Abstract
The immunomodulatory receptor Siglec-3/CD33 influences risk for late-onset Alzheimer's disease (LOAD), an apparently human-specific post-reproductive disease. CD33 generates two splice variants: a full-length CD33M transcript produced primarily by the “LOAD-risk” allele and a shorter CD33m isoform lacking the sialic acid-binding domain produced primarily from the “LOAD-protective” allele. An SNP that modulates CD33 splicing to favor CD33m is associated with enhanced microglial activity. Individuals expressing more protective isoform accumulate less brain β-amyloid and have a lower LOAD risk. How the CD33m isoform increases β-amyloid clearance remains unknown. We report that the protection by the CD33m isoform may not be conferred by what it does but, rather, from what it cannot do. Analysis of blood neutrophils and monocytes and a microglial cell line revealed that unlike CD33M, the CD33m isoform does not localize to cell surfaces; instead, it accumulates in peroxisomes. Cell stimulation and activation did not mobilize CD33m to the surface. Thus, the CD33m isoform may neither interact directly with amyloid plaques nor engage in cell-surface signaling. Rather, production and localization of CD33m in peroxisomes is a way of diminishing the amount of CD33M and enhancing β-amyloid clearance. We confirmed intracellular localization by generating a CD33m-specific monoclonal antibody. Of note, CD33 is the only Siglec with a peroxisome-targeting sequence, and this motif emerged by convergent evolution in toothed whales, the only other mammals with a prolonged post-reproductive lifespan. The CD33 allele that protects post-reproductive individuals from LOAD may have evolved by adaptive loss-of-function, an example of the less-is-more hypothesis.
Keywords: alternative splicing, Alzheimer disease, intracellular trafficking, peroxisome, sialic acid, CD33, Siglec-3
Introduction
CD33 (Siglec-3)4 is the eponymous member of the CD33rSiglec family. CD33rSiglecs recognize sialic acids, the nine carbon-backbone monosaccharides that are often present at the terminal ends of the glycan chain of glycoproteins and glycolipids. Siglecs generally contain several extracellular immunoglobulin domains. A terminal V-set domain binds sialylated ligands and is followed by a varying number of C2-set domains, a single pass transmembrane domain, and an intracellular tail. CD33rSiglecs are commonly expressed on the surface of immune cells and bind extracellular ligands to influence the activation state within the cell (1–3). CD33rSiglecs are classified as inhibitory or activating based on the effect of ligand binding on immune cell activation (1–3). Inhibitory Siglecs carry immunoreceptor tyrosine-based inhibition motif (ITIM) and ITIM-like motifs in their intracellular domain. Ligand binding results in phosphorylation of a tyrosine residue in these motifs, and recruitment of tyrosine phosphatases SHP-1 and SHP-2 initiates a signaling cascade that inhibits the immune response (1–4). Depletion of SHP-1 and SHP-2 leads to an increase in the endocytosis of the CD33 receptor (5). Notably, the functions of CD33rSiglecs evolve rapidly. Mouse and human CD33 are rather different; they are expressed on different cells, bind to different sialic acid probes, and carry different intracellular domains (6).
Several genome-wide association studies found that alleles of CD33 influence the risk of late-onset Alzheimer's disease (LOAD) (7–10). LOAD is a human disease that impairs the cognitive function of post-reproductive individuals, and other primates do not appear to be susceptible (11–13). A new CD33 allele evolved in the human lineage: that is, two human-specific variants (rs3865444A and rs12459419T) are associated with protection against Alzheimer's onset in elderly individuals (14). The protective allele modifies the ratio of two splice-variants (full length CD33M and truncated CD33m) increasing the relative abundance of CD33m (15). CD33m is missing the terminal V-set domain that engages ligands outside the cell (16). The ratio of CD33M and CD33m in protected individuals resembles that of chimpanzees, suggesting that this new allele evolved to compensate for other changes that occurred on the human lineage (14). Because the effects of the derived protective allele are primarily in post-reproductive individuals, it may have evolved by inclusive selection. Preserving the cognitive functions of the elderly increases the survival of their younger relatives and decreases the burden of elder care for the social group (17).
Mutations that improve an existing trait can benefit health, and gain-of-function mutations that prevent disease have been extensively studied. But if an existing function becomes pathological, then losing this function can also prevent disease (18–20). This less-is-more scenario has received relatively little empirical attention (21, 22), perhaps because it opposes the a priori assumption that existing organismal traits are well-suited to their environment or perhaps because the absence of a function is challenging to demonstrate conclusively. We delve into one potential case using the expression and cellular localization of an alternative splice form of CD33 to examine the mechanisms that may explain its protective effect on Alzheimer's disease. Here, we delimit the possible mechanisms of immune cell activation by CD33m. We ask whether the CD33m isoform could engage in any functional interaction with β-amyloid or signaling at the cell surface and whether immune cell activation or ligand binding influences the localization of CD33m within cells. We also trace the evolution of a motif that guides the intracellular localization of CD33m among mammalian species. We also generated and characterized CD33m-specific antibodies that do not react with CD33M.
Results and discussion
Discovery of an intracellular pool of CD33
Previous studies reported very low expression of CD33 on mature granulocytes (6, 23). To determine whether CD33 is expressed at the cell surface or inside the cell, we probed neutrophils using an antibody (HIM3-4) that reacts with C2-set domain of CD33. Notably, we found a large pool of CD33 inside permeabilized neutrophils (Fig. 1A). Neutrophils showed no internal expression of CD45, a control molecule normally expressed on the cell surface (Fig. 1C). We also found an intracellular pool of CD33 in monocytes (Fig. 1A). Unlike neutrophils, monocytes have high expression of CD33 on their cell surface and also express CD33 in an internal compartment (Fig. 1A). Because CD33 expression is relevant for a role of microglia in Alzheimer's dementia, we chose a human microglia cell line CHME-5 to determine the status of CD33. Interestingly, we found large intracellular pool of CD33 in CHME-5 cells. We found no evidence of an intracellular pool of any other CD33rSiglec (Siglec-5/14, Siglec-7, Siglec-8, Siglec-9, Siglec-10, and Siglec-11/16) in neutrophils (Fig. 1B).
Figure 1.
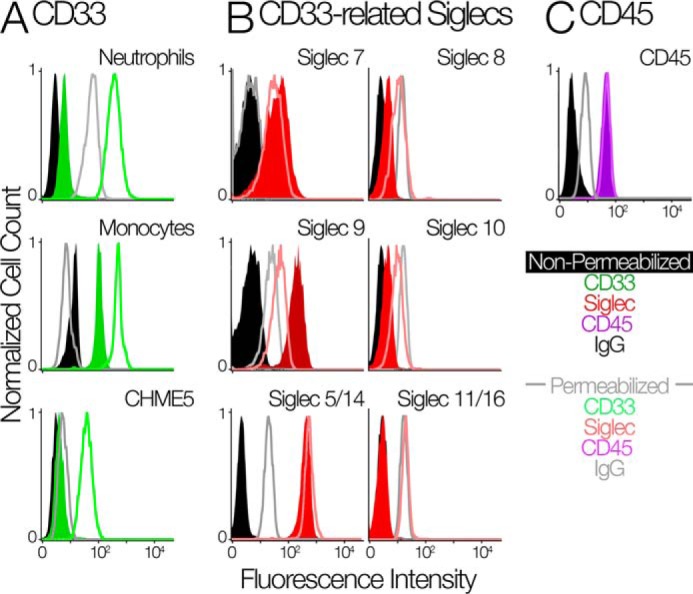
Human innate immune cells contain an intracellular pool of CD33. A, the flow cytometry analysis of CD33 on the cell surface and intracellular in neutrophils (top panel), monocytes (middle panel), and CHME5 cell line (bottom panel). Cells were measured with their membranes intact (green-shaded distributions) and permeabilized using cytoperm/cytofix kit (green-unshaded distributions). IgG controls are in black (intact cells) and gray (permeabilized cells). B, flow cytometry analysis of other CD33rSiglecs in identically treated human neutrophils. The amount of each marker was determined by FACS using the indicated antibodies. Neutrophils were measured with their membranes intact (red-shaded distributions) and permeabilized using cytoperm/cytofix kit (red-unshaded distributions). IgG or secondary antibody controls are in black (intact cells) and gray (permeabilized cells). C, flow cytometry analysis of CD45 in identically treated human neutrophils. Cells were measured with their membranes intact (purple-shaded distributions) and permeabilized using cytoperm/cytofix kit (purple-unshaded distributions). IgG controls are in black (intact cells) and gray (permeabilized cells).
CD33m is the primary intracellular variant
We were interested to know if the intracellular pool of CD33 consists of CD33m, CD33M, or both splice variants. Using antibodies specific to both the V-set domain (WM53) and the C2-set domain (HIM3-4), we found that in neutrophils and CHME-5 cells, the intracellular pool consists primarily of the CD33m isoform (Fig. 2, A and B). One possible reason for this difference in trafficking of intracellular pool could be the loss of a disulfide bond between the V-set and the C2-set domain. CD33M is expressed as a disulfide dimer on the cell surface (24). Interdomain disulfide bonds are salient features of the Siglec family (25). CD33 V-set and C2-set domains each have three cysteines residues. Moreover, it has previously been shown that a CD33M mutant (one where the cysteine of the V-set domain is mutated) stays inside the cell and cannot traffic to the cell surface (16). In pursuing this question, we could not get consistent and reproducible Western blots for CD33m despite trying various lysis protocols for neutrophils and CHME-5 cells such as with Nonidet P-40 buffer, RIPA (radioimmune precipitation assay buffer) buffer, 1% SDS lysis, freeze-thaw, and 1% SDS lysis+sonication. Despite all these methods it seems that CD33m in neutrophils and CHME-5 cells is highly prone to degradation immediately upon cell breakage and cannot be consistently detected and studied on Western blots. Therefore, we relied on the immunofluorescence approach.
Figure 2.

The intracellular pool is composed of the CD33m isoform. A, CD33 exists in two splice forms: a full length CD33M and a truncated CD33m that is lacking the V-set domain. Human alleles that predispose individuals to Alzheimer's are spliced primarily into the CD33M form, alleles that protect from Alzheimer's are spliced primarily into CD33m. B, the left panel shows the schematic regarding the reactivity of different CD33 antibodies that can partially discriminate the two forms. WM53 targets the V-set domain and labels only CD33M (blue). HIM3-4 targets the C2set domain and labels both CD33M and CD33m (green). The middle panel shows the flow cytometry analysis of CD33M and CD33m+M using the antibodies WM53 and HIM3-4 with intact and permeabilized cells. The right panel shows the quantification of mean fluorescence intensity of CD33M and CD33m+M in intact and permeabilized cells. The line shows mean fluorescence intensity. **, p < 0.01, n = 4 for CD33m+M and n = 3 for CD33M.
Subcellular localization of the intracellular CD33 pool
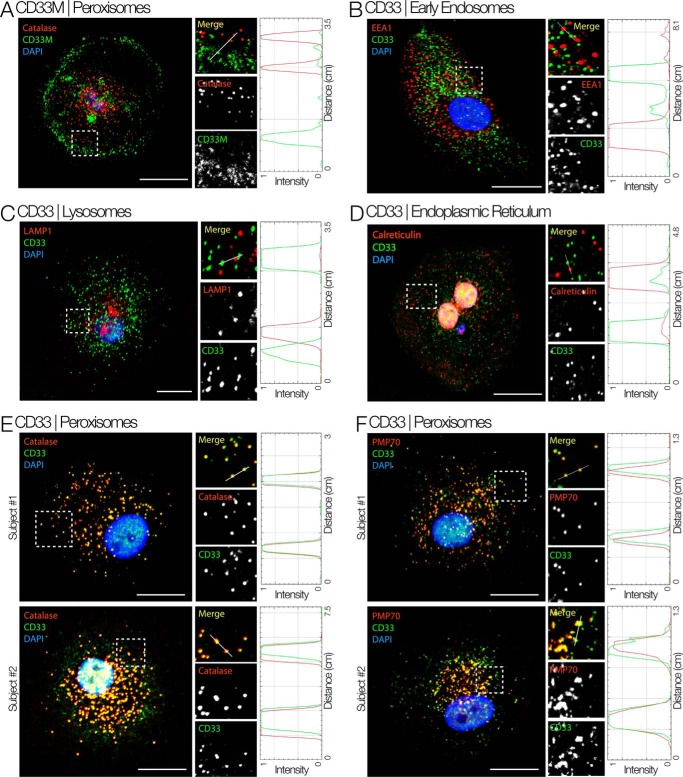
We used immunofluorescence to determine the cellular compartment where the intracellular pool of CD33 is located. We chose monocyte-derived macrophages to do these studies because of their distinct morphology (spread-out, with a large cytoplasmic pool), which allows estimation of co-localization with organelle markers with higher degrees of confidence. Immunostaining with the C2-set specific antibody (HIM3-4 clone, which detects both CD33M and CD33m isoforms) confirmed the intracellular localization of CD33 on punctate structures, and these showed little or no co-localization with early endosomes (marked by EEA1, Fig. 3B), lysosomes (marked by LAMP1, Fig. 3C) or the ER (marked by Calreticulin, Fig. 3D). Instead, CD33-positive punctate structures show clear co-localization with two peroxisome markers, Catalase and PMP70 (Fig. 3, E and F). To determine whether the observed peroxisome-localized pool of CD33 is the full length or truncated isoform, we stained macrophages with an antibody that exclusively detects the full length CD33M isoform (WM53). This CD33M-detecting antibody revealed a pool of CD33 on the cell surface and little or none in the intracellular pool (Fig. 3A). Thus, CD33M localizes predominantly at the cell surface, and the intracellular pool in peroxisomes is primarily composed of the CD33m isoform. We can also eliminate the possibility that the truncated CD33m is present as an aggregate inside the cells. First, we found it in the peroxisomes, and it carries the motif for trafficking to this organelle (see below). Secondly, we observed almost no co-localization with ER marker calreticulin. An early aggregate of CD33 should have shown co-localization with an ER marker.
Figure 3.
Intracellular CD33m localizes to peroxisomes. Shown is immunofluorescence staining for different cell compartments of macrophages isolated from whole blood using HIM3-4 clone. Boxed areas on left are magnified and displayed as individual channels. RGB profile plots measure the degree of co-localization of red and green pixels on vesicles along the line drawn in each magnified (Merge) panel. A, immunostaining specifically for the CD33M isoform (antibody WM53) shows that the full-length CD33M isoform is mostly located on the plasma membrane. B–F, the intracellular pool of CD33 co-localizes with peroxisome markers (Catalase and PMP70) but not with markers of early endosome (EEA1), lysosome (LAMP1), or endoplasmic reticulum (Calreticulin). Scale bar = 10 μm.
Generation and characterization of specific CD33m antibodies
We generated several clones of antibodies for CD33m, which does not react with CD33M. We further characterized clone A16121H on cell lines and primary cells (Fig. 4A). CHME-5 cells, which contains only CD33m and does not have CD33M, was used for initial characterization (Fig. 4A). We found that all the CD33m protein in this cell line is indeed present as an intracellular pool. U937 cells are known to have large amounts of CD33M and small amounts of CD33m (26). Using this newly generated antibody we found that this CD33m is accumulated inside the cells. Furthermore, we confirmed our findings of the existence of an intracellular pool of CD33m in neutrophils (Fig. 4A). Using clone A16121H of CD33m and catalase, we also confirmed peroxisomal localization of CD33m in monocyte-derived macrophages (Fig. 4B).
Figure 4.
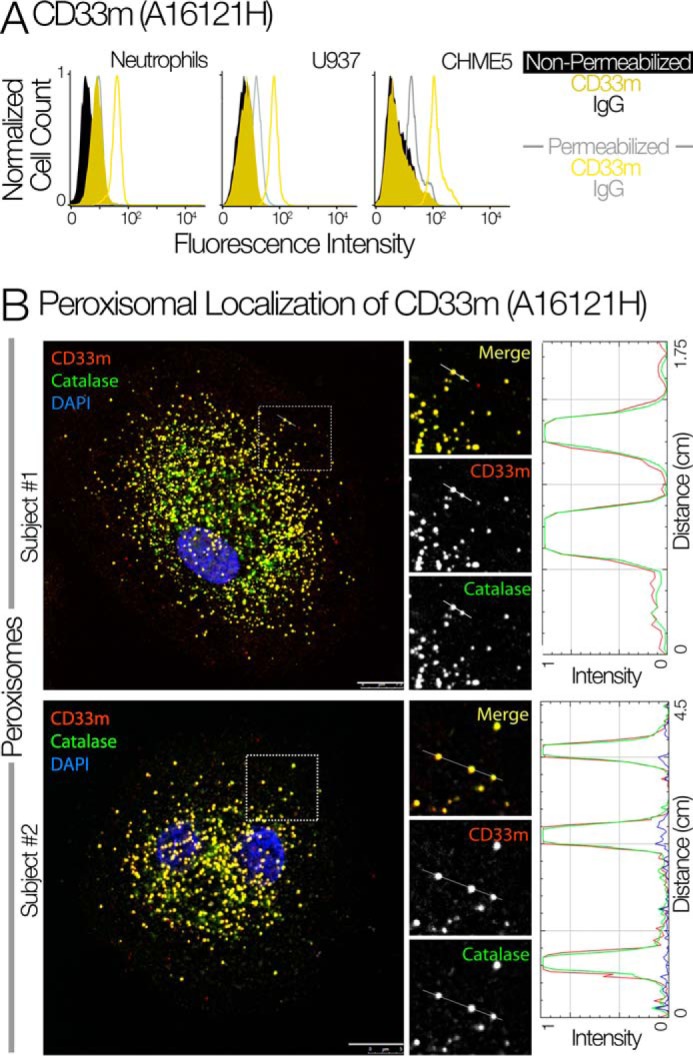
Confirmation of peroxisomal localization of CD33m using isoform-specific antibodies. A, the flow cytometry analysis of CD33m in neutrophils, U937, and CHME-5 cells using the newly generated CD33m antibody (Clone A16121H). B, the immunofluorescence staining of CD33m and catalase showed the peroxisomal targeting of CD33m in monocyte-derived macrophages (n = 2).
Retention of CD33M on the cell surface is independent of interaction with extracellular sialic acid ligands. The monocyte/macrophage-like cell line U937 has robust expression of CD33 that majorly consists of CD33M (26). We hypothesized that all CD33 receptors go to the cell surface and are retained when bound to extracellular sialylated ligands. To test this hypothesis, U937 cells were treated with sialidase to selectively remove sialic acid from the cell surface. However, we found no significant change in the cell surface expression of CD33 (Fig. 5A). This shows that the retention of CD33M on the cell surface and the intracellular localization of CD33m are not a result of their respective ability to bind sialic acids and be retained by extracellular ligands.
Figure 5.
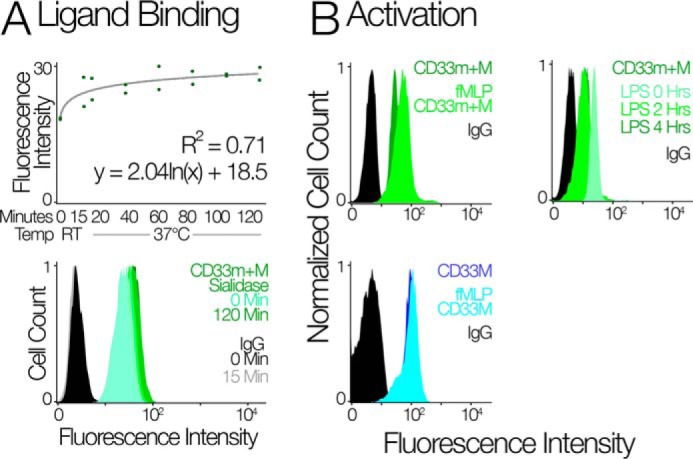
CD33 localization is insensitive to activating stimuli. A, the top panel shows flow cytometry analysis of CD33 on the surface of U937 cells upon sialidase treatment. The scatter plot gives the predicted mean fluorescence intensity based on least squares logarithmic regression. Two replicates were performed for each time step: 0 and 15 min at room temperature and 20, 40, 60, 80, 100, and 120 min at 37 °C. The bottom panel shows the fluorescence intensity of CD33 in U937 cells upon sialidase treatment for different time points. Sialidase treatment with AUS to remove CD33 ligands at the cell surface did not change the distribution of CD33m+M. Thus, the intracellular pool of CD33m is not due to its inability to bind sialic acid ligands. B, the top left panel shows the FACS analysis of CD33 using HIM3-4 clone on neutrophils upon stimulation with fMLP; the top right panel shows the flow cytometry analysis of CD33 on neutrophil upon stimulation with LPS. The bottom panel shows that the flow cytometry analysis of CD33M in neutrophil upon stimulation with fMLP. Treatment of neutrophils with LPS led to a decrease in cell surface CD33, and contrary to this, treatment with fMLP led to partial mobilization of CD33m to the cell surface.
Limited mobilization of intracellular CD33m pool to the cell surface after stimulation.
In some cases cell surface receptors can exist as an intracellular pool and migrate to the cell surface in response to an inflammatory stimulus (27–31). To check the possible mobilization of CD33m to the cell surface, we stimulated/activated the cells with lipopolysaccharide (LPS) or formylmethionyl-leucyl-phenylalanine (fMLP). First, we treated the neutrophils with LPS for 2 and 4 h. We found that LPS stimulation leads to decrease in cell surface expression of CD33 (Fig. 5B, right top panel). Prior studies have also shown this down-regulation of CD33 upon LPS stimulation and that this facilitates inflammation (32). fMLP is the stimulus that activates neutrophils and causes chemotaxis out of the bloodstream (33, 34). If activation motivates CD33m surface expression, we would expect a reduction in the intracellular pool when cells are treated with fMLP. Using different anti-CD33 antibodies (CD33M and CD33m+M) we found there is partial mobilization of CD33 to the surface, and this fraction is CD33m (Fig. 5B, top and bottom left panel).
Phylogenetic origins of the putative peroxisome trafficking motif
We noted that CD33 carries a peroxisome targeting sequence in the cytoplasmic tail, which is absent in all other human CD33rSiglecs (Fig. 6A). Most peroxisomal protein targeting occurs by recognition of a PTS1 motif at the extreme end of the C terminus domain, but in some cases an internal motif can traffic protein to the peroxisomes (35–38). In CD33, the putative motif is present near the C terminus but not at the extreme position. Although we found that only the truncated CD33m variant localizes to peroxisomes, the SKL motif is also present in the full-length CD33M variant. It is possible that the putative PTS1 in the full-length CD33M variant is ineffective due to phospho-modifications on flanking regions of the cytoplasmic tail and/or allosteric competition with endocytic adaptors that mediate clathrin-mediated endocytosis of the cell surface receptor. Comparison of CD33 sequences across mammals showed that predicted PTS1 peroxisome-targeting motifs are present in three groups: Haplorhine primates, Perissodactyl ungulates (in a CD33-like duplicate), and Odontoceti-toothed whales (38). Ancestral state reconstructions indicate that these motifs evolved independently (Fig. 6B). Among mammals, the third amino acid position of the PTS1 motif evolved by positive selection (BEB p = 0.997. PAML M7 versus M8: np = 122, 2ΔL > 356, p < 5 × 10−78; supplemental Figs. S1 and S2), where BEB is Bayes Empirical Bayes, np is the number of parameters, PAML is Phylogenetic Analysis by Maximum Likelihood, and L (in 2ΔL) is likelihood. Odontoceti-toothed whales have an intact peroxisomal targeting sequence that is interesting because they are the only other mammalian group with an extended post-reproductive lifespan (39, 40). As in humans, grandmothering behavior in toothed whales would allow inclusive fitness to select for such alleles that reduce cognitive decline in elderly individuals.
Figure 6.
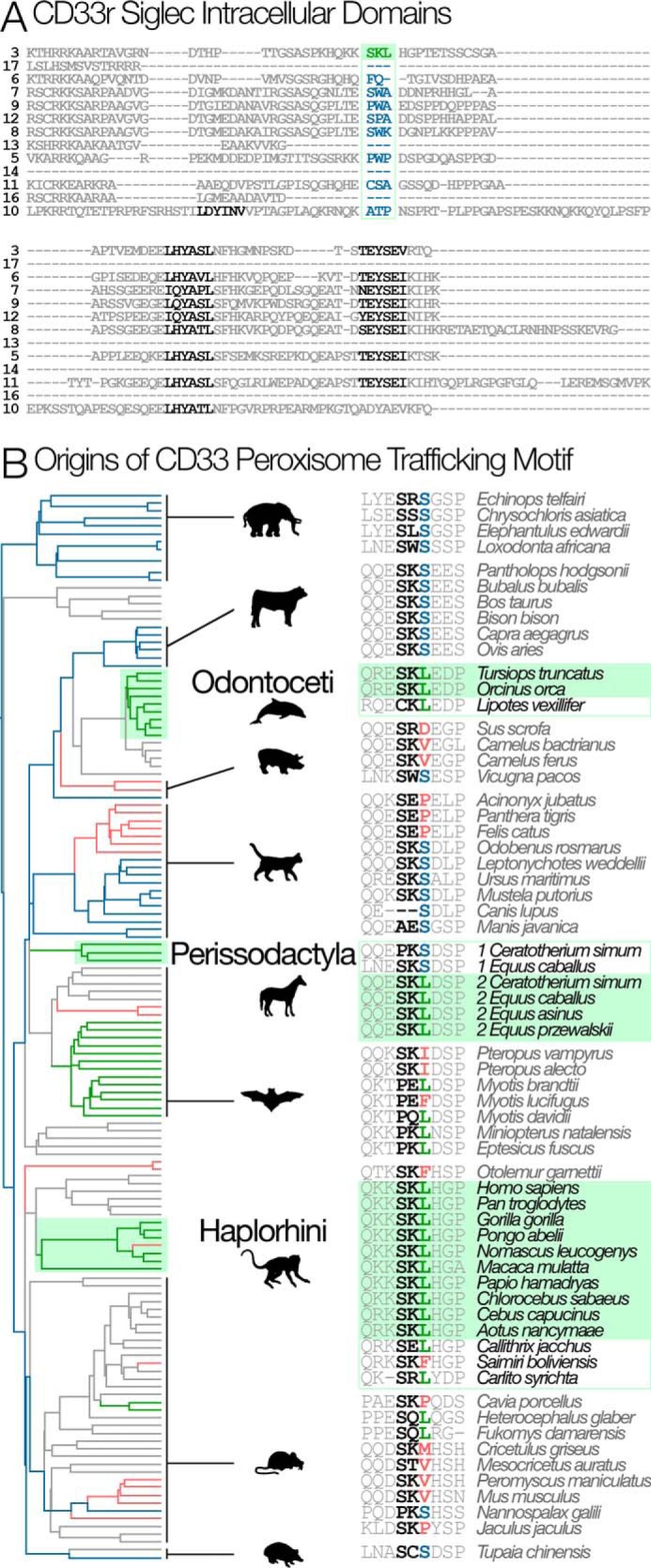
Phylogenetic origins of the peroxisome trafficking motif. A, alignment of the intracellular region of human CD33rSiglecs (Siglec 13 is from chimpanzee). Among human CD33rSiglecs, only CD33 has a peroxisome trafficking motif (SKL: shown in green). ITIM and ITIM-like motifs are shown in black. B, the SKL motif evolved multiple times in parallel (green boxes): Haplorhine primates, Perissodactyl ungulates (in one of two CD33-like duplicates), and Odontoceti-toothed whales. Branches are colored according to the third position of the SKL motif. S is the most likely ancestral state found in clades whose common ancestors existed at the base of the tree (blue branches). L (green branches) exist in several places in the tree and are not always associated with the SKL motif. Other amino acids at this position are colored red and are represented by red branches. Unsampled mammalian clades are shown in gray.
Potential effects of CD33m in the peroxisome
We show the presence of an intracellular pool of CD33 in primary innate immune cells, a finding with many potential clinical implications. The peroxisome is the major site for metabolism of hydrogen peroxide and other reactive oxygen species, and there are clues to how peroxisome functions may influence LOAD (41, 42). Peroxisome dysfunction (either resulting from a peroxisome biogenesis or a single enzyme defect) has been identified in severe demyelinating and neurodegenerative brain diseases (43). Recent evidence suggests that increased oxidative stress during LOAD progression is accompanied by loss of peroxisomes (44). Studies also point to functional and biogenetic relationships linking peroxisomes to mitochondria and suggest peroxisomal proteins as biomarkers/therapeutic targets in pre-symptomatic Alzheimer's disease (45). Much of the literature surrounds the use of agonists against PPARs (peroxisome proliferator-activated receptors) as therapeutic treatments to increase the number of peroxisomes in cells. Recent studies showed that the PPAR-γ agonists play an important role in β-amyloid clearance by the microglia in brain for example, Pioglitazone treatment leads to polarization of microglia in M2 state and enhanced clearance of β-amyloid (46). Many non-steroidal anti-inflammatory drugs (NSAIDs) have also been shown to benefit Alzheimer's disease patients. These effects are associated with PPAR-γ activation in microglia, which leads to down-regulation of cytokines and inducible nitric-oxide synthase expression (47, 48). Whether CD33m contributes to these processes is a complex question that needs further investigation. It may be that CD33m is a decoy receptor that does not require sialic acid ligands for its activity or that its intracellular localization stimulates peroxisome proliferation. Technical difficulties with the immunoprecipitation of intracellular CD33 as mentioned above prevented us from determining whether the CD33m isoform actually takes part in signaling (whether it gets tyrosine-phosphorylated or recruits SHP-1 phosphatase), and if there are functions of CD33m within the peroxisome, they remain to be determined.
Conclusions and perspectives
Prior studies of the two isoforms of CD33 mainly used transfected cell with overexpression and concluded that both forms are found only on the cell surface (26, 49). We studied the cellular localization of two isoforms in native cells and cell lines and instead found that CD33m is virtually undetectable on the cell surface at steady state and, therefore, is unlikely to be engaged in cell surface signaling as previously assumed. Thus, the derived CD33 allele that evolved in humans achieved its protective effect by the loss of an existing signaling function, and this mechanism alone may be sufficient to explain the observed protective effect of the derived CD33 allele. CD33m has lost the ability to suppress microglial activation, and this (along with the concomitant decrease in the abundance of the inhibitory CD33M isoform) may increase overall microglial activity against amyloid plaques in the brain protecting post-reproductive individuals from LOAD. Based on the specific trafficking of CD33m in peroxisome, we could also hypothesize that CD33m in peroxisome might have some other unknown functions that remain to be determined in future studies.
The less-is-more hypothesis suggests that loss-of-function mutations can be adaptive and that they should be common, because mutations that result in a loss-of-function are common (21, 22). There are many gene losses with apparent beneficial effects, but few such mutations are actually recognized as adaptive losses of function (Table 1). For empiricists, the intrinsic difficulty of the less-is-more hypothesis is that it posits a negative. It will not ever be possible to exhaustively explore all potential alternative functions, and indeed this is a challenge for testing any adaptive hypothesis (50). To assess the importance of the less-is-more mode of evolution, we must instead look for examples where an adaptive phenotypic difference can be plausibly explained by effects that can be measured. Here we provide a clear example; CD33m has lost some of the functions of the ancestral CD33M molecule; it cannot bind sialic acids, and its location inside the cell prevents it from signaling to suppress microglial activation. There may of course be other new functions associated with the presence of CD33m in the peroxisome, and if CD33m influences peroxisomal proliferation it may be a promising therapeutic target. But the observed loss of signaling may be sufficient to explain the protective effect of CD33m on LOAD. The adaptive importance of preserving cognitive function in the elderly is clear (14). Thus, the derived allele that protects elderly humans from LOAD disease likely evolved by a less-is-more adaptive loss-of-function.
Table 1.
Loss-of-function mutations that can be beneficial for survival
| Gene | Type of mutation | Disease improved | References |
|---|---|---|---|
| NPC1L1 | Frameshift, non-sense and splice-site mutations | Coronary heart disease | 59 |
| APOC3 | Null mutation | Ischemic vascular disease | 60 |
| SLC30A8 | Non-sense mutations | Type II diabetes | 19 |
| LPA | Splice-site mutations | Cardiovascular diseases | 18 |
| APP | Missense mutation | Alzheimer's disease and age-related cognitive decline | 61 |
| PCSK9 | Non-sense and missense mutations | Coronary heart disease | 20 |
| Caspase12 | Gene deletion | Sepsis | 62 |
| CCR-5 | Deletion leading to frameshift mutation | HIV-1 infection | 63 |
| TYK2 | Missense mutations | Rheumatoid arthritis, systemic lupus erythematosus | 64 |
| IFIH1 | Missense and non-sense mutations | Type 1 diabetes | 65 |
| CARD9 | Splice-site mutation | Inflammatory bowel diseases (IBD) | 66 |
| RNF186 | Non-sense mutation | Ulcerative colitis | 67 |
| CD33 | Increase in non-functional splice variant | Alzheimer's disease and age-related cognitive decline | This paper |
Deciphering the role of CD33m and CD33M in Alzheimer's dementia is also hindered due to non-accessibility of specific antibodies against CD33m that does not react with CD33M. To address this problem we also generated new rat monoclonal CD33m antibodies with no reactivity for CD33M. The new antibody will allow us to study CD33m and CD33M independently and examine potential CD33m functions in the peroxisome.
Experimental procedures
Cells and antibodies
For this study, polymorphonuclear leukocytes (PMN) and monocytes were isolated from healthy donors in accordance with the guidelines issued by the Institutional Review Board, University of California, San Diego (UCSD). PMN isolation was performed with 50 ml of heparinized blood using PolymorphprepTM (Axis Shield, Dundee, Scotland) as per the manufacturer's protocol. A monocyte/macrophage-like cell line U937 was used for sialidase treatment. CD33rSiglec antibodies such as Siglec-5/14 (Clone 1A5, BioLegend Inc., catalog no. 352004), Siglec-7 (Clone 194211, R&D, catalog no. FAB11381A), Siglec-8 (7C9, BioLegend catalog no. 347104), Siglec-10 (Clone 5G6, BioLegend, catalog no. 347604), and Siglec-11/16 (Clone 4C4) were used for FACS analysis.
Flow cytometry of permeabilized and non-permeabilized cells
Cells were stained with anti-CD33 antibodies specific for the different domains as indicated in Fig. 2B. For the non-permeabilization condition, 1 million cells per sample were stained with the indicated antibodies in FACS buffer (1% BSA in PBS with 10 mm EDTA) for 30 min on ice. For the permeabilization condition, cytoperm/cytofix kit (BD Pharmingen, catalog no. 554714) was used. The permeabilization and staining was performed according to the manufacturer.
Immunofluorescence and confocal microscopy
For immunofluorescence staining, 50 ml of blood was isolated from the healthy donors. The peripheral blood mononuclear cell layer was isolated from the blood and split into a 12-well plate in complete media (RPMI-40 + 10%FCS + 1% penicillin/streptomycin) covered with sterile glass coverslips. The medium was replaced after overnight incubation, and new medium containing RPMI-40 + 10%FCS + 100 ng/ml of human M-CSF (BioLegend, catalog no. 574806) was added in order to differentiate monocytes into macrophages. After 5 days of incubation at 37 °C with 5% CO2 the cells were washed 3 times with PBS and fixed with 4% paraformaldehyde at room temperature for 20 min. The cells were permeabilized (0.1% Triton X-100) for 1 h and incubated for 1 h with each primary and then secondary antibodies as described previously (51). Dilutions of antibodies and reagents were as follows: anti-CD33m+M (Clone HIM3-4 Biolegend catalog no. 303304, 1:20), EEA1 (cell signaling, clone C45B10, catalog no. 3288, 1:250), calreticulin (Abcam catalog no. ab2907, 1:1000), LAMP1 (Abcam catalog no. ab24170, 1:1000), Catalase (EMD Millipore catalog no. 219010, 1:200), PMP70 (Abcam catalog no. ab3421, 1:600), and CD33M (Clone WM53, Biolegend, catalog no. 303402, 1:100). Secondary goat anti-mouse-Alexa488 and goat anti-rabbit Alexa594 (1:500) were used. Images were acquired using a Leica CTR4000 Confocal Microscope with a 63× objective. Z-stack images were obtained by imaging ∼4-μm thick sections of cells in all channels. To estimate the degree of co-localization in immunofluorescence assays, ImageJ RGB Profiler plugin was used to determine the intensity fluorescence distribution for corresponding fluorophore (green and red channels). Red-green-blue (RGB) graphic profiles were created by analyzing the distribution and intensity of pixels of these colors along a chosen line using ImageJ software. All individual images were processed using Image J software and assembled for presentation using Photoshop and Illustrator software (Adobe).
Generation of rat monoclonal antibodies against CD33m
The procedure used was similar to that described previously (52) with the following modifications. The immunogen was a Keyhole limpet hemocyanin-conjugated peptide corresponding to the N-terminal sequence unique to CD33m. The hybridomas were screened by enzyme-linked immunoassay on 96-well plates coated with recombinant CD33m-Fc and CD33M-Fc, separately. CD33m-Fc-positive clones that did not show reactivity with CD33M-Fc were selected and subcloned.
Sialidase, LPS, and fMLP treatment
U937 cells were treated with Arthrobacter ureafaciens sialidase (AUS) as described previously (53). Briefly, 20 milliunits of AUS were incubated with 1 million cells per condition at 37 °C for different time points up to 2 h. For the LPS and fMLP treatment, 1000 ng/ml LPS and 100 nm fMLP at 37 °C were used on PMN isolated from human blood. The treatment of LPS was performed for 0, 2, and 4 h, whereas treatment with fMLP was performed for 15 min.
Statistics
GraphPad Prism 5 was used to implement unpaired Student'st tests for analyses involving two groups and an analysis of variance for comparisons of CD33 abundance after sialidase treatment. CD33 sequences were collected from as many mammal species as possible by blast (54). Alignments and a maximum likelihood phylogeny of CD33 were generated using the default settings on the www.phylogeny.fr server5 (38, 55). Models supporting the codon specific tests of selection were estimated using PAML v4.7 (56). MEGA 6.0 (57) was used to reconstruct ancestral states under maximum parsimony and maximum likelihood (JTT substitution model, uniform rates). Peroxisome motifs were predicted using the PTS1 Prediction server (mendel.imp.ac.at/mendeljsp/sat/pts1/PTS1predictor.jsp)5 (58).
Author contributions
S. S. S., S. A. S., P. G, W. J., and A. Varki wrote the manuscript. S. S. S., A. Verhagen, S. A. S., V. S., F. A.-S., and P. G. performed the experiments. All the authors read the manuscript and approved it.
Supplementary Material
Acknowledgments
We thank Dr. Martin Frank for providing models of human CD33m and CD33M. We also thank Dr. Syed Ali Raza (UCSD), Dr. Eillen Tecle (UCSD), and Dr. Flavio Schwarz for the extensive discussion on the manuscript and Sandra Diaz and Nissi Varki for technical help with the work.
This work was supported by National Institutes of Health Grants R01GM32373 and P01HL107150 (to A. V.) and CA160911 and DK099226 (to P. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. 1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- Siglec
- sialic acid-binding immunoglobulin-like lectin
- ITIM
- immunoreceptor tyrosine-based inhibition motif
- LOAD
- late-onset Alzheimer's disease
- CD
- cluster of differentiation
- SHP
- Src homology region 2 domain-containing phosphatase
- PPAR
- peroxisome proliferator-activated receptor
- PTS
- peroxisome targeting sequence
- LPS
- lipopolysaccharide
- fMLP
- N-formylmethionyl-leucyl-phenylalanine
- ER
- endoplasmic reticulum
- AUS
- A. ureafaciens sialidase
- PMN
- polymorphonuclear leukocyte
- FACS
- fluorescence activated cell sorting
- RGB
- red-green-blue.
References
- 1. Macauley M. S., Crocker P. R., and Paulson J. C. (2014) Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 14, 653–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crocker P. R., Paulson J. C., and Varki A. (2007) Siglecs and their roles in the immune system. Nat. Rev. Immunol. 7, 255–266 [DOI] [PubMed] [Google Scholar]
- 3. Schwarz F., Fong J. J., and Varki A. (2015) Human-specific evolutionary changes in the biology of siglecs. Adv. Exp. Med. Biol. 842, 1–16 [DOI] [PubMed] [Google Scholar]
- 4. Varki A., and Angata T. (2006) Siglecs: the major subfamily of I-type lectins. Glycobiology 16, 1R–27R [DOI] [PubMed] [Google Scholar]
- 5. Walter R. B., Raden B. W., Zeng R., Häusermann P., Bernstein I. D., and Cooper J. A. (2008) ITIM-dependent endocytosis of CD33-related Siglecs: role of intracellular domain, tyrosine phosphorylation, and the tyrosine phosphatases, Shp1 and Shp2. J. Leukoc. Biol. 83, 200–211 [DOI] [PubMed] [Google Scholar]
- 6. Brinkman-Van der Linden E. C., Angata T., Reynolds S. A., Powell L. D., Hedrick S. M., and Varki A. (2003) CD33/Siglec-3 binding specificity, expression pattern, and consequences of gene deletion in mice. Mol. Cell. Biol. 23, 4199–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bertram L., Lange C., Mullin K., Parkinson M., Hsiao M., Hogan M. F., Schjeide B. M., Hooli B., Divito J., Ionita I., Jiang H., Laird N., Moscarillo T., Ohlsen K. L., Elliott K., et al. (2008) Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am. J. Hum. Genet. 83, 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J. C., Carrasquillo M. M., Abraham R., Hamshere M. L., Pahwa J. S., Moskvina V., Dowzell K., Jones N., Stretton A., Thomas C., Richards A., et al. (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat. Genet. 43, 429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walker D. G., Whetzel A. M., Serrano G., Sue L. I., Beach T. G., and Lue L. F. (2015) Association of CD33 polymorphism rs3865444 with Alzheimer's disease pathology and CD33 expression in human cerebral cortex. Neurobiol Aging 36, 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Naj A. C., Jun G., Beecham G. W., Wang L. S., Vardarajan B. N., Buros J., Gallins P. J., Buxbaum J. D., Jarvik G. P., Crane P. K., Larson E. B., Bird T. D., Boeve B. F., Graff-Radford N. R., De Jager P. L., et al. (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33, and EPHA1 are associated with late-onset Alzheimer's disease. Nat. Genet. 43, 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gearing M., Rebeck G. W., Hyman B. T., Tigges J., and Mirra S. S. (1994) Neuropathology and apolipoprotein E profile of aged chimpanzees: implications for Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 91, 9382–9386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rosen R. F., Farberg A. S., Gearing M., Dooyema J., Long P. M., Anderson D. C., Davis-Turak J., Coppola G., Geschwind D. H., Paré J. F., Duong T. Q., Hopkins W. D., Preuss T. M., and Walker L. C. (2008) Tauopathy with paired helical filaments in an aged chimpanzee. J. Comp. Neurol. 509, 259–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Finch C. E., and Austad S. N. (2015) Commentary: is Alzheimer's disease uniquely human? Neurobiol Aging 36, 553–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwarz F., Springer S. A., Altheide T. K., Varki N. M., Gagneux P., and Varki A. (2016) Human-specific derived alleles of CD33 and other genes protect against postreproductive cognitive decline. Proc. Natl. Acad. Sci. U.S.A. 113, 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Malik M., Simpson J. F., Parikh I., Wilfred B. R., Fardo D. W., Nelson P. T., and Estus S. (2013) CD33 Alzheimer's risk-altering polymorphism, CD33 expression, and exon 2 splicing. J. Neurosci. 33, 13320–13325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hernández-Caselles T., Martínez-Esparza M., Pérez-Oliva A. B., Quintanilla-Cecconi A. M., García-Alonso A., Alvarez-López D. M., and García-Peñarrubia P. (2006) A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: two isoforms of CD33 are generated by alternative splicing. J. Leukoc. Biol. 79, 46–58 [DOI] [PubMed] [Google Scholar]
- 17. Springer S. A., Schwarz F., Altheide T. K., Varki N. M., Varki A., and Gagneux P. (2016) Reply to Liu and Jiang: Maintenance of postreproductive cognitive capacity by inclusive fitness. Proc. Natl. Acad. Sci. U.S.A. 113, E1591–E1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lim E. T., Würtz P., Havulinna A. S., Palta P., Tukiainen T., Rehnström K., Esko T., Mägi R., Inouye M., Lappalainen T., Chan Y., Salem R. M., Lek M., Flannick J., Sim X., et al. (2014) Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 10, e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flannick J., Thorleifsson G., Beer N. L., Jacobs S. B., Grarup N., Burtt N. P., Mahajan A., Fuchsberger C., Atzmon G., Benediktsson R., Blangero J., Bowden D. W., Brandslund I., Brosnan J., Burslem F., et al. (2014) Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat. Genet. 46, 357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao Z., Tuakli-Wosornu Y., Lagace T. A., Kinch L., Grishin N. V., Horton J. D., Cohen J. C., and Hobbs H. H. (2006) Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am. J. Hum. Genet. 79, 514–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olson M. V. (1999) When less is more: gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 64, 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olson M. V., and Varki A. (2003) Sequencing the chimpanzee genome: insights into human evolution and disease. Nat. Rev. Genet. 4, 20–28 [DOI] [PubMed] [Google Scholar]
- 23. Simmons D., and Seed B. (1988) Isolation of a cDNA encoding CD33, a differentiation antigen of myeloid progenitor cells. J. Immunol. 141, 2797–2800 [PubMed] [Google Scholar]
- 24. Paul S. P., Taylor L. S., Stansbury E. K., and McVicar D. W. (2000) Myeloid specific human CD33 is an inhibitory receptor with differential ITIM function in recruiting the phosphatases SHP-1 and SHP-2. Blood 96, 483–490 [PubMed] [Google Scholar]
- 25. Crocker P. R., Clark E. A., Filbin M., Gordon S., Jones Y., Kehrl J. H., Kelm S., Le Douarin N., Powell L., Roder J., Schnaar R. L., Sgroi D. C., Stamenkovic K., Schauer R., Schachner M., et al. (1998) Siglecs: a family of sialic-acid binding lectins [letter]. Glycobiology 8, v. [DOI] [PubMed] [Google Scholar]
- 26. Pérez-Oliva A. B., Martínez-Esparza M., Vicente-Fernández J. J., Corral-San Miguel R., García-Peñarrubia P., and Hernández-Caselles T. (2011) Epitope mapping, expression and post-translational modifications of two isoforms of CD33 (CD33M and CD33m) on lymphoid and myeloid human cells. Glycobiology 21, 757–770 [DOI] [PubMed] [Google Scholar]
- 27. O'Shea J. J., Brown E. J., Seligmann B. E., Metcalf J. A., Frank M. M., and Gallin J. I. (1985) Evidence for distinct intracellular pools of receptors for C3b and C3bi in human neutrophils. J. Immunol. 134, 2580–2587 [PubMed] [Google Scholar]
- 28. Cognasse F., Hamzeh H., Chavarin P., Acquart S., Genin C., and Garraud O. (2005) Evidence of Toll-like receptor molecules on human platelets. Immunol. Cell Biol. 83, 196–198 [DOI] [PubMed] [Google Scholar]
- 29. Grimsey N. L., Graham E. S., Dragunow M., and Glass M. (2010) Cannabinoid Receptor 1 trafficking and the role of the intracellular pool: implications for therapeutics. Biochem. Pharmacol. 80, 1050–1062 [DOI] [PubMed] [Google Scholar]
- 30. Ajioka R. S., and Kaplan J. (1986) Intracellular pools of transferrin receptors result from constitutive internalization of unoccupied receptors. Proc. Natl. Acad. Sci. U.S.A. 83, 6445–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weigel P. H., and Oka J. A. (1983) The large intracellular pool of asialoglycoprotein receptors functions during the endocytosis of asialoglycoproteins by isolated rat hepatocytes. J. Biol. Chem. 258, 5095–5102 [PubMed] [Google Scholar]
- 32. Lajaunias F., Dayer J. M., and Chizzolini C. (2005) Constitutive repressor activity of CD33 on human monocytes requires sialic acid recognition and phosphoinositide 3-kinase-mediated intracellular signaling. Eur. J. Immunol. 35, 243–251 [DOI] [PubMed] [Google Scholar]
- 33. Carp H. (1982) Mitochondrial N-formylmethionyl proteins as chemoattractants for neutrophils. J. Exp. Med. 155, 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Marasco W. A., Phan S. H., Krutzsch H., Showell H. J., Feltner D. E., Nairn R., Becker E. L., and Ward P. A. (1984) Purification and identification of formyl-methionyl-leucyl-phenylalanine as the major peptide neutrophil chemotactic factor produced by Escherichia coli. J. Biol. Chem. 259, 5430–5439 [PubMed] [Google Scholar]
- 35. de Hoop M. J., and Ab G. (1992) Import of proteins into peroxisomes and other microbodies. Biochem. J. 286, 657–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mohan K. V., and Atreya C. D. (2003) Novel organelle-targeting signals in viral proteins. Bioinformatics 19, 10–13 [DOI] [PubMed] [Google Scholar]
- 37. Brocard C., and Hartig A. (2006) Peroxisome targeting signal 1: is it really a simple tripeptide. Biochim. Biophys. Acta 1763, 1565–1573 [DOI] [PubMed] [Google Scholar]
- 38. Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J. F., Guindon S., Lefort V., Lescot M., Claverie J. M., and Gascuel O. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Croft D. P., Brent L. J., Franks D. W., and Cant M. A. (2015) The evolution of prolonged life after reproduction. Trends Ecol. Evol. 30, 407–416 [DOI] [PubMed] [Google Scholar]
- 40. Jones O. R., Scheuerlein A., Salguero-Gómez R., Camarda C. G., Schaible R., Casper B. B., Dahlgren J. P., Ehrlén J., García M. B., Menges E. S., Quintana-Ascencio P. F., Caswell H., Baudisch A., and Vaupel J. W. (2014) Diversity of ageing across the tree of life. Nature 505, 169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Terlecky S. R., Terlecky L. J., and Giordano C. R. (2012) Peroxisomes, oxidative stress, and inflammation. World J. Biol. Chem. 3, 93–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith J. J., and Aitchison J. D. (2013) Peroxisomes take shape. Nat. Rev. Mol. Cell Biol. 14, 803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Trompier D., Vejux A., Zarrouk A., Gondcaille C., Geillon F., Nury T., Savary S., and Lizard G. (2014) Brain peroxisomes. Biochimie 98, 102–110 [DOI] [PubMed] [Google Scholar]
- 44. Porcellotti S., Fanelli F., Fracassi A., Sepe S., Cecconi F., Bernardi C., Cimini A., Cerù M. P., and Moreno S. (2015) Oxidative stress during the progression of β-amyloid pathology in the neocortex of the Tg2576 mouse model of Alzheimer's disease. Oxid. Med. Cell Longev. 2015, 967203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fanelli F., Sepe S., D'Amelio M., Bernardi C., Cristiano L., Cimini A., Cecconi F., Ceru' M. P., and Moreno S. (2013) Age-dependent roles of peroxisomes in the hippocampus of a transgenic mouse model of Alzheimer's disease. Mol. Neurodegener. 8, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mandrekar-Colucci S., Karlo J. C., and Landreth G. E. (2012) Mechanisms underlying the rapid peroxisome proliferator-activated receptor-γ-mediated amyloid clearance and reversal of cognitive deficits in a murine model of Alzheimer's disease. J. Neurosci. 32, 10117–10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. in t' Veld B. A., Ruitenberg A., Hofman A., Launer L. J., van Duijn C. M., Stijnen T., Breteler M. M., and Stricker B. H. (2001) Nonsteroidal antiinflammatory drugs and the risk of Alzheimer's disease. N. Engl. J. Med. 345, 1515–1521 [DOI] [PubMed] [Google Scholar]
- 48. Landreth G. E., and Heneka M. T. (2001) Anti-inflammatory actions of peroxisome proliferator-activated receptor gamma agonists in Alzheimer's disease. Neurobiol Aging 22, 937–944 [DOI] [PubMed] [Google Scholar]
- 49. Griciuc A., Serrano-Pozo A., Parrado A. R., Lesinski A. N., Asselin C. N., Mullin K., Hooli B., Choi S. H., Hyman B. T., and Tanzi R. E. (2013) Alzheimer's disease risk gene CD33 inhibits microglial uptake of amyloid β. Neuron 78, 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Springer S. A., Manhart M., and Morozov A. V. (2016) Separating Spandrels from Phenotypic Targets of Selection in Adaptive Molecular Evolution, pp. 309–325, Springer International Publishing, Cham, Switzerland [Google Scholar]
- 51. Ghosh P., Garcia-Marcos M., Bornheimer S. J., and Farquhar M. G. (2008) Activation of Gαi3 triggers cell migration via regulation of GIV. J. Cell Biol. 182, 381–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Siddiqui S., Schwarz F., Springer S., Khedri Z., Yu H., Deng L., Verhagen A., Naito-Matsui Y., Jiang W., Kim D., Zhou J., Ding B., Chen X., Varki N., and Varki A. (2017) Studies on the detection, expression, glycosylation, dimerization, and ligand binding properties of mouse Siglec-E. J. Biol. Chem. 292, 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Razi N., and Varki A. (1999) Cryptic sialic acid binding lectins on human blood leukocytes can be unmasked by sialidase treatment or cellular activation. Glycobiology 9, 1225–1234 [DOI] [PubMed] [Google Scholar]
- 54. Altschul S. F., Gish W., Miller W., Myers E. W., and Lipman D. J. (1990) Basic local alignment search tool. J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 55. Dereeper A., Audic S., Claverie J. M., and Blanc G. (2010) BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang Z. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 [DOI] [PubMed] [Google Scholar]
- 57. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Neuberger G., Maurer-Stroh S., Eisenhaber B., Hartig A., and Eisenhaber F. (2003) Prediction of peroxisomal targeting signal 1 containing proteins from amino acid sequence. J. Mol. Biol. 328, 581–592 [DOI] [PubMed] [Google Scholar]
- 59. Myocardial Infarction Genetics Consortium Investigators, Stitziel N. O., Won H. H., Morrison A. C., Peloso G. M., Do R., Lange L. A., Fontanillas P., Gupta N., Duga S., Goel A., Farrall M., Saleheen D., Ferrario P., König I., Asselta R., et al. (2014) Inactivating mutations in NPC1L1 and protection from coronary heart disease. N. Engl. J. Med. 371, 2072–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jørgensen A. B., Frikke-Schmidt R., Nordestgaard B. G., and Tybjærg-Hansen A. (2014) Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N. Engl. J. Med. 371, 32–41 [DOI] [PubMed] [Google Scholar]
- 61. Jonsson T., Atwal J. K., Steinberg S., Snaedal J., Jonsson P. V., Bjornsson S., Stefansson H., Sulem P., Gudbjartsson D., Maloney J., Hoyte K., Gustafson A., Liu Y., Lu Y., Bhangale T., et al. (2012) A mutation in APP protects against Alzheimer's disease and age-related cognitive decline. Nature 488, 96–99 [DOI] [PubMed] [Google Scholar]
- 62. Saleh M., Mathison J. C., Wolinski M. K., Bensinger S. J., Fitzgerald P., Droin N., Ulevitch R. J., Green D. R., and Nicholson D. W. (2006) Enhanced bacterial clearance and sepsis resistance in caspase-12-deficient mice. Nature 440, 1064–1068 [DOI] [PubMed] [Google Scholar]
- 63. Samson M., Libert F., Doranz B. J., Rucker J., Liesnard C., Farber C. M., Saragosti S., Lapoumeroulie C., Cognaux J., Forceille C., Muyldermans G., Verhofstede C., Burtonboy G., Georges M., Imai T., et al. (1996) Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382, 722–725 [DOI] [PubMed] [Google Scholar]
- 64. Diogo D., Bastarache L., Liao K. P., Graham R. R., Fulton R. S., Greenberg J. D., Eyre S., Bowes J., Cui J., Lee A., Pappas D. A., Kremer J. M., Barton A., Coenen M. J., Franke B., et al. (2015) TYK2 protein-coding variants protect against rheumatoid arthritis and autoimmunity, with no evidence of major pleiotropic effects on non-autoimmune complex traits. PLoS ONE 10, e0122271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nejentsev S., Walker N., Riches D., Egholm M., and Todd J. A. (2009) Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science 324, 387–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rivas M. A., Beaudoin M., Gardet A., Stevens C., Sharma Y., Zhang C. K., Boucher G., Ripke S., Ellinghaus D., Burtt N., Fennell T., Kirby A., Latiano A., Goyette P., Green T., et al. (2011) Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 43, 1066–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rivas M. A., Graham D., Sulem P., Stevens C., Desch A. N., Goyette P., Gudbjartsson D., Jonsdottir I., Thorsteinsdottir U., Degenhardt F., Mucha S., Kurki M. I., Li D., D'Amato M., Annese V., et al. (2016) A protein-truncating R179X variant in RNF186 confers protection against ulcerative colitis. Nat Commun 7, 12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.