methanol,10% (v/v) glacial acetic acid; dried for 2 h at 80 °C; and exposed to a storage phosphor screen. In the two-step in vitro import/export assays, radiolabeled proteins were first incubated with the primed PNS for 30 min at 37 °C in the presence of AMP-PNP. The reactions were then diluted to 1 ml with SEMK buffer (0.25 m sucrose, 80 mm KCl, 20 mm MOPS-KOH, pH 7.2, 1 mm EDTA-NaOH, pH 8.0), and the organelles were isolated by centrifugation at 11,300 × g for 15 min at 4 °C. The organelles were resuspended in import buffer and incubated in the presence of either 5 mm ATP or 5 mm AMP-PNP for 15 min at 37 °C. The organelle suspensions were then diluted to 1 ml with SEMK buffer and centrifuged again at 11,300 × g for 15 min at 4 °C. Organelle and soluble fractions were analyzed by SDS-PAGE/Western blotting/autoradiography.
Digestion with Genenase I
PK-treated organelles from an in vitro assay programmed with 35S-labeled PEX5(1–197;C11A) or 0.25 μl of the RRL containing 35S-labeled PEX5(1–197;C11A) were digested with 2 μg of Genenase I (New England Biolabs; Ref. 62) for 30 min at 23 °C in a buffer containing 0.25 m sucrose, 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1% (w/v) Triton X-100, 1 mm EDTA-NaOH, pH 8.0, 1 mm DTT, 50 μg/ml PMSF, and 1:300 (v/v) mammalian protease inhibitor mixture (Sigma). For mass spectrometry analyses, 1 μg of recombinant PEX5(1–197;C11A) in 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA-NaOH, pH 8.0, 1 mm DTT was digested with 0.25 μg of Genenase I for 3 h 30 min at 23 °C (the results of these analyses are presented in supplemental Figs. S3 and Fig. S4).
Mass spectrometry analyses
Mass spectra were acquired in a MALDI mass spectrometer (4800 Plus MALDI TOF/TOD Analyzer, SCIEX) using the 4000 Series Explorer v3.7.0 (build 1) SCIEX software at the i3S Proteomics Core Facility (Portugal). For peptide molecular mass determination, samples were diluted 10-fold in 10 mg/ml α-cyano-4-hydroxycinnamic acid (50% acetonitrile, 0.1% TFA), and 1 μl was spotted in a MALDI sample plate and allowed to dry. Spectra were acquired in reflector positive mode for the mass range m/z 1000–3000 with and without internal standards (angiotensin I, DRVYIHPFHL, m/z 1296.68; ACTH(1–17), SYSMEHFRWGKPVGKKR, m/z 2093.09; and ACTH(18–39), RPVKVYPNGAEDESAEAFPLEF, m/z 2465.20). For protein molecular mass determination, samples were diluted 10-fold in 10 mg/ml sinapic acid matrix (50% acetonitrile, 0.1% TFA), and 1 μl was spotted in the MALDI sample plate and allowed to dry. Mass spectra were acquired in linear positive mode for the mass window m/z 15,000–35,000. Mass spectra were internally calibrated with horse apomyoglobin (m/z 16,952). For determination of protein cleavage location, data analysis was performed with FindPept software (Ref. 84; http://web.expasy.org/findpept/). The maximum allowed error was 10 ppm for peptide masses.
Density gradient centrifugation
A PK-treated PNS-based in vitro reaction (4.8 mg of PNS protein) was diluted with SE buffer (0.25 m sucrose, 20 mm MOPS-KOH, pH 7.2, 1 mm EDTA-NaOH, pH 8.0) to 1.6 ml and loaded onto the top of a HistodenzTM step gradient comprising 1.5 ml of 45% (w/v), 5.5 ml of 28% (w/v), and 1 ml of 20% (w/v) Histodenz in 5 mm MOPS-KOH, pH 7.2, 1 mm EDTA-NaOH, pH 8.0. The gradient was centrifuged at 82,500 × g for 3 h at 4 °C in a 70.1 Ti rotor (Beckman), and 12 fractions were collected from the bottom of the gradient. After precipitation of proteins with 10% (w/v) trichloroacetic acid, ¼ of each fraction was analyzed by SDS-PAGE/Western blotting/autoradiography.
Antibodies
Antibodies directed to catalase (RDI-CATALASEabr, Research Diagnostics, Inc.), KDEL (ab12223, Abcam), cytochrome c (556433, BD PharmingenTM), sterol carrier protein x (19182-1-AP, ProteinTechTM), and tetra-His (34670, Qiagen) were purchased. The antibody directed to PEX14 was described before (13). The antibody directed to PEX13 was produced in rabbits using recombinant histidine-tagged PEX13(236-403)(SH3) (51) by AMS Biotechnology. Antibodies were detected using goat alkaline phosphatase-conjugated anti-rabbit or anti-mouse antibodies (A9919 and A2429, respectively, Sigma).
Miscellaneous
35S-Labeled proteins were synthesized using the TnT® T7 quick coupled transcription/translation kit (Promega) according to the manufacturer's instructions in the presence of [35S]methionine (specific activity, >1000 Ci/mmol; PerkinElmer Life Sciences). Unless otherwise noted, 35S-labeled proteins were expressed as N-terminally histidine-tagged fusion proteins from pET-28-derived plasmids. Synthesis yields from these plasmids were significantly larger than those obtained with pGEM-4-derived plasmids used in previous works. For radiolabeled PEX5(C11A), a semiquantitative Western blot analysis using the recombinant protein as a standard and the tetra-His antibody revealed yields of about 20 ng/μl RRL. Yields were 2–3-fold larger for PEX5(1–197;C11A/K) and PEX5(1–324;C11A/K) as assessed by quantitative autoradiography using radiolabeled PEX5(C11A) as a standard. Endogenous rat liver PEX5 was detected in Western blots by blot overlay using radiolabeled PEX14 as described before (59). Sonication and fractionation of organelles were done exactly as described (27). Autoradiography data stored on phosphor screens were captured using a Storm 860 phosphorimaging system (GE Healthcare). Digital images were analyzed using ImageQuant® version 5.0 software (GE Healthcare). Peak fitting of densitometric profiles was done using Fityk curve fitting and data analysis software (85). To estimate ratios of binding sites at saturation for PEX5(1–324;C11A):PEX5(C11A) and PEX5(1–197;C11A):PEX5(C11A), data from four technical replicates were fitted to a dose-response one-site specific binding curve equation, y = Bmax × x/(EC50 + x), using Prism® version 7.03 software (GraphPad Software). In this equation, y is the methionine- and PNS protein-normalized autoradiography signal of PK-resistant protein bands, x is the volume of RRL containing the radiolabeled protein, Bmax is the maximal response at infinite volume of RRL, and EC50 is the volume of RRL yielding a half-maximal response. Note that because the actual concentrations of radiolabeled proteins in the RRL are not known the absolute EC50 and Bmax values have no meaning per se. However, Bmax ratios provide the relative binding stoichiometries of the different PEX5 species.
Author contributions
All the authors planned the experiments. A. F. D., T. A. R., A. G. P., A. B.-B., and T. F. performed the experiments. A. F. D., T. A. R., and T. F. performed the ImageQuant, Fityk, and GraphPad analyses. A. F. D. and J. E. A. wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Marc Fransen (KU Leuven) for critical reading of the manuscript and Dr. Miguel Soares (Laboratório de Apoio à Investigação em Medicina Molecular, Faculdade de Medicina da Universidade do Porto) for all the help with the use of the Storm 860 phosphorimaging system.
This work was supported in part by Fundo Europeu de Desenvolvimento Regional (FEDER) funds through the COMPETE 2020-Operational Programme for Competitiveness and Internationalization (POCI), Portugal 2020 and by Portuguese funds through Fundação para a Ciência e a Tecnologia/Ministério da Ciência, Tecnologia e Inovação in the framework of the projects “Institute for Research and Innovation in Health Sciences” (Grant POCI-01-0145-FEDER-007274) and “The molecular mechanisms of peroxisome biogenesis” (Grant PTDC/BEX-BCM/2311/2014) and through Norte 2020-Programa Operacional Regional do Norte under the application of the “Porto Neurosciences and Neurologic Disease Research Initiative at i3S” (Grant NORTE-01-0145-FEDER-000008). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Figs. S1–S5 and Table S1.
- PTS
- peroxisomal targeting signal
- DTM
- docking/translocation module
- PEX5
- peroxisomal biogenesis factor 5
- NDPEX14
- protein comprising the first 80 amino acid residues of PEX14
- PIM
- peroxisomal matrix protein import machinery
- PK
- proteinase K
- PNS
- postnuclear supernatant
- PTS1
- PTS type 1
- PTS2
- PTS type 2
- REM
- receptor export module
- RRL
- rabbit reticulocyte lysate
- TPRs
- tetratricopeptide repeats (protein comprising amino acids residues 315–639 of PEX5)
- Ub-PEX5
- monoubiquitinated PEX5
- AMP-PNP
- adenosine 5′-(β,γ-imido)triphosphate
- TEV
- tobacco etch virus
- CI
- confidence interval.
References
- 1. Lazarow P. B., and Fujiki Y. (1985) Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1, 489–530 [DOI] [PubMed] [Google Scholar]
- 2. Brocard C., and Hartig A. (2006) Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573 [DOI] [PubMed] [Google Scholar]
- 3. Gould S. J., Keller G. A., Hosken N., Wilkinson J., and Subramani S. (1989) A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108, 1657–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Swinkels B. W., Gould S. J., Bodnar A. G., Rachubinski R. A., and Subramani S. (1991) A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 10, 3255–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kunze M., Neuberger G., Maurer-Stroh S., Ma J., Eck T., Braverman N., Schmid J. A., Eisenhaber F., and Berger J. (2011) Structural requirements for interaction of peroxisomal targeting signal 2 and its receptor PEX7. J. Biol. Chem. 286, 45048–45062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Francisco T., Rodrigues T. A., Pinto M. P., Carvalho A. F., Azevedo J. E., and Grou C. P. (2014) Ubiquitin in the peroxisomal protein import pathway. Biochimie 98, 29–35 [DOI] [PubMed] [Google Scholar]
- 7. Marzioch M., Erdmann R., Veenhuis M., and Kunau W. H. (1994) PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 13, 4908–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dodt G., and Gould S. J. (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135, 1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dodt G., Braverman N., Wong C., Moser A., Moser H. W., Watkins P., Valle D., and Gould S. J. (1995) Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat. Genet. 9, 115–125 [DOI] [PubMed] [Google Scholar]
- 10. Fransen M., Brees C., Baumgart E., Vanhooren J. C., Baes M., Mannaerts G. P., and Van Veldhoven P. P. (1995) Identification and characterization of the putative human peroxisomal C-terminal targeting signal import receptor. J. Biol. Chem. 270, 7731–7736 [DOI] [PubMed] [Google Scholar]
- 11. Otera H., Okumoto K., Tateishi K., Ikoma Y., Matsuda E., Nishimura M., Tsukamoto T., Osumi T., Ohashi K., Higuchi O., and Fujiki Y. (1998) Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2: studies with PEX5-defective CHO cell mutants. Mol. Cell. Biol. 18, 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Braverman N., Dodt G., Gould S. J., and Valle D. (1998) An isoform of pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum. Mol. Genet. 7, 1195–1205 [DOI] [PubMed] [Google Scholar]
- 13. Reguenga C., Oliveira M. E., Gouveia A. M., Sá-Miranda C., and Azevedo J. E. (2001) Characterization of the mammalian peroxisomal import machinery: Pex2p, Pex5p, Pex12p, and Pex14p are subunits of the same protein assembly. J. Biol. Chem. 276, 29935–29942 [DOI] [PubMed] [Google Scholar]
- 14. Agne B., Meindl N. M., Niederhoff K., Einwächter H., Rehling P., Sickmann A., Meyer H. E., Girzalsky W., and Kunau W. H. (2003) Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol. Cell 11, 635–646 [DOI] [PubMed] [Google Scholar]
- 15. Tamura S., Yasutake S., Matsumoto N., and Fujiki Y. (2006) Dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p. J. Biol. Chem. 281, 27693–27704 [DOI] [PubMed] [Google Scholar]
- 16. Grou C. P., Carvalho A. F., Pinto M. P., Wiese S., Piechura H., Meyer H. E., Warscheid B., Sá-Miranda C., and Azevedo J. E. (2008) Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J. Biol. Chem. 283, 14190–14197 [DOI] [PubMed] [Google Scholar]
- 17. Grou C. P., Francisco T., Rodrigues T. A., Freitas M. O., Pinto M. P., Carvalho A. F., Domingues P., Wood S. A., Rodríguez-Borges J. E., Sá-Miranda C., Fransen M., and Azevedo J. E. (2012) Identification of ubiquitin-specific protease 9X (USP9X) as a deubiquitinase acting on the ubiquitin-peroxin 5 (PEX5) thioester conjugate. J. Biol. Chem. 287, 12815–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Williams C., van den Berg M., Sprenger R. R., and Distel B. (2007) A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282, 22534–22543 [DOI] [PubMed] [Google Scholar]
- 19. Miyata N., Okumoto K., Mukai S., Noguchi M., and Fujiki Y. (2012) AWP1/ZFAND6 functions in Pex5 export by interacting with Cys-monoubiquitinated Pex5 and Pex6 AAA ATPase. Traffic 13, 168–183 [DOI] [PubMed] [Google Scholar]
- 20. Liu X., Ma C., and Subramani S. (2012) Recent advances in peroxisomal matrix protein import. Curr. Opin. Cell Biol. 24, 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baker A., Hogg T. L., and Warriner S. L. (2016) Peroxisome protein import: a complex journey. Biochem. Soc. Trans. 44, 783–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Platta H. W., Brinkmeier R., Reidick C., Galiani S., Clausen M. P., and Eggeling C. (2016) Regulation of peroxisomal matrix protein import by ubiquitination. Biochim. Biophys. Acta 1863, 838–849 [DOI] [PubMed] [Google Scholar]
- 23. Kim P. K., and Hettema E. H. (2015) Multiple pathways for protein transport to peroxisomes. J. Mol. Biol. 427, 1176–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Francisco T., Rodrigues T. A., Freitas M. O., Grou C. P., Carvalho A. F., Sá-Miranda C., Pinto M. P., and Azevedo J. E. (2013) A cargo-centered perspective on the PEX5-mediated peroxisomal protein import pathway. J. Biol. Chem. 288, 29151–29159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gouveia A. M., Guimarães C. P., Oliveira M. E., Sá-Miranda C., and Azevedo J. E. (2003) Insertion of Pex5p into the peroxisomal membrane is cargo protein-dependent. J. Biol. Chem. 278, 4389–4392 [DOI] [PubMed] [Google Scholar]
- 26. Alencastre I. S., Rodrigues T. A., Grou C. P., Fransen M., Sá-Miranda C., and Azevedo J. E. (2009) Mapping the cargo protein membrane translocation step into the PEX5 cycling pathway. J. Biol. Chem. 284, 27243–27251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodrigues T. A., Alencastre I. S., Francisco T., Brites P., Fransen M., Grou C. P., and Azevedo J. E. (2014) A PEX7-centered perspective on the peroxisomal targeting signal type 2-mediated protein import pathway. Mol. Cell. Biol. 34, 2917–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gouveia A. M., Reguenga C., Oliveira M. E., Sa-Miranda C., and Azevedo J. E. (2000) Characterization of peroxisomal Pex5p from rat liver. Pex5p in the Pex5p-Pex14p membrane complex is a transmembrane protein. J. Biol. Chem. 275, 32444–32451 [DOI] [PubMed] [Google Scholar]
- 29. Gouveia A. M., Guimaraes C. P., Oliveira M. E., Reguenga C., Sa-Miranda C., and Azevedo J. E. (2003) Characterization of the peroxisomal cycling receptor, Pex5p, using a cell-free in vitro import system. J. Biol. Chem. 278, 226–232 [DOI] [PubMed] [Google Scholar]
- 30. Oliveira M. E., Gouveia A. M., Pinto R. A., Sá-Miranda C., and Azevedo J. E. (2003) The energetics of Pex5p-mediated peroxisomal protein import. J. Biol. Chem. 278, 39483–39488 [DOI] [PubMed] [Google Scholar]
- 31. Azevedo J. E., Costa-Rodrigues J., Guimarães C. P., Oliveira M. E., and Sã-Miranda C. (2004) Protein translocation across the peroxisomal membrane. Cell Biochem. Biophys. 41, 451–468 [DOI] [PubMed] [Google Scholar]
- 32. Neuhaus A., Kooshapur H., Wolf J., Meyer N. H., Madl T., Saidowsky J., Hambruch E., Lazam A., Jung M., Sattler M., Schliebs W., and Erdmann R. (2014) A novel Pex14 protein-interacting site of human Pex5 is critical for matrix protein import into peroxisomes. J. Biol. Chem. 289, 437–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Otera H., Setoguchi K., Hamasaki M., Kumashiro T., Shimizu N., and Fujiki Y. (2002) Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol. Cell. Biol. 22, 1639–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bottger G., Barnett P., Klein A. T., Kragt A., Tabak H. F., and Distel B. (2000) Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner. Mol. Biol. Cell. 11, 3963–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Carvalho A. F., Pinto M. P., Grou C. P., Alencastre I. S., Fransen M., Sá-Miranda C., and Azevedo J. E. (2007) Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282, 31267–31272 [DOI] [PubMed] [Google Scholar]
- 36. Miyata N., and Fujiki Y. (2005) Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol. Cell. Biol. 25, 10822–10832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Platta H. W., Grunau S., Rosenkranz K., Girzalsky W., and Erdmann R. (2005) Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7, 817–822 [DOI] [PubMed] [Google Scholar]
- 38. Grou C. P., Carvalho A. F., Pinto M. P., Huybrechts S. J., Sá-Miranda C., Fransen M., and Azevedo J. E. (2009) Properties of the ubiquitin-pex5p thiol ester conjugate. J. Biol. Chem. 284, 10504–10513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Debelyy M. O., Platta H. W., Saffian D., Hensel A., Thoms S., Meyer H. E., Warscheid B., Girzalsky W., and Erdmann R. (2011) Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J. Biol. Chem. 286, 28223–28234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Freitas M. O., Francisco T., Rodrigues T. A., Lismont C., Domingues P., Pinto M. P., Grou C. P., Fransen M., and Azevedo J. E. (2015) The peroxisomal protein import machinery displays a preference for monomeric substrates. Open Biol. 5, 140236, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Glover J. R., Andrews D. W., and Rachubinski R. A. (1994) Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc. Natl. Acad. Sci. U.S.A. 91, 10541–10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McNew J. A., and Goodman J. M. (1994) An oligomeric protein is imported into peroxisomes in vivo. J. Cell Biol. 127, 1245–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dias A. F., Francisco T., Rodrigues T. A., Grou C. P., and Azevedo J. E. (2016) The first minutes in the life of a peroxisomal matrix protein. Biochim. Biophys. Acta 1863, 814–820 [DOI] [PubMed] [Google Scholar]
- 44. Terlecky S. R., Nuttley W. M., McCollum D., Sock E., and Subramani S. (1995) The Pichia pastoris peroxisomal protein PAS8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO J. 14, 3627–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wiemer E. A., Nuttley W. M., Bertolaet B. L., Li X., Francke U., Wheelock M. J., Anné U. K., Johnson K. R., and Subramani S. (1995) Human peroxisomal targeting signal-1 receptor restores peroxisomal protein import in cells from patients with fatal peroxisomal disorders. J. Cell Biol. 130, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dodt G., Warren D., Becker E., Rehling P., and Gould S. J. (2001) Domain mapping of human PEX5 reveals functional and structural similarities to Saccharomyces cerevisiae Pex18p and Pex21p. J. Biol. Chem. 276, 41769–41781 [DOI] [PubMed] [Google Scholar]
- 47. Grou C. P., Carvalho A. F., Pinto M. P., Alencastre I. S., Rodrigues T. A., Freitas M. O., Francisco T., Sá-Miranda C., and Azevedo J. E. (2009) The peroxisomal protein import machinery—a case report of transient ubiquitination with a new flavor. Cell. Mol. Life Sci. 66, 254–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carvalho A. F., Costa-Rodrigues J., Correia I., Costa Pessoa J., Faria T. Q., Martins C. L., Fransen M., Sá-Miranda C., and Azevedo J. E. (2006) The N-terminal half of the peroxisomal cycling receptor Pex5p is a natively unfolded domain. J. Mol. Biol. 356, 864–875 [DOI] [PubMed] [Google Scholar]
- 49. Erdmann R., and Schliebs W. (2005) Peroxisomal matrix protein import: the transient pore model. Nat. Rev. Mol. Cell Biol. 6, 738–742 [DOI] [PubMed] [Google Scholar]
- 50. Kerssen D., Hambruch E., Klaas W., Platta H. W., de Kruijff B., Erdmann R., Kunau W.-H., and Schliebs W. (2006) Membrane association of the cycling peroxisome import receptor Pex5p. J. Biol. Chem. 281, 27003–27015 [DOI] [PubMed] [Google Scholar]
- 51. Fransen M., Brees C., Ghys K., Amery L., Mannaerts G. P., Ladant D., and Van Veldhoven P. P. (2002) Analysis of mammalian peroxin interactions using a non-transcription-based bacterial two-hybrid assay. Mol. Cell. Proteomics 1, 243–252 [DOI] [PubMed] [Google Scholar]
- 52. Saidowsky J., Dodt G., Kirchberg K., Wegner A., Nastainczyk W., Kunau W. H., and Schliebs W. (2001) The di-aromatic pentapeptide repeats of the human peroxisome import receptor PEX5 are separate high affinity binding sites for the peroxisomal membrane protein PEX14. J. Biol. Chem. 276, 34524–34529 [DOI] [PubMed] [Google Scholar]
- 53. Costa-Rodrigues J., Carvalho A. F., Fransen M., Hambruch E., Schliebs W., Sá-Miranda C., and Azevedo J. E. (2005) Pex5p, the peroxisomal cycling receptor, is a monomeric non-globular protein. J. Biol. Chem. 280, 24404–24411 [DOI] [PubMed] [Google Scholar]
- 54. Urquhart A. J., Kennedy D., Gould S. J., and Crane D. I. (2000) Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J. Biol. Chem. 275, 4127–4136 [DOI] [PubMed] [Google Scholar]
- 55. Fransen M., Terlecky S. R., and Subramani S. (1998) Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc. Natl. Acad. Sci. U.S.A. 95, 8087–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schliebs W., Saidowsky J., Agianian B., Dodt G., Herberg F. W., and Kunau W.-H. (1999) Recombinant human peroxisomal targeting signal receptor PEX5: structural basis for interaction of PEX5 with PEX14. J. Biol. Chem. 274, 5666–5673 [DOI] [PubMed] [Google Scholar]
- 57. Shimizu N., Itoh R., Hirono Y., Otera H., Ghaedi K., Tateishi K., Tamura S., Okumoto K., Harano T., Mukai S., and Fujiki Y. (1999) The peroxin Pex14p. cDNA cloning by functional complementation on a Chinese hamster ovary Cell mutant, characterization, and functional analysis. J. Biol. Chem. 274, 12593–12604 [DOI] [PubMed] [Google Scholar]
- 58. Will G. K., Soukupova M., Hong X., Erdmann K. S., Kiel J. A., Dodt G., Kunau W. H., and Erdmann R. (1999) Identification and characterization of the human orthologue of yeast Pex14p. Mol. Cell. Biol. 19, 2265–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Oliveira M. E., Reguenga C., Gouveia A. M., Guimarães C. P., Schliebs W., Kunau W.-H., Silva M. T., Sá-Miranda C., and Azevedo J. E. (2002) Mammalian Pex14p: membrane topology and characterisation of the Pex14p-Pex14p interaction. Biochim. Biophys. Acta. 1567, 13–22 [DOI] [PubMed] [Google Scholar]
- 60. Celedon J. M., and Cline K. (2012) Stoichiometry for binding and transport by the twin arginine translocation system. J. Cell Biol. 197, 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rodrigues T. A., Francisco T., Dias A. F., Pedrosa A. G., Grou C. P., and Azevedo J. E. (2016) A cell-free organelle-based in vitro system for studying the peroxisomal protein import machinery. Nat. Protoc. 11, 2454–2469 [DOI] [PubMed] [Google Scholar]
- 62. Carter P., Nilsson B., Burnier J. P., Burdick D., and Wells J. A. (1989) Engineering subtilisin BPN′ for site-specific proteolysis. Proteins 6, 240–248 [DOI] [PubMed] [Google Scholar]
- 63. Fujiki Y., Matsuzono Y., Matsuzaki T., and Fransen M. (2006) Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions. Biochim. Biophys. Acta 1763, 1639–1646 [DOI] [PubMed] [Google Scholar]
- 64. Dammai V., and Subramani S. (2001) The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105, 187–196 [DOI] [PubMed] [Google Scholar]
- 65. Rodrigues T. A., Grou C. P., and Azevedo J. E. (2015) Revisiting the intraperoxisomal pathway of mammalian PEX7. Sci. Rep. 5, 11806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kurochkin I. V., Mizuno Y., Konagaya A., Sakaki Y., Schönbach C., and Okazaki Y. (2007) Novel peroxisomal protease Tysnd1 processes PTS1- and PTS2-containing enzymes involved in β-oxidation of fatty acids. EMBO J. 26, 835–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Costa-Rodrigues J., Carvalho A. F., Gouveia A. M., Fransen M., Sá-Miranda C., and Azevedo J. E. (2004) The N terminus of the peroxisomal cycling receptor, Pex5p, is required for redirecting the peroxisome-associated peroxin back to the cytosol. J. Biol. Chem. 279, 46573–46579 [DOI] [PubMed] [Google Scholar]
- 68. Korotkov K. V., Sandkvist M., and Hol W. G. (2012) The type II secretion system: biogenesis, molecular architecture and mechanism. Nat. Rev. Microbiol. 10, 336–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Berks B. C. (2015) The twin-arginine protein translocation pathway. Annu. Rev. Biochem. 84, 843–864 [DOI] [PubMed] [Google Scholar]
- 70. Fronzes R., Christie P. J., and Waksman G. (2009) The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7, 703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yan Z., Yin M., Xu D., Zhu Y., and Li X. (2017) Structural insights into the secretin translocation channel in the type II secretion system. Nat. Struct. Mol. Biol. 24, 177–183 [DOI] [PubMed] [Google Scholar]
- 72. Dal Peraro M., and van der Goot F. G. (2016) Pore-forming toxins: ancient, but never really out of fashion. Nat. Rev. Microbiol. 14, 77–92 [DOI] [PubMed] [Google Scholar]
- 73. Beck M., and Hurt E. (2017) The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 18, 73–89 [DOI] [PubMed] [Google Scholar]
- 74. Meinecke M., Bartsch P., and Wagner R. (2016) Peroxisomal protein import pores. Biochim. Biophys. Acta 1863, 821–827 [DOI] [PubMed] [Google Scholar]
- 75. Azevedo J. E., and Schliebs W. (2006) Pex14p, more than just a docking protein. Biochim. Biophys. Acta 1763, 1574–1584 [DOI] [PubMed] [Google Scholar]
- 76. Freitas M. O., Francisco T., Rodrigues T. A., Alencastre I. S., Pinto M. P., Grou C. P., Carvalho A. F., Fransen M., Sá-Miranda C., and Azevedo J. E. (2011) PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N-terminal domain of PEX14. J. Biol. Chem. 286, 40509–40519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tompa P., and Fuxreiter M. (2008) Fuzzy complexes: polymorphism and structural disorder in protein–protein interactions. Trends Biochem. Sci. 33, 2–8 [DOI] [PubMed] [Google Scholar]
- 78. Sharma R., Raduly Z., Miskei M., and Fuxreiter M. (2015) Fuzzy complexes: specific binding without complete folding. FEBS Lett. 589, 2533–2542 [DOI] [PubMed] [Google Scholar]
- 79. Li X., Romero P., Rani M., Dunker A. K., and Obradovic Z. (1999) Predicting protein disorder for N-, C-, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 10, 30–40 [PubMed] [Google Scholar]
- 80. Romero P., Obradovic Z., and Dunker A. K. (1997) Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inform. Ser. Workshop Genome Inform. 8, 110–124 [PubMed] [Google Scholar]
- 81. Carvalho A. F., Grou C. P., Pinto M. P., Alencastre I. S., Costa-Rodrigues J., Fransen M., Sá-Miranda C., and Azevedo J. E. (2007) Functional characterization of two missense mutations in Pex5p—C11S and N526K. Biochim. Biophys. Acta. 1773, 1141–1148 [DOI] [PubMed] [Google Scholar]
- 82. Pinto M. P., Grou C. P., Alencastre I. S., Oliveira M. E., Sá-Miranda C., Fransen M., and Azevedo J. E. (2006) The import competence of a peroxisomal membrane protein is determined by Pex19p before the docking step. J. Biol. Chem. 281, 34492–34502 [DOI] [PubMed] [Google Scholar]
- 83. Pinto M. P., Grou C. P., Fransen M., Sá-Miranda C., and Azevedo J. E. (2009) The cytosolic domain of PEX3, a protein involved in the biogenesis of peroxisomes, binds membrane lipids. Biochim. Biophys. Acta. 1793, 1669–1675 [DOI] [PubMed] [Google Scholar]
- 84. Gattiker A., Bienvenut W. V., Bairoch A., and Gasteiger E. (2002) FindPept, a tool to identify unmatched masses in peptide mass fingerprinting protein identification. Proteomics 2, 1435–1444 [DOI] [PubMed] [Google Scholar]
- 85. Wojdyr M. (2010) Fityk: a general-purpose peak fitting program. J. Appl. Crystallogr. 43, 1126–1128 [Google Scholar]
- 86. Francisco T., Dias A. F., Pedrosa A. G., Grou C. P., Rodrigues T. A., and Azevedo J. E. (2017) Determining the topology of peroxisomal proteins using protease protection assays. Methods Mol. Biol. 1595, 27–35 [DOI] [PubMed] [Google Scholar]
- 87. Gatto G. J. Jr., Geisbrecht B. V., Gould S. J., and Berg J. M. (2000) Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7, 1091–1095 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.